An Update on the Evolutionary History of Bregs
Abstract
:1. Introduction
2. Evolution of Cell Types
3. B Cell Differentiation and Evolutionary History
3.1. B Cell Main Groups
3.2. B Cell Differentiation
3.3. Evolution of B Cells
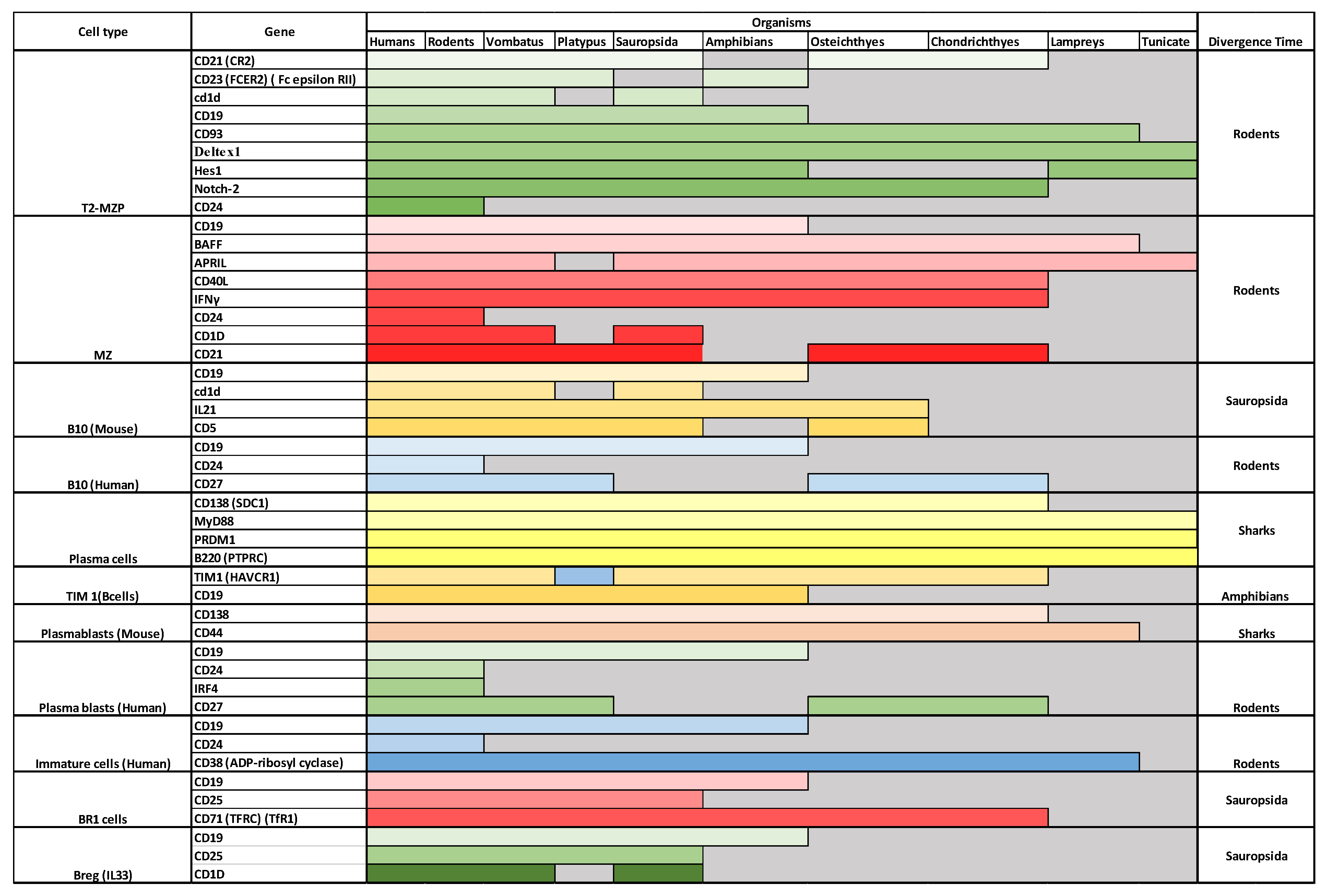
4. Breg Evolutionary History
4.1. Breg Subpopulations
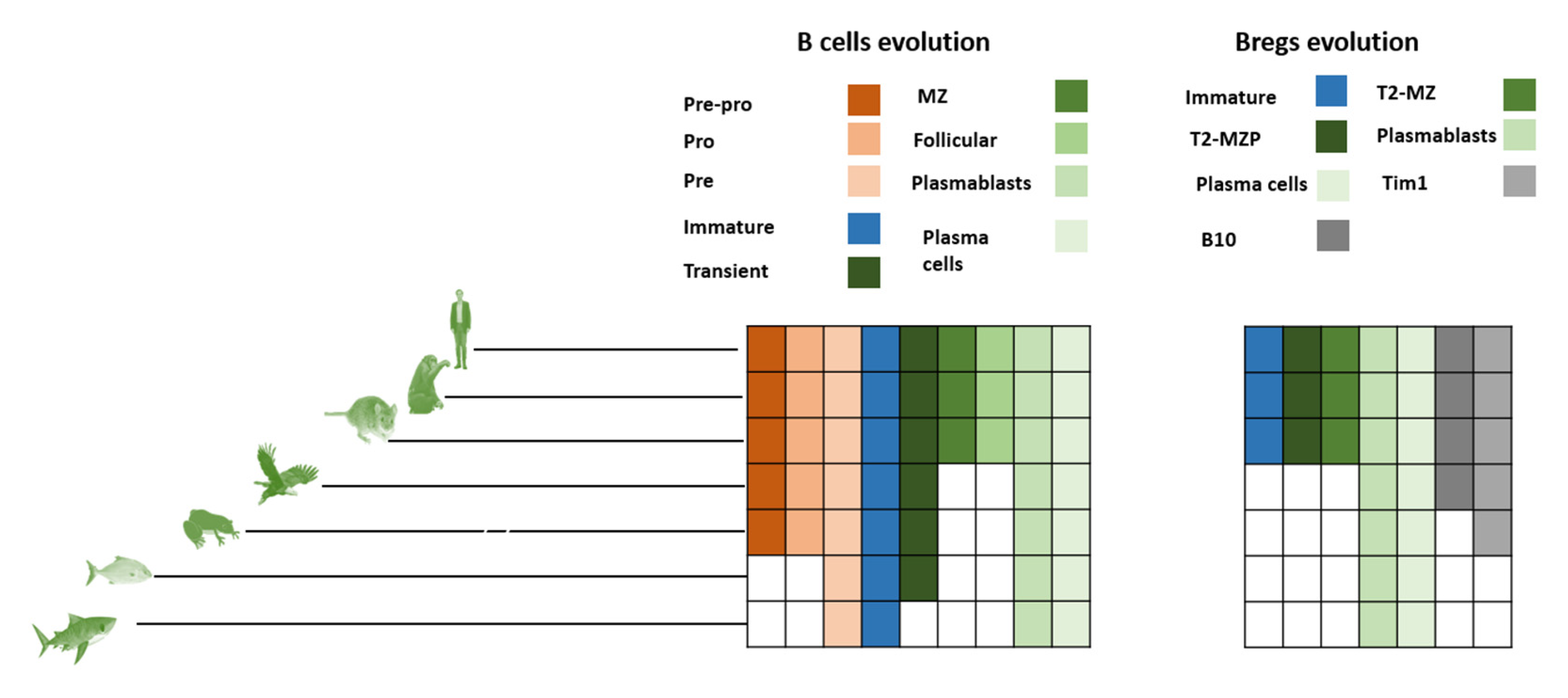
4.2. Evolution of Bregs
4.2.1. Immature Bregs
4.2.2. T2-MZP Bregs
4.2.3. MZ Bregs
4.2.4. Plasma Bregs
4.2.5. Plasmablast Bregs
4.2.6. B10 Bregs
4.2.7. Tim1 Bregs, BR1, and Breg (IL33)
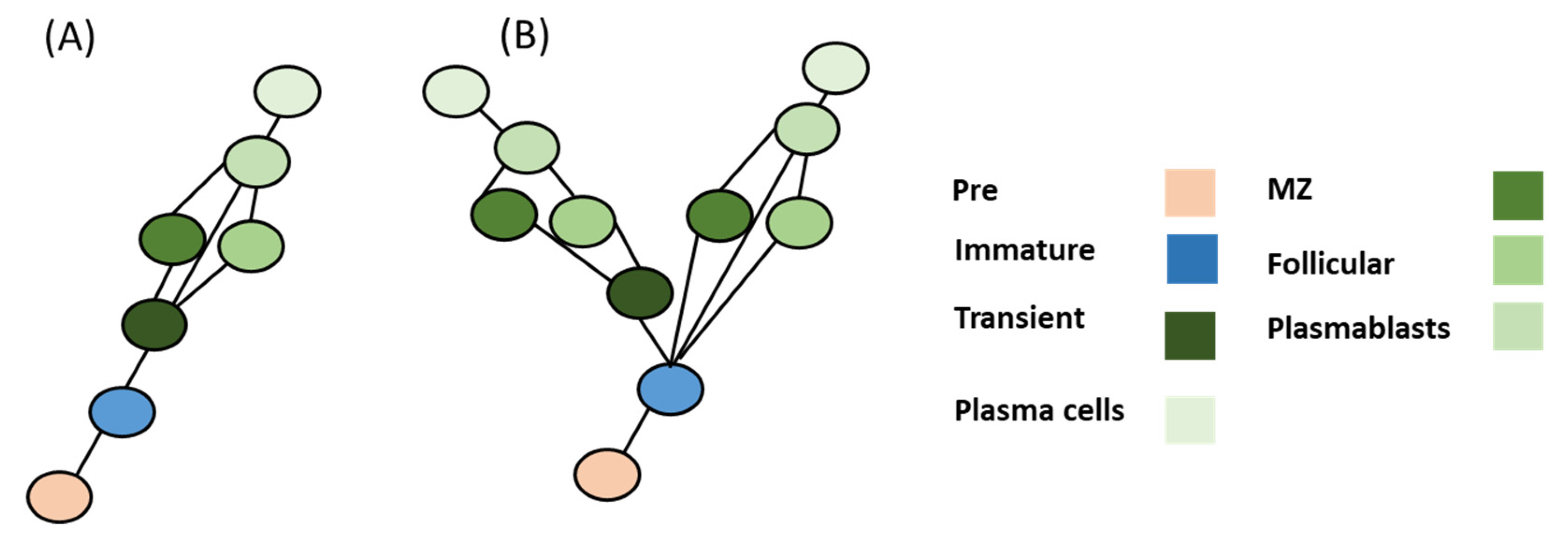
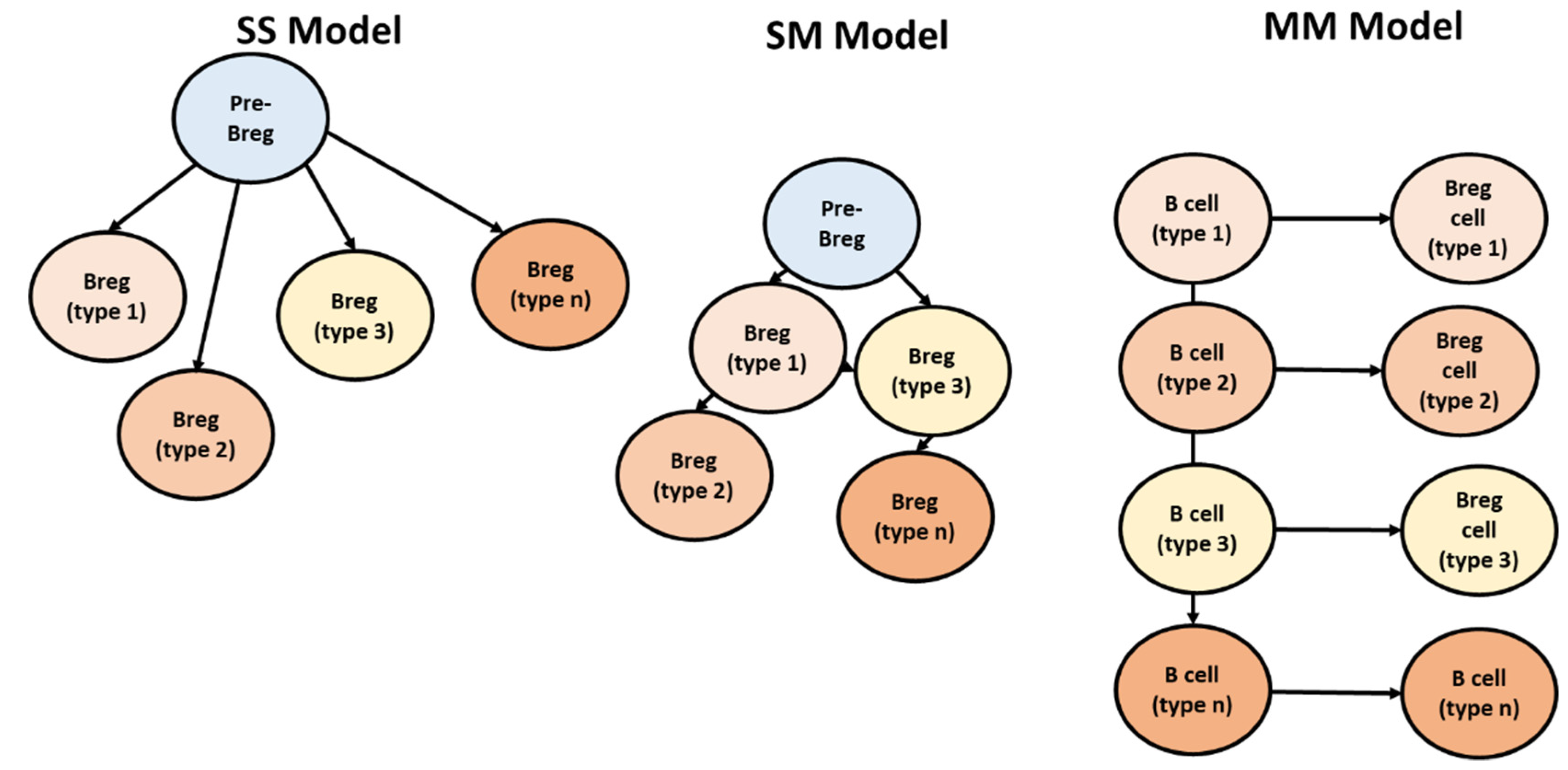
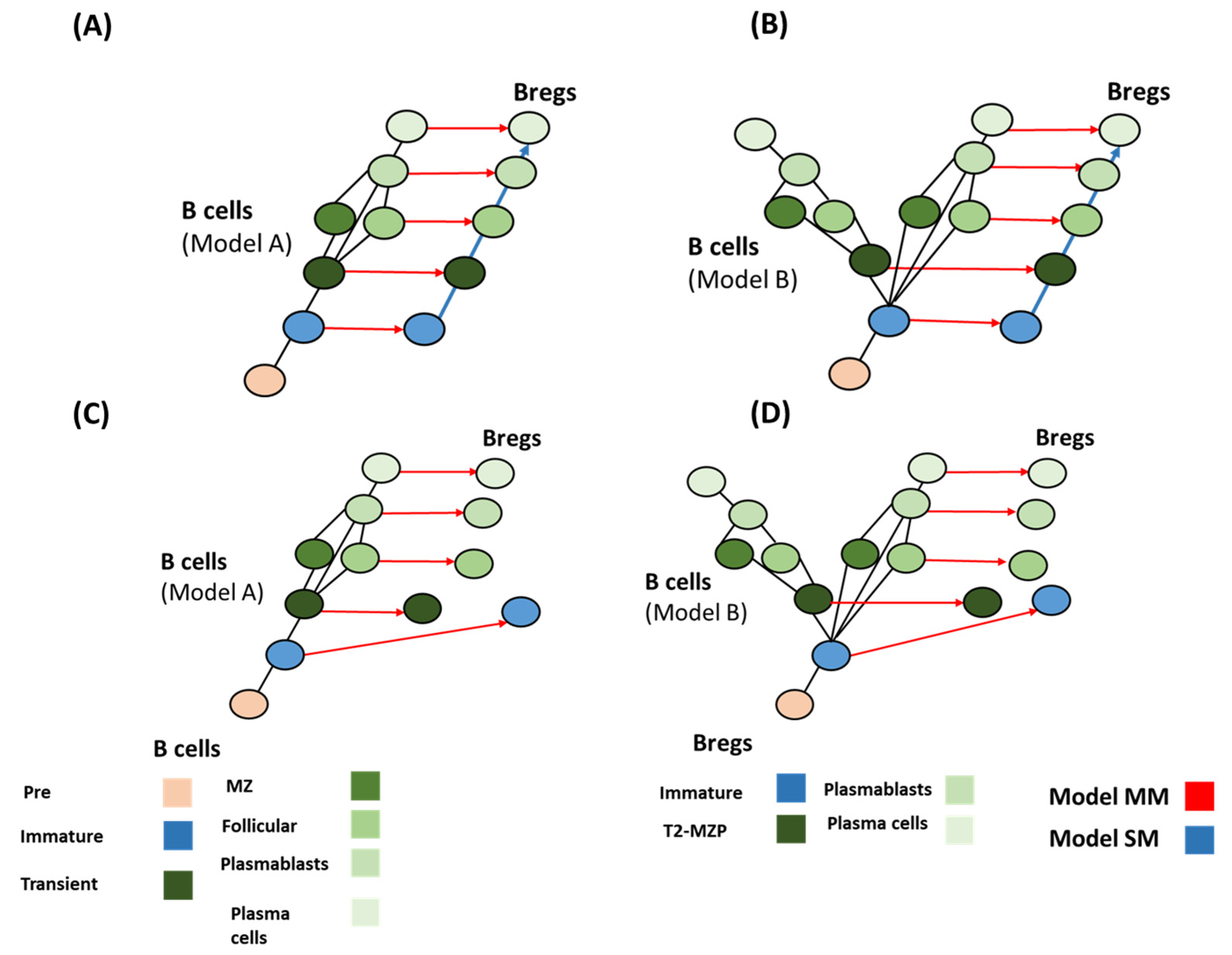
4.3. Breg Development Models
4.4. Deduction of Breg Evolution Models
5. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Guzman-Genuino, R.M.; Hayball, J.D.; Diener, K.R. Regulatory B Cells: Dark Horse in Pregnancy Immunotherapy? J. Mol. Biol. 2021, 433, 166596. [Google Scholar] [CrossRef] [PubMed]
- Fugger, L.; Jensen, L.T.; Rossjohn, J. Challenges, Progress, and Prospects of Developing Therapies to Treat Autoimmune Diseases. Cell 2020, 181, 63–80. [Google Scholar] [CrossRef] [PubMed]
- Cherukuri, A.; Mohib, K.; Rothstein, D.M. Regulatory B cells: TIM-1, transplant tolerance, and rejection. Immunol. Rev. 2021, 299, 31–44. [Google Scholar] [CrossRef] [PubMed]
- Fontenot, J.D.; Gavin, M.A.; Rudensky, A.Y. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat. Immunol. 2003, 4, 330–336. [Google Scholar] [CrossRef] [PubMed]
- Bhaumik, S.; Mickael, M.E.; Moran, M.; Spell, M.; Basu, R. RORγt Promotes Foxp3 Expression by Antagonizing the Effector Program in Colonic Regulatory T Cells. J. Immunol. 2021, 207, 2027–2038. [Google Scholar] [CrossRef] [PubMed]
- Kubick, N.; Flournoy, P.C.H.; Enciu, A.-M.; Manda, G.; Mickael, M.-E. Drugs Modulating CD4+ T Cells Blood–Brain Barrier Interaction in Alzheimer’s Disease. Pharmaceutics 2020, 12, 880. [Google Scholar] [CrossRef] [PubMed]
- Mickael, M.E.; Bhaumik, S.; Basu, R. Retinoid-Related Orphan Receptor RORγt in CD4+ T-Cell–Mediated Intestinal Homeostasis and Inflammation. Am. J. Pathol. 2020, 190, 1984–1999. [Google Scholar] [CrossRef]
- Katz, S.I.; Parker, D.; Turk, J.L. B-cell suppression of delayed hypersensitivity reactions. Nature 1974, 251, 550–551. [Google Scholar] [CrossRef]
