Single-Cell RNA-Seq Analysis Reveals Lung Epithelial Cell Type-Specific Responses to HDM and Regulation by Tet1
Abstract
1. Introduction
2. Methods
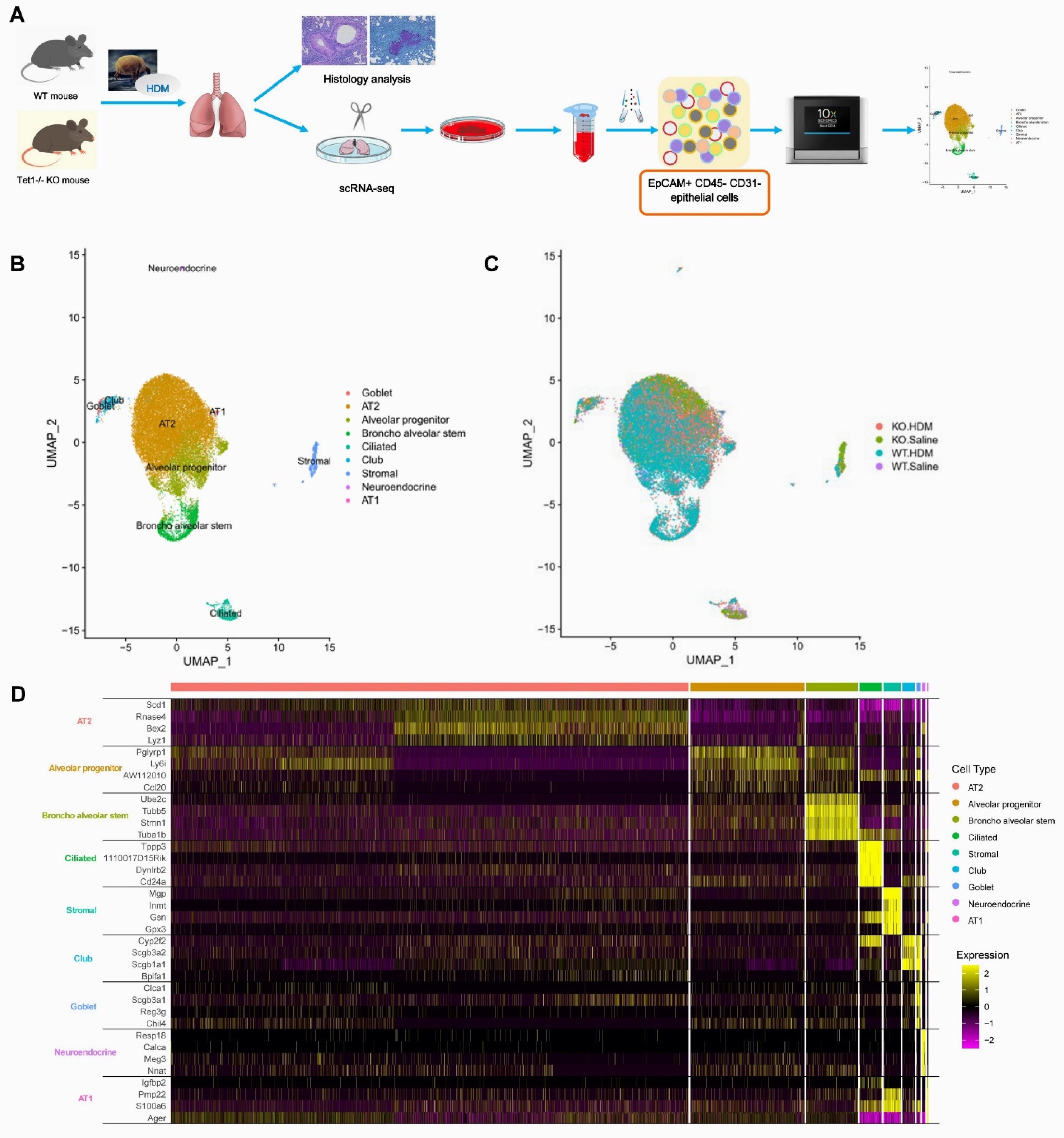
2.1. Establishment of HDM-Induced Airway Inflammation Mouse Model and Isolation of EpCAM+ CD31− CD45− (EpCAM+) Lung Epithelial Cells
2.2. Single-Cell Library Preparation and Sequencing
2.3. Single-Cell RNA-Sequencing Data Analysis
2.4. Transcription Factor Activity Analysis
2.5. Single-Cell Trajectory Analysis
3. Results
3.1. Single-Cell Analysis Identified Nine Cell Types in the EpCAM+ CD45− CD31− Cells from Mouse Lungs
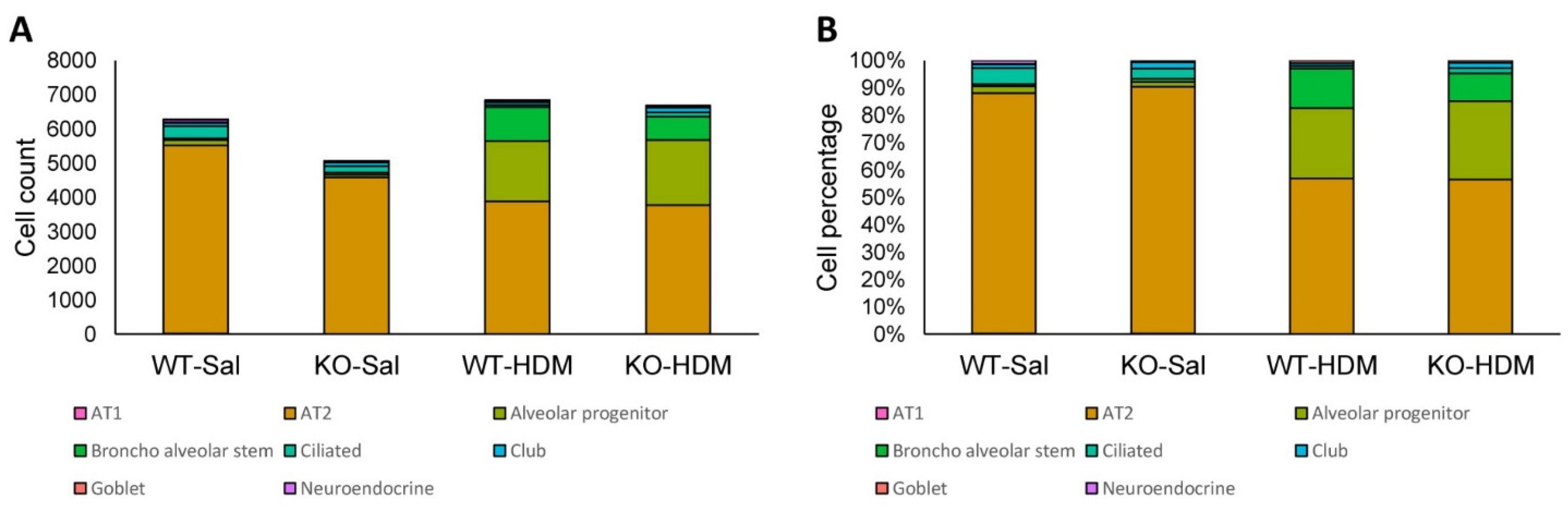
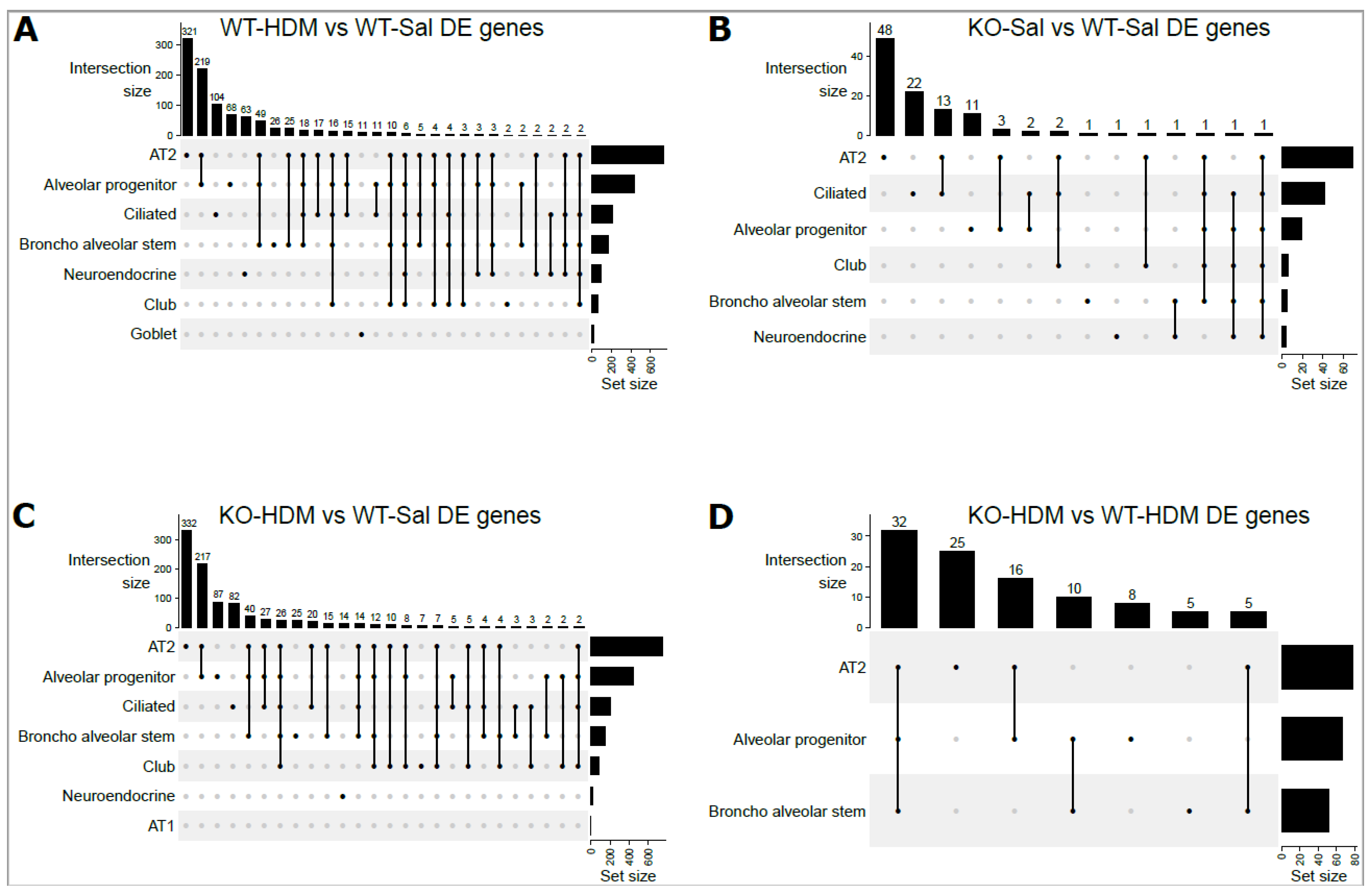
3.2. Exposure to HDM Led to Cell Type-Specific Changes in the EpCAM+ Lung Epithelial Cells
3.3. Tet1 Knockout Led to Cell-Specific Changes in Baseline EpCAM+ Lung Epithelial Cells
3.4. Tet1 Knockout and HDM Exposure Induced Overlapping Cell Type-Specific Changes in EpCAM+ Lung Epithelial Cells
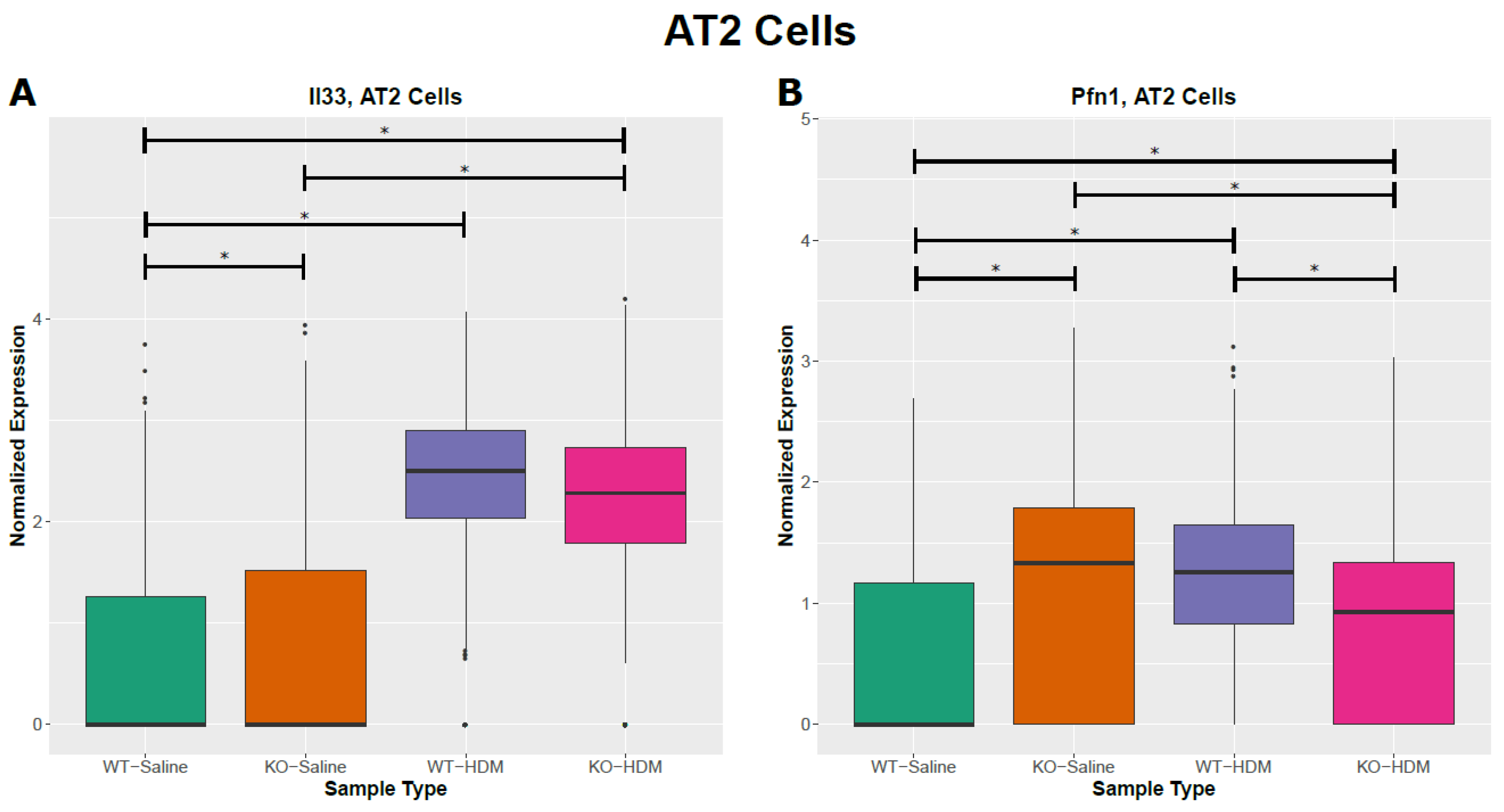
3.5. Tet1 Dysregulates the Single-Cell Transcriptomic Signature of HDM-Induced Lung Inflammation in Mice
3.6. Tet1 Knockout and HDM Exposure Induced Cell Type-Specific Changes in Transcription Factor (TF) Activity in EpCAM+ Lung Epithelial Cells
3.7. HDM, Not Tet1, Promotes the Differentiation of AP and BAS Cells into AT2 Cells
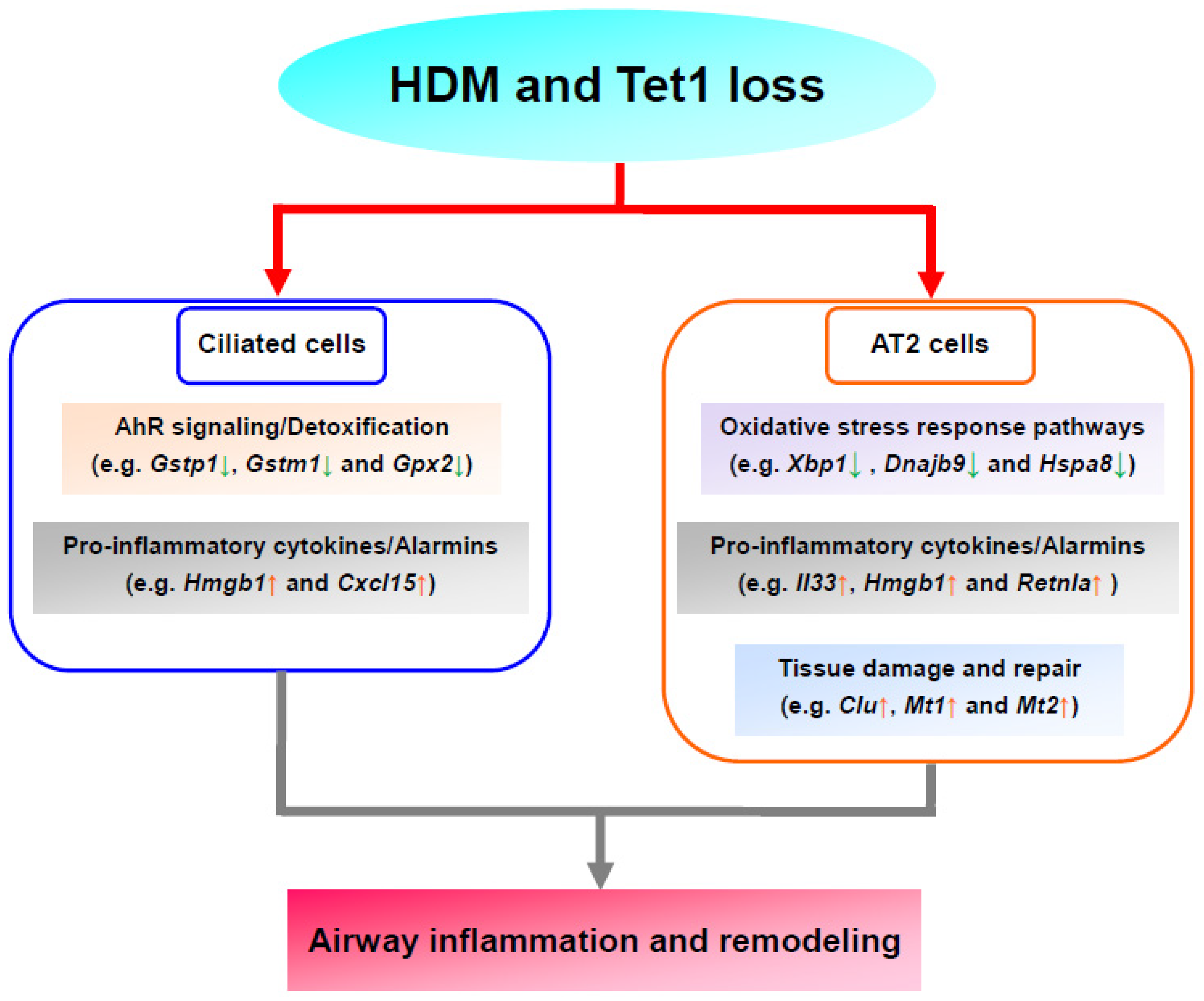
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Scherzer, R.; Grayson, M.H. Heterogeneity and the origins of asthma. Ann. Allergy Asthma Immunol. 2018, 121, 400–405. [Google Scholar] [CrossRef]
- Xu, C.-J.; Söderhäll, C.; Bustamante, M.; Baïz, N.; Gruzieva, O.; Gehring, U.; Mason, D.; Chatzi, L.; Basterrechea, M.; Llop, S.; et al. DNA methylation in childhood asthma: An epigenome-wide meta-analysis. Lancet Respir. Med. 2018, 6, 379–388. [Google Scholar] [CrossRef]
- Lee, Y.; Hwang, Y.-H.; Kim, K.-J.; Park, A.-K.; Paik, M.-J.; Kim, S.H.; Lee, S.U.; Yee, S.-T.; Son, Y.-J. Proteomic and transcriptomic analysis of lung tissue in OVA-challenged mice. Arch. Pharmacal Res. 2018, 41, 87–100. [Google Scholar] [CrossRef]
- Cui, J.; Lv, Z.; Teng, F.; Yi, L.; Tang, W.; Wang, W.; Tulake, W.; Qin, J.; Zhu, X.; Wei, Y.; et al. RNA-Seq Expression Analysis of Chronic Asthmatic Mice with Bu-Shen-Yi-Qi Formula Treatment and Prediction of Regulated Gene Targets of Anti-Airway Remodeling. Evid. -Based Complement. Altern. Med. 2021, 2021, 1–9. [Google Scholar] [CrossRef]
- Zhu, T.; Zhang, X.; Chen, X.; Brown, A.P.; Weirauch, M.T.; Guilbert, T.W.; Hershey, G.K.K.; Biagini, J.M.; Ji, H. Nasal DNA methylation differentiates severe from non-severe asthma in African-American children. Allergy 2021, 76, 1836–1845. [Google Scholar] [CrossRef]
- Forno, E.; Wang, T.; Qi, C.; Yan, Q.; Xu, C.-J.; Boutaoui, N.; Han, Y.-Y.; Weeks, D.; Jiang, Y.; Rosser, F.; et al. DNA methylation in nasal epithelium, atopy, and atopic asthma in children: A genome-wide study. Lancet Respir. Med. 2019, 7, 336–346. [Google Scholar] [CrossRef]
- Qi, C.; Jiang, Y.; Yang, I.V.; Forno, E.; Wang, T.; Vonk, J.M.; Gehring, U.; Smit, H.A.; Milanzi, E.; Carpaij, O.A.; et al. Nasal DNA methylation profiling of asthma and rhinitis. J. Allergy Clin. Immunol. 2020, 145, 1655–1663. [Google Scholar] [CrossRef]
- Yu, Q.; Zhou, B.; Zhang, Y.; Nguyen, E.T.; Du, J.; Glosson, N.L.; Kaplan, M.H. DNA methyltransferase 3a limits the expression of interleukin-13 in T helper 2 cells and allergic airway inflammation. Proc. Natl. Acad. Sci. USA 2012, 109, 541–546. [Google Scholar] [CrossRef]
