Integrated Analysis of mRNAs and Long Non-Coding RNAs Expression of Oviduct That Provides Novel Insights into the Prolificacy Mechanism of Goat (Capra hircus)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethics
2.2. Animals and Sample Collection
2.3. RNA Isolation, Library Preparation, and Sequencing
2.4. Quality Control and Reference Genome Mapping
2.5. lncRNA Identification and Differentially Expression Analysis
2.6. Potential Target Gene Prediction and Networks Construction
2.7. Function Enrichment Analyses
2.8. Quantitative Real-Time PCR Validation of lncRNA and mRNA
3. Results
3.1. Overview of Transcriptome Sequencing Data
3.2. Identification of mRNA and lncRNA in YunShang Black Goat Oviduct Tissue
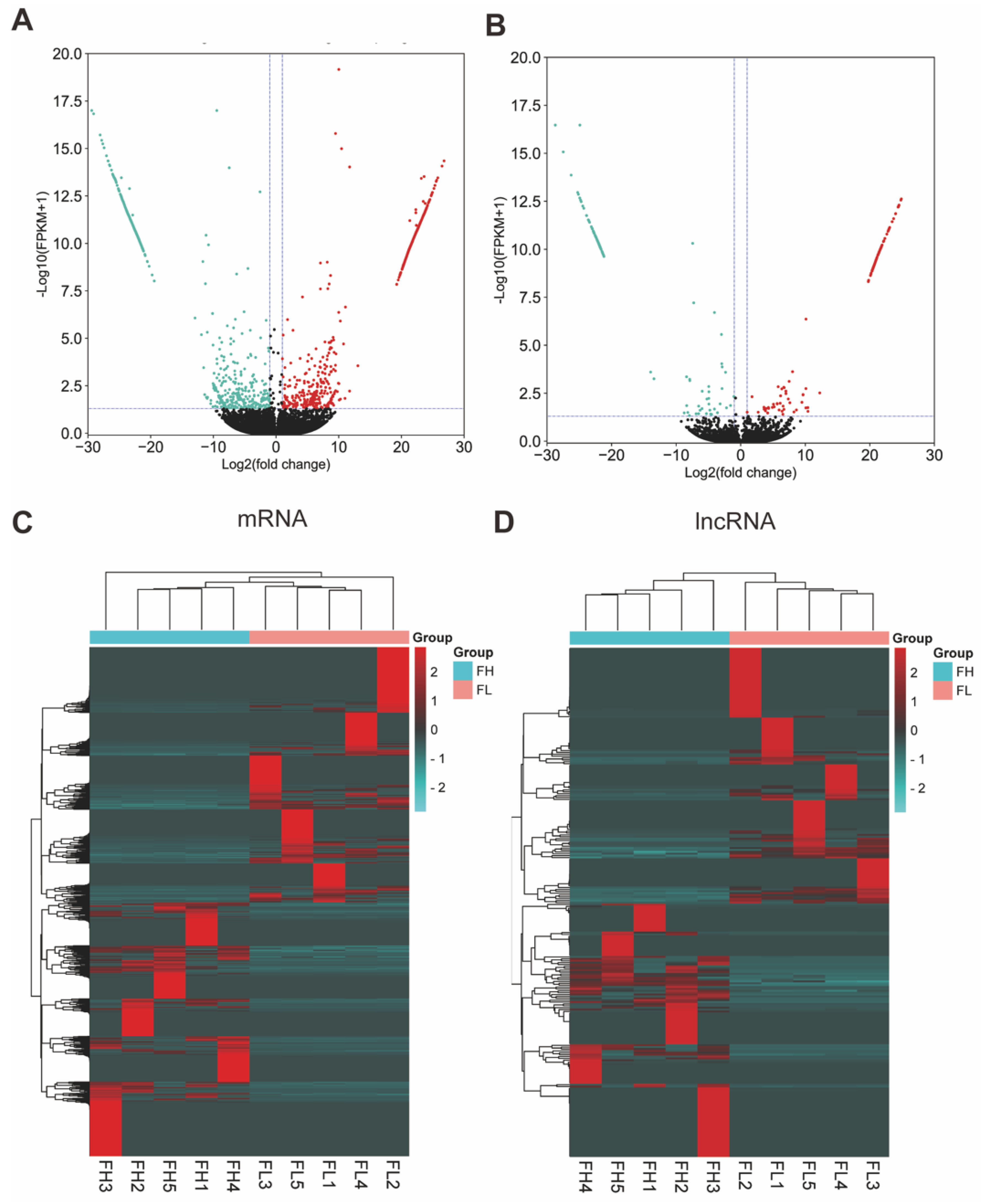
3.3. Analysis of DEG and DEL Expression Profiles in the Goat Oviduct
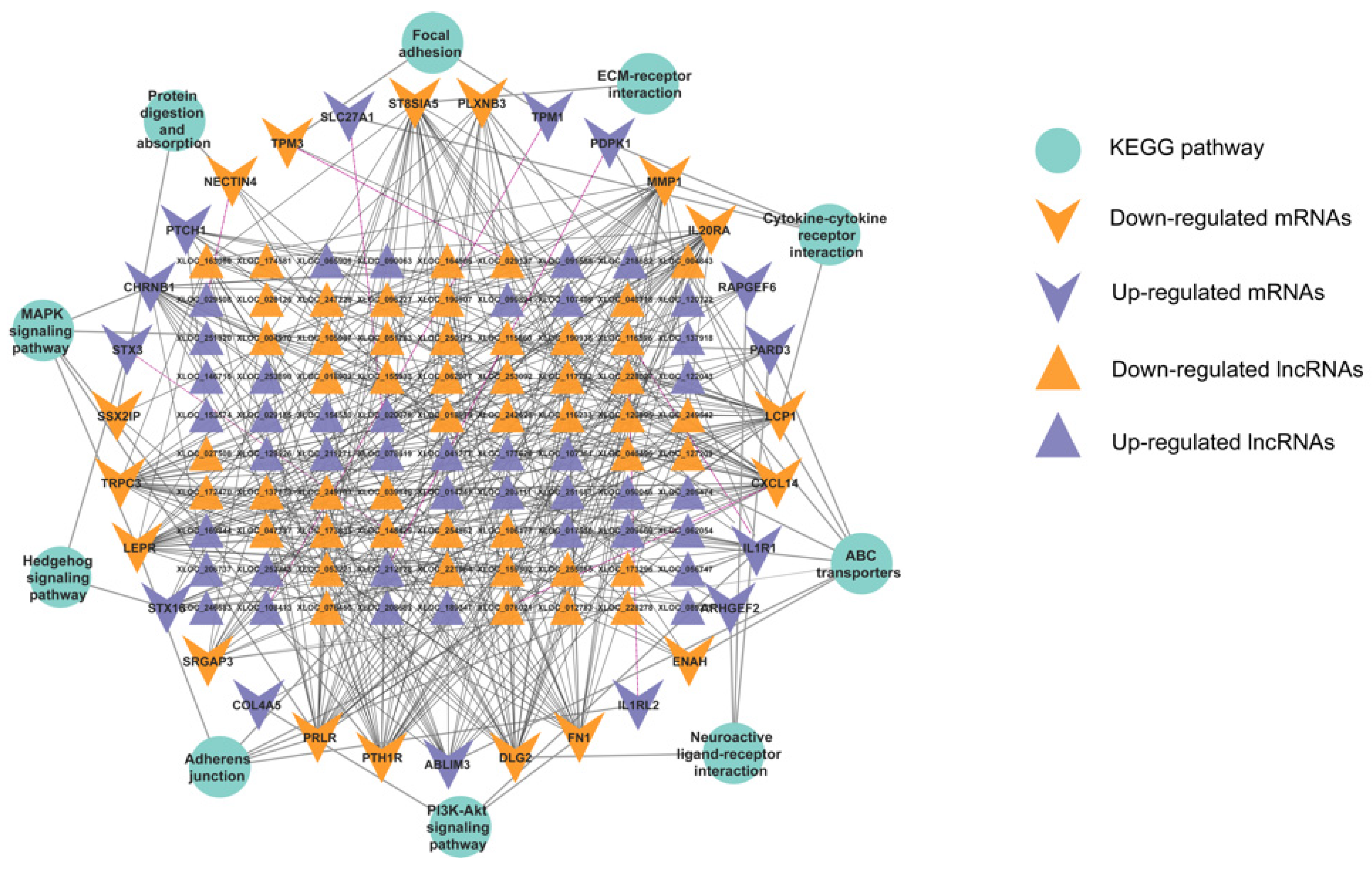
3.4. PPI Analysis of the DEGs Related to Prolificacy Trait
3.5. Functional Analysis of the DEGs
3.6. Analysis of the Targeting Relationship between lncRNAs and mRNAs
3.7. Verification of RNA Expression Profiles with Quantitative Real-Time PCR
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gooki, F.G.; Mohammadabadi, M.R.; Fozi, M.A.; Soflaei, M. Association of biometric traits with growth hormone gene diversity in Raini cashmere goats. Walailak J. Sci. Technol. 2019, 16, 499–508. [Google Scholar] [CrossRef]
- Askari, N.; Mohammad, M.R.; Baghizadeh, A. ISSR markers for assessing DNA polymorphism and genetic characterization of cattle, goat and sheep populations. Iran. J. Biotechnol. 2011, 9, 222–229. [Google Scholar]
- Barazandeh, A.; Mohammadabadi, M.R.; Ghaderi-Zefrehei, M.; Rafeie, F.; Imumorin, I.G. Whole genome comparative analysis of CpG islands in camelid and other mammalian genomes. Mamm. Biol. 2019, 98, 73–79. [Google Scholar] [CrossRef]
- Gholamhoseini, F.G.; Mohammadabadi, M.R.; Fozi, M.A. Polymorphism of the growth hormone gene and its effect on production and reproduction traits in goat. Iran. J. Appl. Anim. Sci. 2018, 8, 653–659. [Google Scholar]
- Noori, A.N.; Behzadi, M.; Mohammadabadi, M.R. Expression pattern of Rheb gene in Jabal Barez Red goat. Indian J. Anim. Sci. 2017, 87, 1375–1378. [Google Scholar]
- Mohammadabadi, M.; Bordbar, F.; Jensen, J.; Du, M.; Guo, W. Key genes regulating skeletal muscle development and growth in farm animals. Animals 2021, 11, 835. [Google Scholar] [CrossRef]
- Hunter, R.H.F. Components of oviduct physiology in eutherian mammals. Biol. Rev. Camb. Philos. Soc. 2012, 87, 244–255. [Google Scholar] [CrossRef]