- Wolf, S.D.; Dittel, B.N.; Hardardottir, F.; Janeway, C.A. Experimental autoimmune encephalomyelitis induction in genetically B cell-deficient mice. J. Exp. Med. 1996, 184, 2271–2278. [Google Scholar] [CrossRef] [Green Version]
- Xie, J.; Shi, C.-W.; Huang, H.-B.; Yang, W.-T.; Jiang, Y.-L.; Ye, L.-P.; Zhao, Q.; Yang, G.-L.; Wang, C.-F. Induction of the IL-10-producing regulatory B cell phenotype following Trichinella spiralis infection. Mol. Immunol. 2021, 133, 86–94. [Google Scholar] [CrossRef]
- Mizoguchi, A.; Mizoguchi, E.; Takedatsu, H.; Blumberg, R.S.; Bhan, A.K. Chronic intestinal inflammatory condition generates IL-10-producing regulatory B cell subset characterized by CD1d upregulation. Immunity 2002, 16, 219–230. [Google Scholar] [CrossRef] [Green Version]
- Zwollo, P.; Cole, S.; Bromage, E.; Kaattari, S. B Cell Heterogeneity in the Teleost Kidney: Evidence for a Maturation Gradient from Anterior to Posterior Kidney. J. Immunol. 2005, 174, 6608–6616. [Google Scholar] [CrossRef] [Green Version]
- Mauri, C.; Menon, M. The expanding family of regulatory B cells. Int. Immunol. 2015, 27, 479–486. [Google Scholar] [CrossRef] [Green Version]
- Arendt, D.; Musser, J.M.; Baker, C.V.H.; Bergman, A.; Cepko, C.; Erwin, D.H.; Pavlicev, M.; Schlosser, G.; Widder, S.; Laubichler, M.D.; et al. The origin and evolution of cell types. Nat. Rev. Genet. 2016, 17, 744–757. [Google Scholar] [CrossRef] [PubMed]
- Winkler, T.H.; Martensson, I.L. The role of the pre-b cell receptor in b cell development, repertoire selection, and tolerance. Front. Immunol. 2018, 9, 2423. [Google Scholar] [CrossRef] [PubMed]
- Vettermann, C.; Schlissel, M.S. Allelic exclusion of immunoglobulin genes: Models and mechanisms. Immunol. Rev. 2010, 237, 22–42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chung, J.B.; Silverman, M.; Monroe, J.G. Transitional B cells: Step by step towards immune competence. Trends Immunol. 2003, 24, 342–348. [Google Scholar] [CrossRef]
- Noviski, M.; Mueller, J.L.; Satterhwaite, A.; Garret-Sinha, L.A.; Brombacher, F.; Zikherman, J. IgM and igD b cell receptors differentially respond to endogenous antigens and control B cell fate. Elife 2018, 7, e35074. [Google Scholar] [CrossRef]
- Kraal, G.; Mebius, R. New Insights into the Cell Biology of the Marginal Zone of the Spleen. Int. Rev. Cytol. 2006, 250, 175–215. [Google Scholar] [CrossRef]
- Ghia, P.; Ten Boekel, E.; Rolink, A.G.; Melchers, F. B-cell development: A comparison between mouse and man. Immunol. Today 1998, 19, 480–485. [Google Scholar] [CrossRef]