- Burleson, J.D.; Siniard, D.; Yadagiri, V.K.; Chen, X.; Weirauch, M.T.; Ruff, B.P.; Brandt, E.; Hershey, G.K.K.; Ji, H. TET1 contributes to allergic airway inflammation and regulates interferon and aryl hydrocarbon receptor signaling pathways in bronchial epithelial cells. Sci. Rep. 2019, 9, 7361. [Google Scholar] [CrossRef]
- Zhang, X.; Chen, X.; Weirauch, M.T.; Zhang, X.; Burleson, J.D.; Brandt, E.; Ji, H. Diesel exhaust and house dust mite allergen lead to common changes in the airway methylome and hydroxymethylome. Environ. Epigenetics 2018, 4, dvy020. [Google Scholar] [CrossRef]
- Somineni, H.K.; Zhang, X.; Biagini Myers, J.M.; Kovacic, M.B.; Ulm, A.; Jurcak, N.; Ryan, P.H.; Hershey, G.; Ji, H. Ten-eleven translocation 1 (TET1) methylation is associated with childhood asthma and traffic-related air pollution. J. Allergy Clin. Immunol. 2016, 137, 797.e795–805.e795. [Google Scholar] [CrossRef]
- Zhu, T.; Brown, A.P.; Ji, H. The Emerging Role of Ten-Eleven Translocation 1 in Epigenetic Responses to Environmental Exposures. Epigenetics Insights 2020, 13, 2516865720910155. [Google Scholar] [CrossRef]
- Zheng, G.X.Y.; Terry, J.M.; Belgrader, P.; Pyvkin, P.; Bent, Z.W.; Wilson, R.; Ziraldo, S.B.; Wheeler, T.D.; McDermott, G.P.; Zhu, J.; et al. Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 2017, 8, 14049. [Google Scholar] [CrossRef]
- Stuart, T.; Butler, A.; Hoffman, P.; Hafemeister, C.; Papalexi, E.; Mauck, W.M., III; Hao, Y.; Stoeckius, M.; Smibert, P.; Satija, R. Comprehensive Integration of Single-Cell Data. Cell 2019, 177, 1888.E21–1902.E21. [Google Scholar] [CrossRef]
- Miettinen, M.; Lindenmayer, A.E.; Chaubal, A. Endothelial cell markers CD31, CD34, and BNH9 antibody to H- and Y-antigens—Evaluation of their specificity and sensitivity in the diagnosis of vascular tumors and comparison with von Willebrand factor. Mod. Pathol. 1994, 7, 82–90. [Google Scholar]
- Scialdone, A.; Natarajan, K.N.; Saraiva, L.; Proserpio, V.; Teichmann, S.; Stegle, O.; Marioni, J.C.; Buettner, F. Computational assignment of cell-cycle stage from single-cell transcriptome data. Methods 2015, 85, 54–61. [Google Scholar] [CrossRef]
- Johansen, N.; Quon, G. scAlign: A tool for alignment, integration, and rare cell identification from scRNA-seq data. Genome Biol. 2019, 20, 166. [Google Scholar] [CrossRef]
- Han, X.; Wang, R.; Zhou, Y.; Fei, L.; Sun, H.; Lai, S.; Saadatpour, A.; Zhou, Z.; Chen, H.; Ye, F.; et al. Mapping the Mouse Cell Atlas by Microwell-Seq. Cell 2018, 172, 1091.e17–1107.e17. [Google Scholar] [CrossRef]
- Schilders, K.A.A.; Eenjes, E.; Van Riet, S.; Poot, A.A.; Stamatialis, D.; Truckenmüller, R.; Hiemstra, P.S.; Rottier, R.J. Regeneration of the lung: Lung stem cells and the development of lung mimicking devices. Respir. Res. 2016, 17, 44. [Google Scholar] [CrossRef]
- Garcia-Alonso, L.; Holland, C.H.; Ibrahim, M.M.; Turei, D.; Saez-Rodriguez, J. Benchmark and integration of resources for the estimation of human transcription factor activities. Genome Res. 2019, 29, 1363–1375. [Google Scholar] [CrossRef]
- Holland, C.H.; Szalai, B.; Saez-Rodriguez, J. Transfer of regulatory knowledge from human to mouse for functional genomics analysis. Biochim. Biophys. Acta Gene Regul Mech. 2019, 1863, 194431. [Google Scholar] [CrossRef] [PubMed]