- Avilés, M.; Gutiérrez-Adán, A.; Coy, P. Oviductal secretions: Will they be key factors for the future ARTs? Mol. Hum. Reprod. 2010, 16, 896–906. [Google Scholar] [CrossRef]
- Vinet, A.; Drouilhet, L.; Bodin, L.; Mulsant, P.; Fabre, S.; Phocas, F. Genetic control of multiple births in low ovulating mammalian species. Mamm. Genome 2012, 23, 727–740. [Google Scholar] [CrossRef]
- Warriach, H.M.; McGill, D.M.; Bush, R.D.; Wynn, P.C.; Chohan, K.R. A review of recent developments in buffalo reproduction—A review. Asian-Australas J. Anim. Sci. 2015, 28, 451–455. [Google Scholar] [CrossRef]
- Stephens, S.M.; Moley, K.H. Follicular origins of modern reproductive endocrinology. Am. J. Physiol. Endocrinol. Metab. 2009, 297, E1235–E1236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mondéjar, I.; Acuña, O.S.; Izquierdo-Rico, M.J.; Coy, P.; Avilés, M. The oviduct: Functional genomic and proteomic approach. Reprod. Domest. Anim. 2012, 47, 22–29. [Google Scholar] [CrossRef] [PubMed]
- Abe, H. The mammalian oviductal epithelium: Regional variations in cytological and functional aspects of the oviductal secretory cells. Histol. Histopathol. 1996, 11, 743–768. [Google Scholar]
- Pan, Z.X.; Zhang, J.B.; Lin, F.; Ma, X.S.; Wang, X.G.; Liu, H.L. Expression profiles of key candidate genes involved in steroidogenesis during follicular atresia in the pig ovary. Mol. Biol. Rep. 2012, 39, 10823–10832. [Google Scholar] [CrossRef] [PubMed]
- Buhi, W.C. Characterization and biological roles of oviduct-specific, oestrogen-dependent glycoprotein. Reproduction 2002, 123, 355–362. [Google Scholar] [CrossRef] [PubMed]
- Killian, G.J. Evidence for the role of oviduct secretions in sperm function, fertilization and embryo development. Anim. Reprod. Sci. 2004, 82, 141–153. [Google Scholar] [CrossRef]
- Acuña, O.S.; Avilés, M.; López-Úbeda, R.; Guillén-Martínez, A.; Soriano-Úbeda, C.; Torrecillas, A.; Coy, P.; Izquierdo-Rico, M.J. Differential gene expression in porcine oviduct during the oestrous cycle. Reprod. Fertil. Dev. 2017, 29, 2387–2399. [Google Scholar] [CrossRef]
- Silva, A.L.; Fuhrich, D.G.; Carson, D.D.; Engel, B.J.; Savaris, R.F. MUC1 expression in fallopian tubes of women with hydrosalpinx. Eur. J. Obs. Gynecol. Reprod. Biol. 2014, 180, 106–110. [Google Scholar] [CrossRef]
- Wilk, R.; Hu, J.; Blotsky, D.; Krause, H.M. Diverse and pervasive subcellular distributions for both coding and long noncoding RNAs. Genes Dev. 2016, 30, 594–609. [Google Scholar] [CrossRef] [Green Version]