- Sanz, I.; Wei, C.; Jenks, S.A.; Cashman, K.S.; Tipton, C.; Woodruff, M.C.; Hom, J.; Lee, F.E.-H. Challenges and opportunities for consistent classification of human b cell and plasma cell populations. Front. Immunol. 2019, 10, 2458. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsai, D.Y.; Hung, K.H.; Chang, C.W.; Lin, K.I. Regulatory mechanisms of B cell responses and the implication in B cell-related diseases. J. Biomed. Sci. 2019, 26, 64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Von Muenchow, L.; Tsapogas, P.; Alberti-Servera, L.; Capoferri, G.; Doelz, M.; Rolink, H.; Bosco, N.; Ceredig, R.; Rolink, A.G. Pro-B cells propagated in stromal cell-free cultures reconstitute functional B-cell compartments in immunodeficient mice. Eur. J. Immunol. 2017, 47, 394–405. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patton, D.T.; Plumb, A.W.; Redpath, S.A.; Osborne, L.C.; Perona-Wright, G.; Abraham, N. The development and survival but not function of follicular B cells is dependent on IL-7Rα Tyr449 signaling. PLoS ONE 2014, 9, e88771. [Google Scholar] [CrossRef] [PubMed]
- Allman, D.; Li, J.; Hardy, R.R. Commitment to the B lymphoid lineage occurs before D(H)-J(H) recombination. J. Exp. Med. 1999, 189, 735–740. [Google Scholar] [CrossRef] [PubMed]
- Wentink, M.W.J.; Kalina, T.; Perez-Andres, M.; del Pino Molina, L.; IJspeert, H.; Kavelaars, F.G.; Lankester, A.C.; Lecrevisse, Q.; van Dongen, J.J.M.; Orfao, A.; et al. Delineating Human B Cell Precursor Development With Genetically Identified PID Cases as a Model. Front. Immunol. 2019, 10, 2680. [Google Scholar] [CrossRef]
- Halverson, R.; Torres, R.M.; Pelanda, R. Receptor editing is the main mechanism of B cell tolerance toward membrane antigens. Nat. Immunol. 2004, 5, 645–660. [Google Scholar] [CrossRef]
- Martin, V.G.; Wu, Y.-C.B.; Townsend, C.L.; Lu, G.H.C.; O’Hare, J.S.; Mozeika, A.; Coolen, A.C.C.; Kipling, D.; Fraternali, F.; Dunn-Walters, D.K. Transitional B cells in early human B cell development-Time to revisit the paradigm? Front. Immunol. 2016, 7, 546. [Google Scholar] [CrossRef] [Green Version]
- Shahaf, G.; Zisman-Rozen, S.; Benhamou, D.; Melamed, D.; Mehr, R. B cell development in the bone marrow is regulated by homeostatic feedback exerted by mature B cells. Front. Immunol. 2016, 7, 77. [Google Scholar] [CrossRef] [Green Version]