- Holland, C.H.; Tanevski, J.; Perales-Patón, J.; Gleixner, J.; Kumar, M.P.; Mereu, E.; Joughin, B.A.; Stegle, O.; Lauffenburger, D.A.; Heyn, H.; et al. Robustness and applicability of transcription factor and pathway analysis tools on single-cell RNA-seq data. Genome Biol. 2020, 21, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, M.J.; Shen, Y.; Giorgi, F.M.; Lachmann, A.; Ding, B.B.; Ye, B.H.; Califano, A. Functional characterization of somatic mutations in cancer using network-based inference of protein activity. Nat. Genet. 2016, 48, 838–847. [Google Scholar] [CrossRef]
- Trapnell, C.; Cacchiarelli, D.; Grimsby, J.; Pokharel, P.; Li, S.; A Morse, M.; Lennon, N.J.; Livak, K.J.; Mikkelsen, T.S.; Rinn, J.L. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 2014, 32, 381–386. [Google Scholar] [CrossRef]
- Hong, H.; Liao, S.; Chen, F.; Yang, Q.; Wang, D.-Y. Role of IL-25, IL-33, and TSLP in triggering united airway diseases toward type 2 inflammation. Allergy 2020, 75, 2794–2804. [Google Scholar] [CrossRef] [PubMed]
- Kalchiem-Dekel, O.; Yao, X.; Barochia, A.V.; Kaler, M.; Figueroa, D.M.; Karkowsky, W.B.; Gordon, E.M.; Gao, M.; Fergusson, M.M.; Qu, X.; et al. Apolipoprotein E Signals via TLR4 to Induce CXCL5 Secretion by Asthmatic Airway Epithelial Cells. Am. J. Respir. Cell Mol. Biol. 2020, 63, 185–197. [Google Scholar] [CrossRef] [PubMed]
- Hong, G.H.; Kwon, H.-S.; Lee, K.Y.; Ha, E.H.; Moon, K.-A.; Kim, S.W.; Oh, W.; Kim, T.-B.; Moon, H.-B.; Cho, Y.S. hMSCs suppress neutrophil-dominant airway inflammation in a murine model of asthma. Exp. Mol. Med. 2017, 49, e288. [Google Scholar] [CrossRef]
- Busse, P.J.; Birmingham, J.M.; Calatroni, A.; Manzi, J.; Goryachokovsky, A.; Fontela, G.; Federman, A.D.; Wisnivesky, J.P. Effect of aging on sputum inflammation and asthma control. J. Allergy Clin. Immunol. 2017, 139, 1808–1818.e1806. [Google Scholar] [CrossRef]
- Sokulsky, L.A.; Garcia-Netto, K.; Nguyen, T.H.; Girkin, J.L.N.; Collison, A.; Mattes, J.; Kaiko, G.; Liu, C.; Bartlett, N.W.; Yang, M.; et al. A Critical Role for the CXCL3/CXCL5/CXCR2 Neutrophilic Chemotactic Axis in the Regulation of Type 2 Responses in a Model of Rhinoviral-Induced Asthma Exacerbation. J. Immunol. 2020, 205, 2468–2478. [Google Scholar] [CrossRef]
- Choi, Y.; Kim, Y.M.; Lee, H.R.; Mun, J.; Sim, S.; Lee, D.; Le Pham, D.; Kim, S.; Shin, Y.S.; Lee, S.; et al. Eosinophil extracellular traps activate type 2 innate lymphoid cells through stimulating airway epithelium in severe asthma. Allergy 2020, 75, 95–103. [Google Scholar] [CrossRef]
- Vedel-Krogh, S.; Rasmussen, K.L.; Nordestgaard, B.G.; Nielsen, S.F. Complement C3 and allergic asthma: A cohort study of the general population. Eur. Respir. J. 2021, 57, 2000645. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Cleary, R.A.; Wang, T.; Li, J.; Tang, D.D. The Association of Cortactin with Profilin-1 Is Critical for Smooth Muscle Contraction. J. Biol. Chem. 2014, 289, 14157–14169. [Google Scholar] [CrossRef] [PubMed]
- Whitsett, J.A. Airway Epithelial Differentiation and Mucociliary Clearance. Ann. Am. Thorac. Soc. 2018, 15 (Suppl. S3), S143–S148. [Google Scholar] [CrossRef] [PubMed]
- Tanabe, T.; Rubin, B.K. Airway Goblet Cells Secrete Pro-Inflammatory Cytokines, Chemokines, and Growth Factors. Chest 2016, 149, 714–720. [Google Scholar] [CrossRef] [PubMed]
- Hellings, P.W.; Steelant, B. Epithelial barriers in allergy and asthma. J. Allergy Clin. Immunol. 2020, 145, 1499–1509. [Google Scholar] [CrossRef]