- Fatica, A.; Bozzoni, I. Long non-coding RNAs: New players in cell differentiation and development. Nat. Rev. Genet. 2014, 15, 7–21. [Google Scholar] [CrossRef]
- Mohammad, F.; Mondal, T.; Guseva, N.; Pandey, G.K.; Kanduri, C. Kcnq1ot1 noncoding RNA mediates transcriptional gene silencing by interacting with Dnmt1. Development 2010, 137, 2493–2499. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lian, Z.Q.; Zou, X.; Han, Y.R.; Deng, M.; Sun, B.L.; Guo, Y.Q.; Zhou, L.; Liu, G.B.; Liu, D.W.; Li, Y.K. Role of mRNAs and long non-coding RNAs in regulating the litter size trait in Chuanzhong black goats. Reprod. Domest. Anim. 2020, 55, 486–495. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Li, W.J.; Feng, P.; Liu, Y.Y.; Meng, P.; Chu, B.; Zhao, J.L.; Li, Y.X.; Zhang, Y.; Liu, J. Lnc5926 is essential for early embryonic development in goats through regulation of ZSCAN4 and EIF1AX. Theriogenology 2022, 180, 87–93. [Google Scholar] [CrossRef]
- Taylor, D.H.; Chu, E.T.; Spektor, R.; Soloway, P.D. Long non-coding RNA regulation of reproduction and development. Mol. Reprod. Dev. 2015, 82, 932–956. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Cao, Y.X.; Xu, X.F.; Xiang, H.F.; Zhang, Z.G.; Chen, B.L.; Hao, Y.; Wei, Z.L.; Zhou, P.; Chen, D.W. Increased new lncRNA-mRNA gene pair levels in human cumulus cells correlate with oocyte maturation and embryo development. Reprod. Sci. 2015, 22, 1008–1014. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Qi, B.; Xie, J.; Wu, X.Q.; Ling, Y.H.; Cao, X.Y.; Kong, F.; Xin, J.; Jiang, X.; Wu, Q.Q.; et al. Filtered reproductive long non-coding RNAs by genome-wide analyses of goat ovary at different estrus periods. BMC Genom. 2018, 19, 866. [Google Scholar] [CrossRef]
- Hong, L.J.; Hu, Q.; Zang, X.P.; Xie, Y.S.; Zhou, C.; Zou, X.; Li, Y.K.; Deng, M.; Guo, Y.Q.; Liu, G.B.; et al. Analysis and screening of reproductive long non-coding RNAs through genome-wide analyses of goat endometrium during the pre-attachment phase. Front. Genet. 2020, 11, 568017. [Google Scholar] [CrossRef]
- Zhao, Z.F.; Zou, X.; Lu, T.T.; Deng, M.; Li, Y.K.; Guo, Y.Q.; Sun, B.L.; Liu, G.B.; Liu, D.W. Identification of mRNAs and lncRNAs involved in the regulation of follicle development in goat. Front. Genet. 2020, 11, 589076. [Google Scholar] [CrossRef]