- Michelle, D.; Peñaranda, M.; Jensen, I.; Tollersrud, L.G.; Bruun, J.A.; Jørgensen, J.B. Profiling the Atlantic salmon IgM+ B cell surface proteome: Novel information on teleost fish B cell protein repertoire and identification of potential B cell markers. Front. Immunol. 2019, 10, 37. [Google Scholar] [CrossRef] [Green Version]
- Page, D.M.; Wittamer, V.; Bertrand, J.Y.; Lewis, K.L.; Pratt, D.N.; Delgado, N.; Schale, S.E.; McGue, C.; Jacobsen, B.H.; Doty, A.; et al. An evolutionarily conserved program of B-cell development and activation in zebrafish. Blood J. Am. Soc. Hematol. 2013, 122, e1–e11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sunyer, J.O. Evolutionary and functional relationships of B cells from fish and mammals: Insights into their novel roles in phagocytosis and presentation of particulate antigen. Infect. Disord. Drug Targets 2012, 12, 200–212. [Google Scholar] [CrossRef]
- Bemark, M. Translating transitions-How to decipher peripheral human B cell development. J. Biomed. Res. 2015, 29, 264. [Google Scholar] [CrossRef] [Green Version]
- Fillatreau, S.; Six, A.; Magadan, S.; Castro, R.; Sunyer, J.O.; Boudinot, P. The astonishing diversity of Ig classes and B cell repertoires in teleost fish. Front. Immunol. 2013, 4, 28. [Google Scholar] [CrossRef] [Green Version]
- Kubick, N.; Klimovich, P.; Flournoy, P.H.; Bienkowska, I.; Lazarczyk, M.; Sacharczuk, M.; Bhaumik, S.; Mickael, M.-E.; Basu, R. Interleukins and Interleukin Receptors Evolutionary History and Origin in Relation to CD4+ T Cell Evolution. Genes 2021, 12, 813. [Google Scholar] [CrossRef]
- Little, T.J.; Hultmark, D.; Read, A.F. Invertebrate immunity and the limits of mechanistic immunology. Nat. Immunol. 2005, 6, 651–654. [Google Scholar] [CrossRef]
- Parra, D.; Takizawa, F.; Sunyer, J.O. Evolution of B cell immunity. Annu. Rev. Anim. Biosci. 2013, 1, 65–97. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, N.C.; Rise, M.L.; Christian, S.L. A Comparison of the Innate and Adaptive Immune Systems in Cartilaginous Fish, Ray-Finned Fish, and Lobe-Finned Fish. Front. Immunol. 2019, 10, 2292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tafalla, C.; González, L.; Castro, R.; Granja, A.G. B Cell-Activating Factor Regulates Different Aspects of B Cell Functionality and Is Produced by a Subset of Splenic B Cells in Teleost Fish. Front. Immunol. 2017, 8, 295. [Google Scholar] [CrossRef] [Green Version]