- Zhu, T.; Chen, Z.; Chen, G.; Wang, D.; Tang, S.; Deng, H.; Wang, J.; Li, S.; Lan, J.; Tong, J.; et al. Curcumin Attenuates Asthmatic Airway Inflammation and Mucus Hypersecretion Involving a PPARgamma-Dependent NF-kappaB Signaling Pathway In Vivo and In Vitro. Mediat. Inflamm. 2019, 2019, 4927430. [Google Scholar] [CrossRef]
- Patil, R.H.; Naveen Kumar, M.; Kiran Kumar, K.M.; Nagesh, R.; Kavya, K.; Babu, R.; Ramesh, G.T.; Sharma, S.C. Dexamethasone inhibits inflammatory response via down regulation of AP-1 transcription factor in human lung epithelial cells. Gene 2018, 645, 85–94. [Google Scholar] [CrossRef]
- Guo, M.; Liu, Y.; Han, X.; Han, F.; Zhu, J.; Zhu, S.; Chen, B. Tobacco smoking aggravates airway inflammation by upregulating endothelin-2 and activating the c-Jun amino terminal kinase pathway in asthma. Int. Immunopharmacol. 2019, 77, 105916. [Google Scholar] [CrossRef]
- Zeng, Z.; Wang, L.; Ma, W.; Zheng, R.; Zhang, H.; Zeng, X.; Zhang, H.; Zhang, W. Inhibiting the Notch signaling pathway suppresses Th17-associated airway hyperresponsiveness in obese asthmatic mice. Lab. Investig. 2019, 99, 1784–1794. [Google Scholar] [CrossRef]
- Feng, J.; Zhang, C.; Wang, Z.; Li, Q.; Li, J.; Wang, H. Association betweenCD14gene promoter polymorphisms with serum total-IgE and eosinophil levels in atopic and non-atopic asthma patients in a Chinese Han population. J. Asthma 2016, 53, 119–124. [Google Scholar] [CrossRef]
- Hachim, M.Y.; Elemam, N.M.; Ramakrishnan, R.K.; Salameh, L.; Olivenstein, R.; Hachim, I.Y.; Venkatachalam, T.; Mahboub, B.; Al Heialy, S.; Halwani, R.; et al. Blood and Salivary Amphiregulin Levels as Biomarkers for Asthma. Front. Med. 2020, 7, 561866. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, N.; Li, X.; Seibold, M.A.; Jarjour, N.N.; Denlinger, L.C.; Castro, M.; Coverstone, A.M.; Teague, W.G.; Boomer, J.; Bleecker, E.R.; et al. The effect of BPIFA1/SPLUNC1 genetic variation on its expression and function in asthmatic airway epithelium. JCI Insight. 2019, 4, e127237. [Google Scholar] [CrossRef] [PubMed]
- Wong, S.L.; To, J.; Santos, J.; Allam, V.S.R.R.; Dalton, J.P.; Jordjevic, S.P.; Donnelly, S.; Padula, M.P.; Sukkar, M.B. Proteomic Analysis of Extracellular HMGB1 Identifies Binding Partners and Exposes Its Potential Role in Airway Epithelial Cell Homeostasis. J. Proteome Res. 2018, 17, 33–45. [Google Scholar] [CrossRef] [PubMed]
- Singhania, A.; Wallington, J.C.; Smith, C.G.; Horowitz, D.; Staples, K.J.; Howarth, P.H.; Gadola, S.D.; Djukanović, R.; Woelk, C.; Hinks, T.S.C. Multitissue Transcriptomics Delineates the Diversity of Airway T Cell Functions in Asthma. Am. J. Respir. Cell Mol. Biol. 2018, 58, 261–270. [Google Scholar] [CrossRef]
- Lkhagvadorj, K.; Meyer, K.F.; Verweij, L.P.; Kooistra, W.; Reinders-Luinge, M.; Dijkhuizen, H.W.; de Graaf, I.A.M.; Plösch, T.; Hylkema, M.N. Prenatal smoke exposure induces persistent Cyp2a5 methylation and increases nicotine metabolism in the liver of neonatal and adult male offspring. Epigenetics 2020, 15, 1370–1385. [Google Scholar] [CrossRef]
- A Sokulsky, L.; Goggins, B.; Sherwin, S.; Eyers, F.; E Kaiko, G.; Board, P.G.; Keely, S.; Yang, M.; Foster, P.S. GSTO1-1 is an upstream suppressor of M2 macrophage skewing and HIF-1α-induced eosinophilic airway inflammation. Clin. Exp. Allergy. 2020, 50, 609–624. [Google Scholar] [CrossRef]