- Wang, L.J.; Yang, X.; Zhu, Y.H.; Zhan, S.Y.; Chao, Z.; Zhong, T.; Guo, J.Z.; Wang, Y.; Li, L.; Zhang, H.P. Genome-wide identification and characterization of long noncoding RNAs of brown to white adipose tissue transformation in goats. Cells 2019, 8, 904. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.K.; Xu, X.P.; Deng, M.; Zou, X.; Zhao, Z.F.; Huang, S.X.; Liu, D.W.; Liu, G.B. Identification and Comparative analysis of long non-coding RNAs in high- and low-fecundity goat ovaries during estrus. Front. Genet. 2021, 12, 648158. [Google Scholar] [CrossRef]
- Molaei Moghbeli, S.; Barazandeh, A.; Vatankhah, M.; Mohammadabadi, M. Genetics and non-genetics parameters of body weight for post-weaning traits in Raini cashmere goats. Trop. Anim. Health Prod. 2013, 45, 1519–1524. [Google Scholar] [CrossRef] [PubMed]
- Mohammadabadi, M.R.; Asadollahpour, H. Leptin gene expression in Raini Cashmere goat using Real Time PCR. Agric. Biotechnol. J. 2021, 13, 197–214. [Google Scholar]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kovaka, S.; Zimin, A.V.; Pertea, G.M.; Razaghi, R.; Salzberg, S.L.; Pertea, M. Transcriptome assembly from long-read RNA-seq alignments with StringTie2. Genome Biol. 2019, 20, 278. [Google Scholar] [CrossRef] [Green Version]
- Trapnell., C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef] [Green Version]
- Sun, L.; Luo, H.T.; Bu, D.C.; Zhao, G.G.; Yu, K.T.; Zhang, C.H.; Liu, Y.N.; Chen, R.S.; Zhao, Y. Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts. Nucleic Acids Res. 2013, 41, e166. [Google Scholar] [CrossRef]
- Kang, Y.J.; Yang, D.C.; Kong, L.; Hou, M.; Meng, Y.Q.; Wei, L.; Gao, G. CPC2: A fast and accurate coding potential calculator based on sequence intrinsic features. Nucleic Acids Res. 2017, 45, W12–W16. [Google Scholar] [CrossRef] [Green Version]
- Li, A.M.; Zhang, J.Y.; Zhou, Z.Y. PLEK: A tool for predicting long non-coding RNAs and messenger RNAs based on an improved k-mer scheme. BMC Bioinform. 2014, 15, 311. [Google Scholar] [CrossRef] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Hess, A.P.; Talbi, S.; Hamilton, A.E.; Baston-Buest, D.M.; Nyegaard, M.; Irwin, J.C.; Barragan, F.; Kruessel, J.S.; Germeyer, A.; Giudice, L.C. The human oviduct transcriptome reveals an anti-inflammatory, anti-angiogenic, secretory and matrix-stable environment during embryo transit. Reprod. Biomed Online 2013, 27, 423–435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kolle, S.; Hughes, B.; Steele, H. Early embryo-maternal communication in the oviduct: A review. Mol. Reprod. Dev. 2020, 87, 650–662. [Google Scholar] [CrossRef] [PubMed]