- Schouten, J.; Clister, T.; Bruce, A.; Epp, L.; Zwollo, P. Sockeye salmon retain immunoglobulin-secreting plasma cells throughout their spawning journey and post-spawning. Dev. Comp. Immunol. 2013, 40, 202–209. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.I.; Rothstein, D.M.; Markmann, J.F. Role of B cells in tolerance induction. Curr. Opin. Organ Transplant. 2015, 20, 369. [Google Scholar] [CrossRef] [Green Version]
- Matsumoto, M.; Baba, A.; Yokota, T.; Nishikawa, H.; Ohkawa, Y.; Kayama, H.; Kallies, A.; Nutt, S.L.; Sakaguchi, S.; Takeda, K.; et al. Interleukin-10-producing plasmablasts exert regulatory function in autoimmune inflammation. Immunity 2014, 41, 1040–1051. [Google Scholar] [CrossRef] [Green Version]
- Tedder, T.F. B10 Cells: A Functionally Defined Regulatory B Cell Subset. J. Immunol. 2015, 194, 1395–1401. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blair, P.A.; Norena, L.Y.; Flores-Borja, F.; Rawlings, D.J.; Isenberg, D.A.; Ehrenstein, M.R.; Mauri, C. CD19+CD24hiCD38hi B Cells Exhibit Regulatory Capacity in Healthy Individuals but Are Functionally Impaired in Systemic Lupus Erythematosus Patients. Immunity 2010, 32, 129–140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neves, P.; Lampropoulou, V.; Calderon-Gomez, E.; Roch, T.; Stervbo, U.; Shen, P.; Kuhl, A.A.; Loddenkemper, C.; Haury, M.; Nedospasov, S.A.; et al. Signaling via the MyD88 adaptor protein in B cells suppresses protective immunity during salmonella typhimurium infection. Immunity 2010, 33, 777–790. [Google Scholar] [CrossRef] [Green Version]
- Shen, P.; Roch, T.; Lampropoulou, V.; O’Conor, R.A.; Stervbo, U.; Hilgenberg, E.; Ries, S.; Dang, V.D.; Jaimes, Y.; Daridon, C.; et al. IL-35-producing B cells are critical regulators of immunity during autoimmune and infectious diseases. Nature 2014, 507, 366–370. [Google Scholar] [CrossRef] [Green Version]
- Carter, N.A.; Vasconcellos, R.; Rosser, E.C.; Tulone, C.; Munoz-Suano, A.; Karmanaka, M.; Ehrenstein, M.R.; Flavell, R.A.; Mauri, C. Mice Lacking Endogenous IL-10–Producing Regulatory B Cells Develop Exacerbated Disease and Present with an Increased Frequency of Th1/Th17 but a Decrease in Regulatory T Cells. J. Immunol. 2011, 186, 5569–5579. [Google Scholar] [CrossRef] [Green Version]
- Evans, J.G.; Chavez-Rueda, K.A.; Eddaoudi, A.; Meyer-Bahlburg, A.; Rawlings, D.J.; Ehrenstein, M.R.; Mauri, C. Novel Suppressive Function of Transitional 2 B Cells in Experimental Arthritis. J. Immunol. 2007, 178, 7868–7878. [Google Scholar] [CrossRef] [PubMed]