- Lin, T.-J.; Karmaus, W.J.; Chen, M.-L.; Hsu, J.-C.; Wang, I.-J. Interactions Between Bisphenol A Exposure andGSTP1Polymorphisms in Childhood Asthma. Allergy Asthma Immunol. Res. 2018, 10, 172–179. [Google Scholar] [CrossRef]
- Choi, H.; Song, W.-M.; Wang, M.; Sram, R.J.; Zhang, B. Benzo[a]pyrene is associated with dysregulated myelo-lymphoid hematopoiesis in asthmatic children. Environ. Int. 2019, 128, 218–232. [Google Scholar] [CrossRef]
- Poulain-Godefroy, O.; Boute, M.; Carrard, J.; Alvarez-Simon, D.; Tsicopoulos, A.; de Nadai, P. The Aryl Hydrocarbon Receptor in Asthma: Friend or Foe? Int. J. Mol. Sci. 2020, 21, 8797. [Google Scholar] [CrossRef]
- Shang, L.; Wang, L.; Shi, X.; Wang, N.; Zhao, L.; Wang, J.; Liu, C. HMGB1 was negatively regulated by HSF1 and mediated the TLR4/MyD88/NF-kappaB signal pathway in asthma. Life Sci. 2020, 241, 117120. [Google Scholar] [CrossRef]
- Simpson, J.; Loh, Z.; Ullah, M.A.; Lynch, J.P.; Werder, R.B.; Collinson, N.; Zhang, V.; Dondelinger, Y.; Bertrand, M.; Everard, M.; et al. Respiratory Syncytial Virus Infection Promotes Necroptosis and HMGB1 Release by Airway Epithelial Cells. Am. J. Respir. Crit. Care Med. 2020, 201, 1358–1371. [Google Scholar] [CrossRef] [PubMed]
- Todd, J.L.; Kelly, F.L.; Nagler, A.; Banner, K.; Pavlisko, E.N.; Belperio, J.A.; Brass, D.; Weigt, S.S.; Palmer, S. Amphiregulin contributes to airway remodeling in chronic allograft dysfunction after lung transplantation. Am. J. Transplant. 2020, 20, 825–833. [Google Scholar] [CrossRef] [PubMed]
- Nordgren, T.; Heires, A.J.; Bailey, K.L.; Katafiasz, D.M.; Toews, M.L.; Wichman, C.S.; Romberger, D.J. Docosahexaenoic acid enhances amphiregulin-mediated bronchial epithelial cell repair processes following organic dust exposure. Am. J. Physiol. Cell. Mol. Physiol. 2018, 314, L421–L431. [Google Scholar] [CrossRef] [PubMed]
- Cui, H.; Banerjee, S.; Guo, S.; Xie, N.; Ge, J.; Jiang, D.; Zörnig, M.; Thannickal, V.J.; Liu, G. Long noncoding RNA Malat1 regulates differential activation of macrophages and response to lung injury. JCI Insight 2019, 4, e124522. [Google Scholar] [CrossRef]
- Lin, L.; Li, Q.; Hao, W.; Zhang, Y.; Zhao, L.; Han, W. Upregulation of LncRNA Malat1 Induced Proliferation and Migration of Airway Smooth Muscle Cells via miR-150-eIF4E/Akt Signaling. Front. Physiol. 2019, 10, 1337. [Google Scholar] [CrossRef]
- Rohde, D.; Schön, C.; Boerries, M.; Didrihsone, I.; Ritterhoff, J.; Kubatzky, K.F.; Völkers, M.; Herzog, N.; Mähler, M.; Tsoporis, J.N.; et al. S100A1 is released from ischemic cardiomyocytes and signals myocardial damage via Toll-like receptor 4. EMBO Mol. Med. 2014, 6, 778–794. [Google Scholar] [CrossRef]
- Brinks, H.; Rohde, D.; Voelkers, M.; Qiu, G.; Pleger, S.T.; Herzog, N.; Rabinowitz, J.; Ruhparwar, A.; Silvestry, S.; Lerchenmüller, C.; et al. S100A1 Genetically Targeted Therapy Reverses Dysfunction of Human Failing Cardiomyocytes. J. Am. Coll. Cardiol. 2011, 58, 966–973. [Google Scholar] [CrossRef]
- Ma, W.-Y.; Song, R.-J.; Xu, B.-B.; Xu, Y.; Wang, X.-X.; Sun, H.-Y.; Li, S.-N.; Liu, S.-Z.; Yu, M.-X.; Yang, F.; et al. Melatonin promotes cardiomyocyte proliferation and heart repair in mice with myocardial infarction via miR-143-3p/Yap/Ctnnd1 signaling pathway. Acta Pharmacol. Sin. 2021, 42, 921–931. [Google Scholar] [CrossRef]
- Kim, H.G.; Huang, M.; Xin, Y.; Zhang, Y.; Zhang, X.; Wang, G.; Liu, S.; Wan, J.; Ahmadi, A.R.; Sun, Z.; et al. The epigenetic regulator SIRT6 protects the liver from alcohol-induced tissue injury by reducing oxidative stress in mice. J. Hepatol. 2019, 71, 960–969. [Google Scholar] [CrossRef]