- Cerny, K.L.; Garrett, E.; Walton, A.J.; Anderson, L.H.; Bridges, P.J. A transcriptomal analysis of bovine oviductal epithelial cells collected during the follicular phase versus the luteal phase of the estrous cycle. Reprod. Biol. Endocrinol. 2015, 13, 84. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maillo, V.; de Frutos, C.; O’Gaora, P.; Forde, N.; Burns, G.W.; Spencer, T.E.; Gutierrez-Adan, A.; Lonergan, P.; Rizos, D. Spatial differences in gene expression in the bovine oviduct. Reproduction 2016, 152, 37–46. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flegel, C.; Manteniotis, S.; Osthold, S.; Hatt, H.; Gisselmann, G. Expression profile of ectopic olfactory receptors determined by deep sequencing. PLoS ONE 2013, 8, e55368. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, W.; He, X.Y.; Di, R.; Wang, X.Y.; Chu, M.X. Transcriptome analysis revealed long non-coding RNAs associated with mRNAs in sheep thyroid gland under different photoperiods. Genes 2022, 13, 606. [Google Scholar] [CrossRef]
- Yang, W.N.; Wang, L.J.; Wang, F.L.; Yuan, S.Q. Roles of AMP-Activated Protein Kinase (AMPK) in Mammalian Reproduction. Front. Cell Dev. Biol. 2020, 8, 593005. [Google Scholar] [CrossRef]
- Andrade, G.M.; da Silveira, J.C.; Perrini, C.; Del Collado, M.; Gebremedhn, S.; Tesfaye, D.; Meirelles, F.V.; Perecin, F. The role of the PI3K-Akt signaling pathway in the developmental competence of bovine oocytes. PLoS ONE 2017, 12, e0185045. [Google Scholar] [CrossRef]
- Zhou, S.; Yan, W.; Shen, W.; Cheng, J.; Xi, Y.Y.; Yuan, S.Z.; Fu, F.F.; Ding, T.; Luo, A.Y.; Wang, S.X. Low expression of SEMA6C accelerates the primordial follicle activation in the neonatal mouse ovary. J. Cell. Mol. Med. 2018, 22, 486–496. [Google Scholar] [CrossRef] [Green Version]
- Luvoni, G.C.; Chigioni, S.; Allievi, E.; Macis, D. Meiosis resumption of canine oocytes cultured in the isolated oviduct. Reprod. Domest. Anim. 2003, 38, 410–414. [Google Scholar] [CrossRef]
- Kim, J.M.; Park, J.E.; Yoo, I.; Han, J.; Kim, N.; Lim, W.J.; Cho, E.S.; Choi, B.; Choi, S.; Kim, T.H.; et al. Integrated transcriptomes throughout swine oestrous cycle reveal dynamic changes in reproductive tissues interacting networks. Sci. Rep. 2018, 8, 5436. [Google Scholar] [CrossRef] [PubMed]
- Bryan, J.; Muñoz, A.; Zhang, X.N.; Düfer, M.; Drews, G.; Krippeit-Drews, P.; Aguilar-Bryan, L. ABCC8 and ABCC9: ABC transporters that regulate K+ channels. Pflug. Arch. 2007, 453, 703–718. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McCartney, B.M.; Näthke, I.S. Cell regulation by the Apc protein Apc as master regulator of epithelia. Curr. Opin. Cell Biol. 2008, 20, 186–193. [Google Scholar] [CrossRef] [PubMed]