- Blair, P.A.; Ehrenstein, M.R.; Mauri, C. Immunoregulatory potential of T2-MZP B cells. Future Rheumatol. 2008, 3, 79. [Google Scholar] [CrossRef]
- Su, T.T.; Guo, B.; Wei, B.; Braun, J.; Rawlings, D.J. Signaling in transitional type 2 B cells is critical for peripheral B-cell development. Immunol. Rev. 2004, 197, 161–178. [Google Scholar] [CrossRef]
- Cerutti, A.; Cols, M.; Puga, I. Marginal zone B cells: Virtues of innate-like antibody-producing lymphocytes. Nat. Rev. Immunol. 2013, 13, 118–132. [Google Scholar] [CrossRef] [Green Version]
- O’garra, A.; Chang, R.; Go, N.; Hastings, R.; Haughton, G.; Howard, M. Ly-1 B (B-1) cells are the main source of B cell-derived interleukin 10. Eur. J. Immunol. 1992, 22, 711–717. [Google Scholar] [CrossRef]
- Iwata, Y.; Matsushita, T.; Horikawa, M.; DiLillo, D.J.; Yanaba, K.; Venturi, G.M.; Szabolsc, P.M.; Bernstein, S.H.; Magro, C.M.; Williams, A.D.; et al. Characterization of a rare IL-10-competent B-cell subset in humans that parallels mouse regulatory B10 cells. Blood J. Am. Soc. Hematol. 2011, 117, 530–541. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yanaba, K.; Bouaziz, J.-D.; Haas, K.M.; Poe, J.C.; Fujimoto, M.; Tedder, T.F. A Regulatory B Cell Subset with a Unique CD1dhiCD5+ Phenotype Controls T Cell-Dependent Inflammatory Responses. Immunity 2008, 28, 639–650. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suzuki-Yamazaki, N.; Yanobu-Takanashi, R.; Okamura, T.; Takaki, S. IL-10 production in murine IgM+CD138hi cells is driven by Blimp-1 and downregulated in class-switched cells. Eur. J. Immunol. 2017, 47, 493–503. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Shen, C.; Liu, Y.; Li, Y.; Sun, L.; Jiao, L.; Jiao, W.; Xiao, J.; Shen, C.; Qi, H.; et al. Impaired function of CD5+CD19+CD1dhi B10 cells on IgE secretion in an atopic dermatitis-like mouse model. PLoS ONE 2015, 10, e0132173. [Google Scholar] [CrossRef] [PubMed]
- Yoshizaki, A.; Miyagaki, T.; DiLillo, D.J.; Matsushita, T.; Horikawa, M.; Kountikov, E.I.; Spolski, R.; Poe, J.C.; Leonard, W.J.; Tedder, T.F. Regulatory B cells control T-cell autoimmunity through IL-21-dependent cognate interactions. Nature 2012, 491, 264–268. [Google Scholar] [CrossRef]
- Ding, Q.; Yeung, M.; Camirand, G.; Zeng, Q.; Akiba, H.; Yagita, H.; Chalasani, G.; Sayegh, M.H.; Najafian, N.; Rothstein, D.M. Regulatory B cells are identified by expression of TIM-1 and can be induced through TIM-1 ligation to promote tolerance in mice. J. Clin. Investig. 2011, 121, 3645–3656. [Google Scholar] [CrossRef] [Green Version]