- Rawlins, E.L.; Okubo, T.; Xue, Y.; Brass, D.M.; Auten, R.L.; Hasegawa, H.; Wang, F.; Hogan, B.L. The Role of Scgb1a1+ Clara Cells in the Long-Term Maintenance and Repair of Lung Airway, but Not Alveolar, Epithelium. Cell Stem Cell 2009, 4, 525–534. [Google Scholar] [CrossRef]
- Brasier, A.R. Therapeutic targets for inflammation-mediated airway remodeling in chronic lung disease. Expert Rev. Respir. Med. 2018, 12, 931–939. [Google Scholar] [CrossRef] [PubMed]
- Habiel, D.M.; Camelo, A.; Espindola, M.; Burwell, T.; Hanna, R.; Miranda, E.; Carruthers, A.; Bell, M.; Coelho, A.L.; Liu, H.; et al. Divergent roles for Clusterin in Lung Injury and Repair. Sci. Rep. 2017, 7, 15444. [Google Scholar] [CrossRef] [PubMed]
- Hong, J.; Kim, M.N.; Kim, E.G.; Lee, J.; Kim, H.; Kim, S.; Lee, S.; Kim, Y.; Kim, K.; Sohn, M. Clusterin Deficiency Exacerbates Hyperoxia-Induced Acute Lung Injury. Cells 2021, 10, 944. [Google Scholar] [CrossRef] [PubMed]
- Lauzon-Joset, J.-F.; Langlois, A.; Lai, L.J.; Santerre, K.; Lee-Gosselin, A.; Bossé, Y.; Marsolais, D.; Bissonnette, E.Y. Lung CD200 Receptor Activation Abrogates Airway Hyperresponsiveness in Experimental Asthma. Am. J. Respir. Cell Mol. Biol. 2015, 53, 276–284. [Google Scholar] [CrossRef] [PubMed]
- Baos, S.; Calzada, D.; Cremades-Jimeno, L.; de Pedro, M.; Sastre, J.; Picado, C.; Quiralte, J.; Florido, F.; Lahoz, C.; Cárdaba, B. Discriminatory Molecular Biomarkers of Allergic and Nonallergic Asthma and Its Severity. Front. Immunol. 2019, 10, 1051. [Google Scholar] [CrossRef] [PubMed]
- Cardenas, A.; Sordillo, J.E.; Rifas-Shiman, S.L.; Chung, W.; Liang, L.; Coull, B.A.; Hivert, M.-F.; Lai, P.S.; Forno, E.; Celedón, J.C.; et al. The nasal methylome as a biomarker of asthma and airway inflammation in children. Nat. Commun. 2019, 10, 3095. [Google Scholar] [CrossRef]
- Xu, Y.; Wang, Y.; Besnard, V.; Ikegami, M.; Wert, S.E.; Heffner, C.; Murray, S.A.; Donahue, L.R.; Whitsett, J.A. Transcriptional Programs Controlling Perinatal Lung Maturation. PLoS ONE 2012, 7, e37046. [Google Scholar] [CrossRef]
- Little, D.R.; Lynch, A.M.; Yan, Y.; Akiyama, H.; Kimura, S.; Chen, J. Differential chromatin binding of the lung lineage transcription factor NKX2-1 resolves opposing murine alveolar cell fates in vivo. Nat. Commun. 2021, 12, 1–18. [Google Scholar] [CrossRef]
- Zhou, P.; Gu, F.; Zhang, L.; Akerberg, B.N.; Ma, Q.; Li, K.; He, A.; Lin, Z.; Stevens, S.M.; Zhou, B.; et al. Mapping cell type-specific transcriptional enhancers using high affinity, lineage-specific Ep300 bioChIP-seq. eLife 2017, 6, 166. [Google Scholar] [CrossRef]
- Hohl, M.; Thiel, G. Cell type-specific regulation of RE-1 silencing transcription factor (REST) target genes. Eur. J. Neurosci. 2005, 22, 2216–2230. [Google Scholar] [CrossRef]
- Lemeille, S.; Paschaki, M.; Baas, D.; Morlé, L.; Duteyrat, J.-L.; Ait-Lounis, A.; Barras, E.; Soulavie, F.; Jerber, J.; Thomas, J.; et al. Interplay of RFX transcription factors 1, 2 and 3 in motile ciliogenesis. Nucleic Acids Res. 2020, 48, 9019–9036. [Google Scholar] [CrossRef] [PubMed]
- Patir, A.; Fraser, A.M.; Barnett, M.W.; McTeir, L.; Rainger, J.; Davey, M.G.; Freeman, T.C. The transcriptional signature associated with human motile cilia. Sci. Rep. 2020, 10, 1081. [Google Scholar] [CrossRef] [PubMed]