- Stricker, S.A. Comparative biology of calcium signaling during fertilization and egg activation in animals. Dev. Biol. 1999, 211, 157–176. [Google Scholar] [CrossRef] [PubMed]
- Coy, P.; García-Vázquez, F.A.; Visconti, P.E.; Avilés, M. Roles of the oviduct in mammalian fertilization. Reproduction 2012, 144, 649–660. [Google Scholar] [CrossRef] [Green Version]
- Demott, R.P.; Suarez, S.S. Hyperactivated sperm progress in the mouse oviduct. Biol. Reprod. 1992, 46, 779–785. [Google Scholar] [CrossRef]
- Panizza, E.; Zhang, L.; Fontana, J.M.; Hamada, K.; Svensson, D.; Akkuratov, E.E.; Scott, L.; Mikoshiba, K.; Brismar, H.; Lehtiö, J.; et al. Ouabain-regulated phosphoproteome reveals molecular mechanisms for Na(+), K(+)-ATPase control of cell adhesion, proliferation, and survival. Faseb J. 2019, 33, 10193–10206. [Google Scholar] [CrossRef] [Green Version]
- Medvedev, S.; Stein, P.; Schultz, R.M. Specificity of calcium/calmodulin-dependent protein kinases in mouse egg activation. Cell Cycle 2014, 13, 1482–1488. [Google Scholar] [CrossRef] [Green Version]
- Tan, L.Z.; Song, Y.; Nelson, J.; Yu, Y.P.; Luo, J.H. Integrin alpha7 binds tissue inhibitor of metalloproteinase 3 to suppress growth of prostate cancer cells. Am. J. Pathol. 2013, 183, 831–840. [Google Scholar] [CrossRef] [Green Version]
- Ge, J.C.; Wang, Y.X.; Chen, Z.B.; Chen, D.F. Integrin alpha 7 correlates with poor clinical outcomes, and it regulates cell proliferation, apoptosis and stemness via PTK2-PI3K-Akt signaling pathway in hepatocellular carcinoma. Cell Signal 2020, 66, 109465. [Google Scholar] [CrossRef]
- Artini, P.G.; Tatone, C.; Sperduti, S.; D’Aurora, M.; Franchi, S.; Emidio, G.D.; Ciriminna, R.; Vento, M.; Pietro, C.D.; Stuppia, L.; et al. Cumulus cells surrounding oocytes with high developmental competence exhibit down-regulation of phosphoinositol 1,3 kinase/protein kinase B (PI3K/AKT) signalling genes involved in proliferation and survival. Hum. Reprod. 2017, 32, 2474–2484. [Google Scholar] [CrossRef] [PubMed]
- Hall, S.E.; Upton, R.M.O.; McLaughlin, E.A.; Sutherland, J.M. Phosphoinositide 3-kinase/protein kinase B (PI3K/AKT) and Janus kinase/signal transducer and activator of transcription (JAK/STAT) follicular signalling is conserved in the mare ovary. Reprod. Fertil. Dev. 2018, 30, 624–633. [Google Scholar] [CrossRef] [PubMed]