- Aravena, O.; Ferrier, A.; Menon, M.; Mauri, C.; Agullion, J.C.; Soto, L.; Catalan, D. TIM-1 defines a human regulatory B cell population that is altered in frequency and function in systemic sclerosis patients. Arthritis Res. Ther. 2017, 19, 8. [Google Scholar] [CrossRef] [Green Version]
- Mickael, M.E.; Rajput, A.; Steyn, J.; Wiemerslage, L.; Bürglin, T. An optimised phylogenetic method sheds more light on the main branching events of rhodopsin-like superfamily. Comp. Biochem. Physiol. Part D Genom. Proteom. 2016, 20, 85–94. [Google Scholar] [CrossRef]
- Mickael, M.-E.; Kubick, N.; Klimovich, P.; Henckel, P.; Bieńkowska, I.; Sacharczuk, M. Paracellular and Transcellular Leukocytes Diapedesis Are Divergent but Interconnected Evolutionary Events. Genes 2021, 12, 254. [Google Scholar] [CrossRef] [PubMed]
- Kubick, N.; Brösamle, D.; Mickael, M.E. Molecular Evolution and Functional Divergence of the IgLON Family. Evol. Bioinforma. 2018, 14, 1176934318775081. [Google Scholar] [CrossRef] [PubMed] [Green Version]
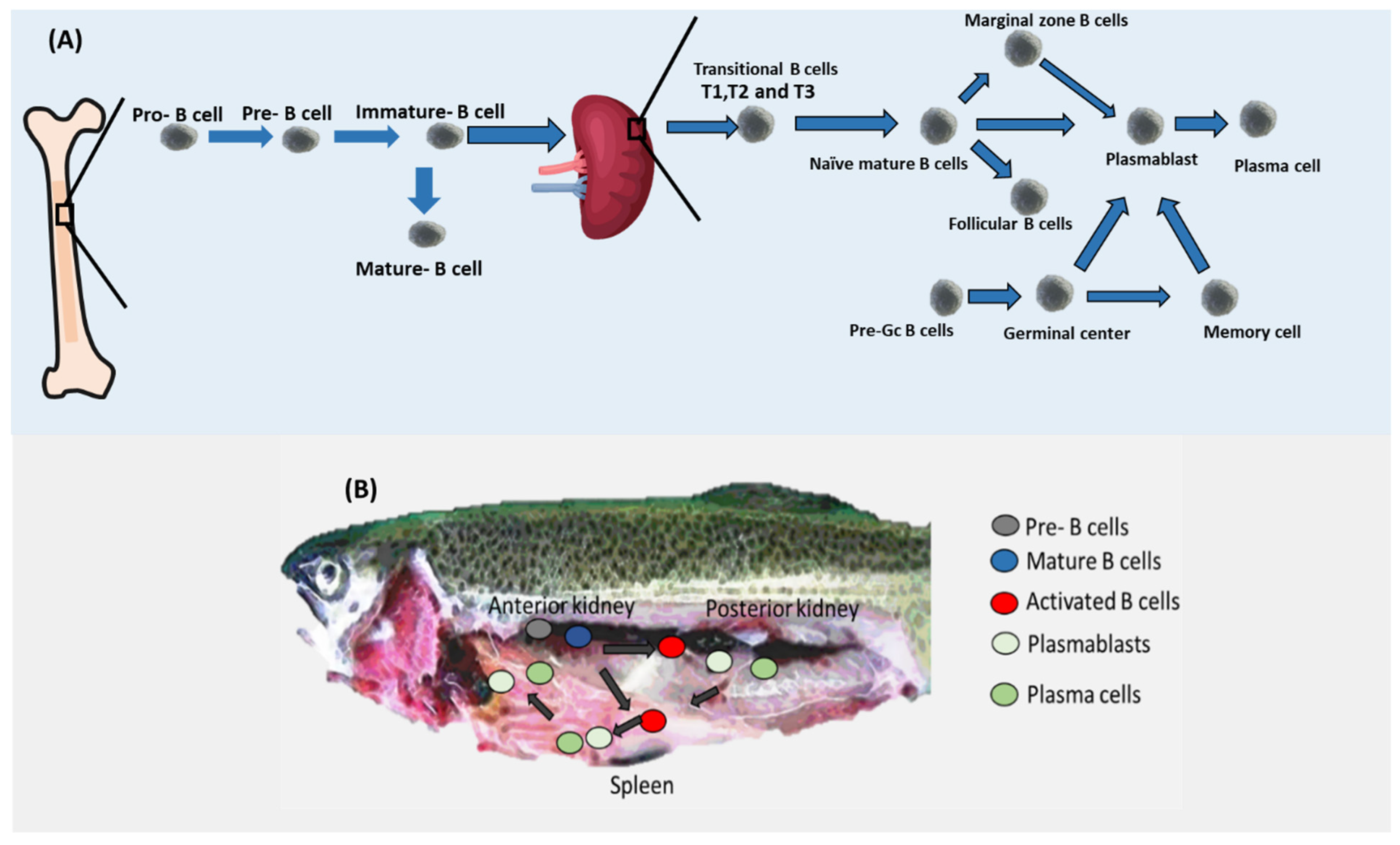
| Cell Type | Humans | Mice | Fish |
|---|---|---|---|
| B cell commitment | Pax5 | Pax5 | Pax5 |
| Pre-pro | CD117 CD10 CD34+ CD38+ Pax5+ | Lin– B220/CD45 R+ CD19− CD24low CD43+ C1q R1/CD93+ CD117/c-kit− CXCR4+ Flt-3/Flk-2+ IL-7 Ra+ IgM | Not reported in fish |
| Pro-B cells | (CD19+ CD10+ CD34+ IgM−) | Lin– B220/CD45R+ CD19+ CD24+ CD43+ CD117low IL7 Rα+ IgM− | Not reported in fish |
| Pre-B cells | (CD19+ CD10+ CD34− IgM−) | Lin− B220/CD45R+ CD19+ CD24+ CD43− IL-7Rα+ IgM−. | B220 [12] |
| Immature B cells | CD10+ CD19+ CD20+ CD21+ CD24+ CD27− CD38+ CD40+ CD93+ IL4Rα+ IL7 Rα− [33] | CD45R+ CD19+ CD24+ CD43− CD93+ IgD− IgM+ | |
| Transitional cells | CD10+ CD5+ CD19+ CD20+ CD21+ CD23+ CD24+ CD27− CD38+ CD93+ TACI+ | CD19+ CD10+ CD34+ CD24+ IgM+ | Not reported in fish |
| Naïve mature B cells | CD19+ CD38+ and IgD+ [21] | pax5 [12,34] | |
| Follicular B cells | CD10− CD19+ CD20+ CD21+ CD22+ CD23+ CD24low CD27− CD38low CXCR5+ TACI+. | CD45R+ CD1dmid CD19mid CD21low CD23+ CD43− CXCR5+ IgMlow IgDhigh. | Reported missing in fish |
| Marginal zone B cells | CD1c+ CD19+ CD20+ CD21+ CD27+ FCRL3 + TACI+ | CD45R+ CD19+ CD21+ CD27+ CD40+. | Not reported in fish |
| Plasmablasts | BCMA+ CD19low CD27high CD38+ CD93+ CD138− | CD45Rlow CD19+ CD27high CD38+ CD138+ | [34] (blimp or pax5) |