- Besnard, V.; Wert, S.E.; Kaestner, K.H.; Whitsett, J.A. Stage-specific regulation of respiratory epithelial cell differentiation by Foxa1. Am. J. Physiol. Cell. Mol. Physiol. 2005, 289, L750–L759. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mümmler, C.; Burgy, O.; Hermann, S.; Mutze, K.; Günther, A.; Königshoff, M. Cell-specific expression of runt-related transcription factor 2 contributes to pulmonary fibrosis. FASEB J. 2018, 32, 703–716. [Google Scholar] [CrossRef] [PubMed]
- Paranjapye, A.; Mutolo, M.J.; Ebron, J.S.; Leir, S.-H.; Harris, A. The FOXA1 transcriptional network coordinates key functions of primary human airway epithelial cells. Am. J. Physiol. Cell. Mol. Physiol. 2020, 319, L126–L136. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Wang, K.; Li, T.; Chen, J.; Xie, D.; Chang, X.; Yao, J.; Wu, J.; Zhou, Q.; Jia, Y.; et al. Hypoxia-induced TET1 facilitates trophoblast cell migration and invasion through HIF1α signaling pathway. Sci. Rep. 2017, 7, 807. [Google Scholar] [CrossRef]
- Ali, M.M.; Phillips, S.A.; Mahmoud, A.M. HIF1alpha/TET1 Pathway Mediates Hypoxia-Induced Adipocytokine Promoter Hypomethylation in Human Adipocytes. Cells 2020, 9, 134. [Google Scholar] [CrossRef]
- Johnsen, O.; Skammelsrud, N.; Luna, L.; Nishizawa, M.; Prydz, H.; Kolsto, A.B. Small Maf proteins interact with the human transcription factor TCF11/Nrf1/LCR-F1. Nucleic Acids Res. 1996, 24, 4289–4297. [Google Scholar] [CrossRef]
- Steffen, J.; Seeger, M.; Koch, A.; Krüger, E. Proteasomal Degradation Is Transcriptionally Controlled by TCF11 via an ERAD-Dependent Feedback Loop. Mol. Cell 2010, 40, 147–158. [Google Scholar] [CrossRef]
- Radhakrishnan, S.K.; den Besten, W.; Deshaies, R.J. p97-Dependent retrotranslocation and proteolytic processing govern formation of active Nrf1 upon proteasome inhibition. Elife 2014, 3, e01856. [Google Scholar] [CrossRef]
- Tonelli, C.; Chio, I.I.C.; Tuveson, D.A. Transcriptional Regulation by Nrf2. Antioxid. Redox Signal. 2018, 29, 1727–1745. [Google Scholar] [CrossRef] [PubMed]
- Kawakita, N.; Toba, H.; Miyoshi, K.; Sakamoto, S.; Matsumoto, D.; Takashima, M.; Aoyama, M.; Inoue, S.; Morimoto, M.; Nishino, T.; et al. Bronchioalveolar stem cells derived from mouse-induced pluripotent stem cells promote airway epithelium regeneration. Stem Cell Res. Ther. 2020, 11, 430. [Google Scholar] [CrossRef] [PubMed]
- Hogan, B.L.; Barkauskas, C.E.; Chapman, H.A.; Epstein, J.A.; Jain, R.; Hsia, C.C.; Niklason, L.; Calle, E.; Le, A.; Randell, S.H.; et al. Repair and Regeneration of the Respiratory System: Complexity, Plasticity, and Mechanisms of Lung Stem Cell Function. Cell Stem Cell 2014, 15, 123–138. [Google Scholar] [CrossRef] [PubMed]
- Gon, Y.; Hashimoto, S. Role of airway epithelial barrier dysfunction in pathogenesis of asthma. Allergol. Int. 2018, 67, 12–17. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Yue, H.; Jiang, T.; Gao, J.; Shi, Y.; Shi, B.; Wu, X.; Gou, X. IL-31 plays dual roles in lung inflammation in an OVA-induced murine asthma model. Biol. Open 2019, 8, bio036244. [Google Scholar] [CrossRef]
- Flayer, C.H.; Ge, M.Q.; Hwang, J.W.; Kokalari, B.; Redai, I.G.; Jiang, Z.; Haczku, A. Ozone Inhalation Attenuated the Effects of Budesonide on Aspergillus fumigatus-Induced Airway Inflammation and Hyperreactivity in Mice. Front. Immunol. 2019, 10, 2173. [Google Scholar] [CrossRef] [PubMed]
- Golebski, K.; Luiten, S.; Van Egmond, D.; De Groot, E.; Röschmann, K.I.L.; Fokkens, W.J.; Van Drunen, C.M. High Degree of Overlap between Responses to a Virus and to the House Dust Mite Allergen in Airway Epithelial Cells. PLoS ONE 2014, 9, e87768. [Google Scholar] [CrossRef]