- Bazer, F.W.; Song, G.H.; Kim, J.Y.; Dunlap, K.A.; Satterfield, M.C.; Johnson, G.A.; Burghardt, R.C.; Wu, G.Y. Uterine biology in pigs and sheep. J. Anim. Sci. Biotechnol. 2012, 3, 23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Russell, D.L.; Salustri, A. Extracellular matrix of the cumulus-oocyte complex. Semin. Reprod. Med. 2006, 24, 217–227. [Google Scholar] [CrossRef]
- Gao, X.X.; Ye, J.; Yang, C.; Luo, L.; Liu, Y.; Ding, J.P.; Zhang, Y.H.; Ling, Y.H.; Huang, W.P.; Zhang, X.R.; et al. RNA-seq analysis of lncRNA-controlled developmental gene expression during puberty in goat & rat. BMC Genet. 2018, 19, 19. [Google Scholar]
- Morozumi, Y.; Boussouar, F.; Tan, M.; Chaikuad, A.; Jamshidikia, M.; Colak, G.; He, H.; Nie, L.; Petosa, C.; de Dieuleveult, M.; et al. Atad2 is a generalist facilitator of chromatin dynamics in embryonic stem cells. J. Mol. Cell. Biol. 2016, 8, 349–362. [Google Scholar] [CrossRef] [Green Version]
- Hughes, J.; Dawson, R.; Tea, M.; McAninch, D.; Piltz, S.; Jackson, D.; Stewart, L.; Ricos, M.G.; Dibbens, L.M.; Harvey, N.L.; et al. Knockout of the epilepsy gene Depdc5 in mice causes severe embryonic dysmorphology with hyperactivity of mTORC1 signalling. Sci. Rep. 2017, 7, 12618. [Google Scholar] [CrossRef] [Green Version]
- Walder, R.Y.; Yang, B.; Stokes, J.B.; Kirby, P.A.; Cao, X.; Shi, P.; Searby, C.C.; Husted, R.F.; Sheffield, V.C. Mice defective in Trpm6 show embryonic mortality and neural tube defects. Hum. Mol. Genet. 2009, 18, 4367–4375. [Google Scholar] [CrossRef] [Green Version]
- Kulus, M.; Kranc, W.; Jeseta, M.; Sujka-Kordowska, P.; Konwerska, A.; Ciesiolka, S.; Celichowski, P.; Moncrieff, L.; Kocherova, I.; Jozkowiak, M.; et al. Cortical granule distribution and expression pattern of genes regulating cellular component size, morphogenesis, and potential to differentiation are related to oocyte developmental competence and maturational capacity in vivo and in vitro. Genes 2020, 11, 815. [Google Scholar] [CrossRef]
- Azmal, S.A.; Bhuiyan, A.A.; Omar, A.I.; Ma, S.; Sun, C.H.; Han, Z.D.; Zhang, M.K.; Zhao, S.H.; Li, S.J. Novel polymorphisms in RAPGEF6 gene associated with egg-laying rate in chinese Jing Hong chicken using genome-wide SNP scan. Genes 2019, 10, 384. [Google Scholar] [CrossRef] [Green Version]
- Fang, E.; Zhang, H.; Yin, W.L.; Wang, C.Y.; Liu, Y.L.; Li, Y.Z.; Wang, L.L.; Wu, Y.; Zhang, R.Z.; Zou, C.D.; et al. CPEB3 deficiency in mice affect ovarian follicle development and causes premature ovarian insufficiency. Cell Death Dis. 2021, 13, 21. [Google Scholar]