| Plasma cells | BCMA+ BLIMP1+ CD19low CD20low CD27high CD38high CD138+ CXCR4+ | CD45Rlow BLIMP1+ CD19− CD27high, CD38low CXCR4high CD138+. | [34] BLIMP1+ |
| Class | Type of Bregs | Human | Mice |
|---|---|---|---|
| Nomenclature related to classical B2 cells | Immature Bregs | CD19+CD24hiCD38hi [44] | - |
| T2-MZP | - | CD19+ CD21hi CD23hi CD24hi | |
| MZ cells | - | CD19+CD21hi CD23− | |
| Plasma blasts | CD27intCD38hi [42] | CD138+CD44hi [42] | |
| Plasma cells | - | CD138hiIgM+TACI+CXCR4+CD1dhiTim1int | |
| Br1 cells | CD19+ CD25hi CD71hi | - | |
| Tim1 B cells | - | Tim1+ CD19+ | |
| B10 | CD19+CD24hiCD27+ [53] | CD19+ CD5+ CD1dhi [54] |
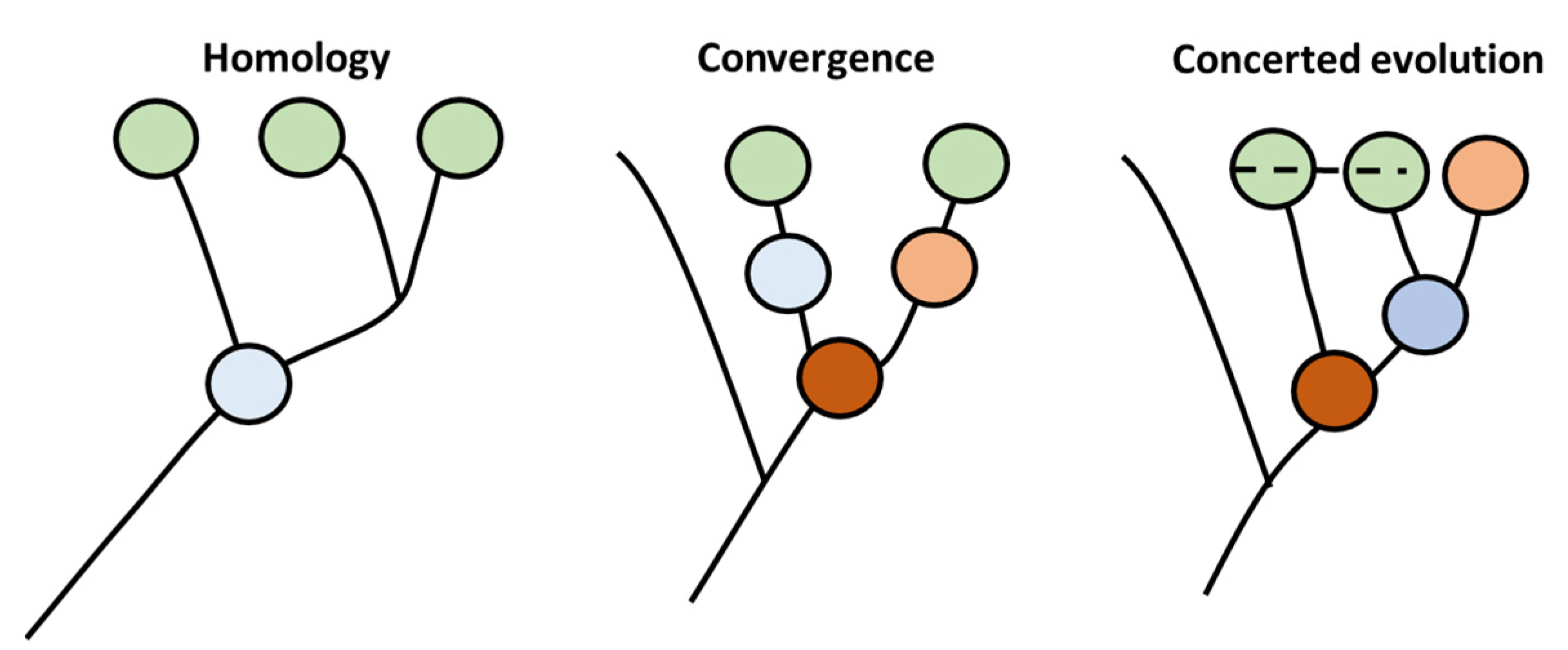
| Assumption 1 | Assumption 2 | Model | Homology | Convergence | Concerted |
|---|---|---|---|---|---|
| Transient B cell stages found in sharks | Immature Bregs exist in sharks | SS | Incompatible | Incompatible | Incompatible |
| SM | Compatible | Incompatible | Incompatible | ||
| MM | Compatible | Incompatible | Incompatible | ||
| Immature Bregs do not exist in sharks | SS | Incompatible | Incompatible | Incompatible | |
| SM | Compatible | Incompatible | Incompatible | ||
| MM | Compatible | Incompatible | Incompatible | ||
| Transient B cell stages found in sharks | Immature Bregs exist in sharks | SS | Incompatible | Incompatible | Incompatible |
| SM | Compatible | Incompatible | Incompatible | ||
| MM | Incompatible | Incompatible | Incompatible | ||
| Immature Bregs do not exist in sharks | SS | Incompatible | Incompatible | Incompatible | |
| SM | Compatible | Incompatible | Incompatible | ||
| MM | Incompatible | Incompatible | Incompatible |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mickael, M.-E.; Bieńkowska, I.; Sacharczuk, M. An Update on the Evolutionary History of Bregs. Genes 2022, 13, 890. https://doi.org/10.3390/genes13050890
Mickael M-E, Bieńkowska I, Sacharczuk M. An Update on the Evolutionary History of Bregs. Genes. 2022; 13(5):890. https://doi.org/10.3390/genes13050890
Chicago/Turabian StyleMickael, Michel-Edwar, Irmina Bieńkowska, and Mariusz Sacharczuk. 2022. "An Update on the Evolutionary History of Bregs" Genes 13, no. 5: 890. https://doi.org/10.3390/genes13050890
APA StyleMickael, M.-E., Bieńkowska, I., & Sacharczuk, M. (2022). An Update on the Evolutionary History of Bregs. Genes, 13(5), 890. https://doi.org/10.3390/genes13050890