- Lv, X.B.; Liu, C.Y.; Wang, Z.; Sun, Y.P.; Xiong, Y.; Lei, Q.Y.; Guan, K.L. PARD3 induces TAZ activation and cell growth by promoting LATS1 and PP1 interaction. EMBO Rep. 2015, 16, 975–985. [Google Scholar] [CrossRef] [PubMed] [Green Version]




| Sample | Clean Reads | Clean Base (bp) | Length | Q20 (%) | Q30 (%) | GC (%) | Total Mapped (%) |
|---|---|---|---|---|---|---|---|
| FL-1 | 52,752,779 | 15,825,833,700 | 150 | 98.35 | 94.75 | 51.45 | 97.13 |
| FL-2 | 60,824,479 | 18,247,343,700 | 150 | 98.15 | 94.60 | 50.70 | 96.86 |
| FL-3 | 53,704,715 | 16,111,414,500 | 150 | 98.20 | 94.60 | 50.10 | 96.86 |
| FL-4 | 51,703,403 | 15,511,020,900 | 150 | 98.45 | 95.05 | 50.10 | 97.35 |
| FL-5 | 54,659,450 | 16,397,835,000 | 150 | 98.35 | 95.00 | 48.55 | 96.40 |
| FH-1 | 56,457,768 | 16,937,330,400 | 150 | 98.50 | 95.25 | 51.15 | 96.90 |
| FH-2 | 51,119,834 | 15,335,950,200 | 150 | 98.08 | 94.20 | 50.00 | 96.40 |
| FH-3 | 67,474,481 | 20,242,344,300 | 150 | 97.80 | 93.50 | 51.15 | 96.99 |
| FH-4 | 52,753,848 | 15,826,154,400 | 150 | 98.25 | 94.40 | 49.25 | 97.18 |
| FH-5 | 50,538,996 | 15,161,698,800 | 150 | 98.50 | 95.10 | 51.25 | 97.18 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, Z.; Zhang, Z.; Liu, Y.; Ren, C.; He, X.; Jiang, Y.; Ouyang, Y.; Hong, Q.; Chu, M. Integrated Analysis of mRNAs and Long Non-Coding RNAs Expression of Oviduct That Provides Novel Insights into the Prolificacy Mechanism of Goat (Capra hircus). Genes 2022, 13, 1031. https://doi.org/10.3390/genes13061031
Sun Z, Zhang Z, Liu Y, Ren C, He X, Jiang Y, Ouyang Y, Hong Q, Chu M. Integrated Analysis of mRNAs and Long Non-Coding RNAs Expression of Oviduct That Provides Novel Insights into the Prolificacy Mechanism of Goat (Capra hircus). Genes. 2022; 13(6):1031. https://doi.org/10.3390/genes13061031
Chicago/Turabian StyleSun, Zhipeng, Zijun Zhang, Yufang Liu, Chunhuan Ren, Xiaoyun He, Yanting Jiang, Yina Ouyang, Qionghua Hong, and Mingxing Chu. 2022. "Integrated Analysis of mRNAs and Long Non-Coding RNAs Expression of Oviduct That Provides Novel Insights into the Prolificacy Mechanism of Goat (Capra hircus)" Genes 13, no. 6: 1031. https://doi.org/10.3390/genes13061031
APA StyleSun, Z., Zhang, Z., Liu, Y., Ren, C., He, X., Jiang, Y., Ouyang, Y., Hong, Q., & Chu, M. (2022). Integrated Analysis of mRNAs and Long Non-Coding RNAs Expression of Oviduct That Provides Novel Insights into the Prolificacy Mechanism of Goat (Capra hircus). Genes, 13(6), 1031. https://doi.org/10.3390/genes13061031