- Liu, B.; Sun, H.; Wang, J.; Liu, H.; Zhao, C. Potential role for EZH2 in promotion of asthma through suppression of miR-34b transcription by inhibition of FOXO3. Lab. Investig. 2021, 101, 998–1010. [Google Scholar] [CrossRef]
- Ravanetti, L.; Dijkhuis, A.; Dekker, T.; Pineros, Y.S.S.; Ravi, A.; Dierdorp, B.S.; Erjefält, J.S.; Mori, M.; Pavlidis, S.; Adcock, I.M.; et al. IL-33 drives influenza-induced asthma exacerbations by halting innate and adaptive antiviral immunity. J. Allergy Clin. Immunol. 2019, 143, 1355–1370.e1316. [Google Scholar] [CrossRef]
- Li, R.; Wang, J.; Zhu, F.; Li, R.; Liu, B.; Xu, W.; He, G.; Cao, H.; Wang, Y.; Yang, J. HMGB1 regulates T helper 2 and T helper17 cell differentiation both directly and indirectly in asthmatic mice. Mol. Immunol. 2018, 97, 45–55. [Google Scholar] [CrossRef]
- Fribley, A.; Zhang, K.; Kaufman, R.J. Regulation of Apoptosis by the Unfolded Protein Response. In Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 2009; Volume 559, pp. 191–204. [Google Scholar]
- Pathinayake, P.S.; Hsu, A.C.-Y.; Waters, D.W.; Hansbro, P.; Wood, L.G.; Wark, P. Understanding the Unfolded Protein Response in the Pathogenesis of Asthma. Front. Immunol. 2018, 9, 175. [Google Scholar] [CrossRef] [PubMed]
- Yaghi, A.; Dolovich, M.B. Airway Epithelial Cell Cilia and Obstructive Lung Disease. Cells 2016, 5, 40. [Google Scholar] [CrossRef] [PubMed]
- Thomas, B.; Rutman, A.; Hirst, R.A.; Haldar, P.; Wardlaw, A.; Bankart, J.; Brightling, C.; O’Callaghan, C. Ciliary dysfunction and ultrastructural abnormalities are features of severe asthma. J. Allergy Clin. Immunol. 2010, 126, 722.e2–729.e2. [Google Scholar] [CrossRef] [PubMed]
- Aizawa, H.; Koarai, A.; Shishikura, Y.; Yanagisawa, S.; Yamaya, M.; Sugiura, H.; Numakura, T.; Yamada, M.; Ichikawa, T.; Fujino, N.; et al. Oxidative stress enhances the expression of IL-33 in human airway epithelial cells. Respir Res. 2018, 19, 52. [Google Scholar] [CrossRef]
- Wan, H.; Dingle, S.; Xu, Y.; Besnard, V.; Kaestner, K.H.; Ang, S.-L.; Wert, S.; Stahlman, M.T.; Whitsett, J.A. Compensatory Roles of Foxa1 and Foxa2 during Lung Morphogenesis. J. Biol. Chem. 2005, 280, 13809–13816. [Google Scholar] [CrossRef]
- Sun, Z.; Xu, X.; He, J.; Murray, A.; Sun, M.; Wei, X.; Wang, X.; McCoig, E.; Xie, E.; Jiang, X.; et al. EGR1 recruits TET1 to shape the brain methylome during development and upon neuronal activity. Nat. Commun. 2019, 10, 3892. [Google Scholar] [CrossRef]
- Yang, Y.A.; Zhao, J.C.; Fong, K.-W.; Kim, J.; Li, S.; Song, C.-X.; Song, B.; Zheng, B.; He, C.; Yu, J. FOXA1 potentiates lineage-specific enhancer activation through modulating TET1 expression and function. Nucleic Acids Res. 2016, 44, 8153–8164. [Google Scholar] [CrossRef]
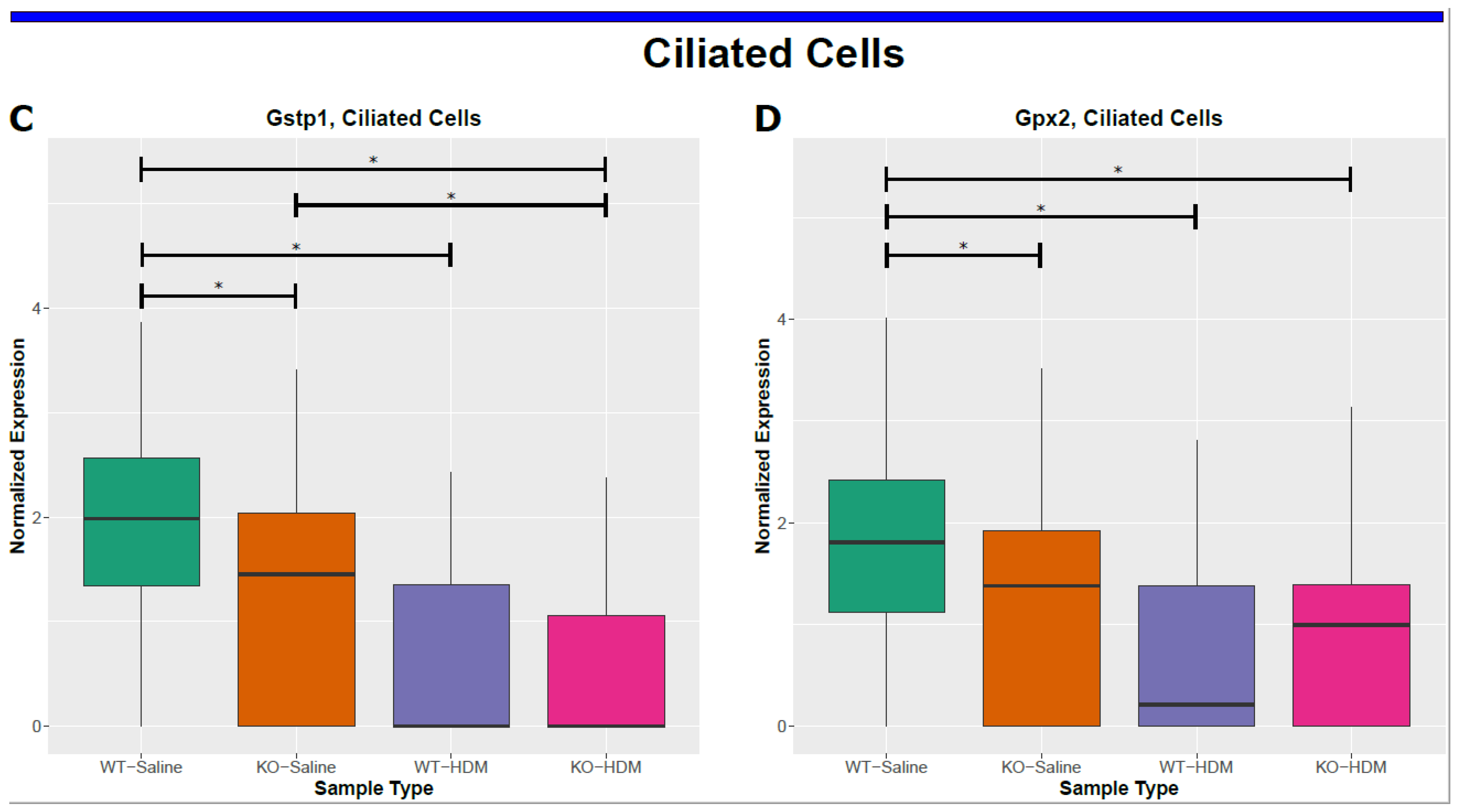
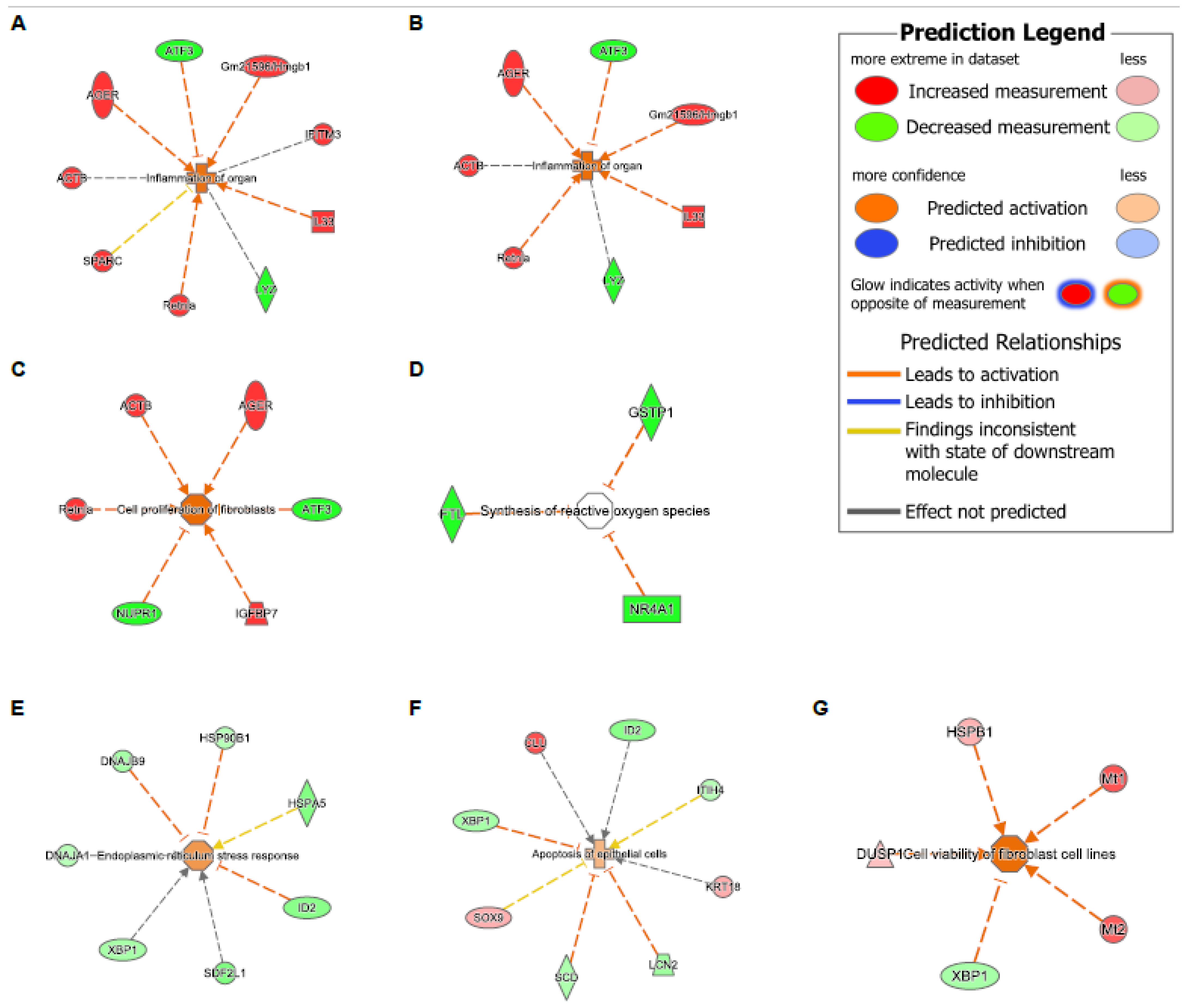
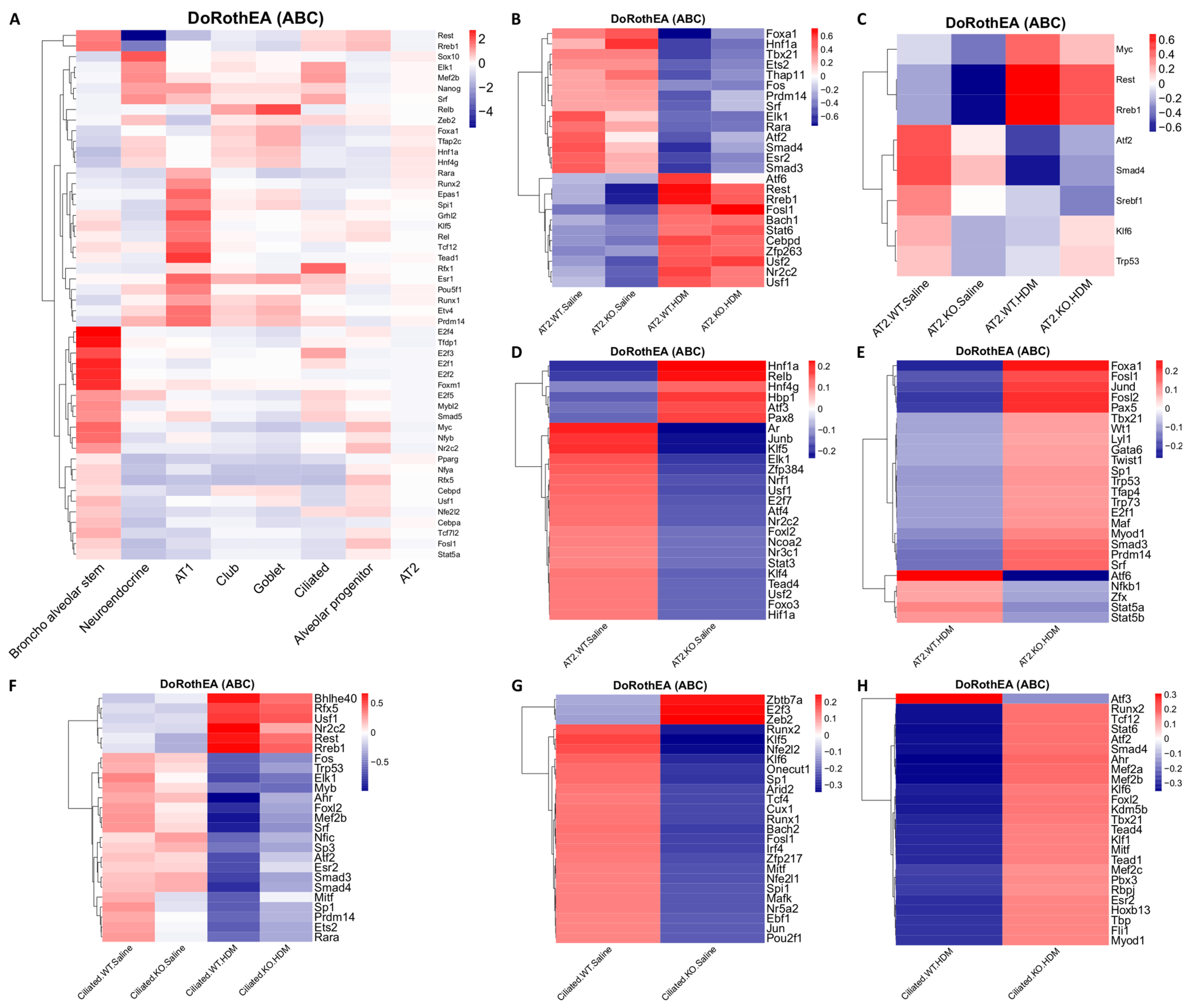
| Comparisons | WT-HDM vs. WT-Sal | KO-Sal vs. WT-Sal | KO-HDM vs. WT-Sal | Overlap of Three Comparisons (Same Direction) | KO-HDM vs. WT-HDM |
| Bulk cell analysis | |||||
| EpCAM+ cells | 830 | 98 | 817 | 48 (36) | 72 |
| Individual cell types | |||||
| AT1 | 0 | 0 | 1 | 0 | 0 |
| AT2 | 732 | 70 | 745 | 39 (30) | 78 |
| Alveolar progenitor (AP) | 431 | 18 | 445 | 4 (2) | 65 |
| Broncho alveolar stem (BAS) | 172 | 4 | 154 | 1(1) | 51 |
| Ciliated | 210 | 41 | 200 | 5 (5) | 0 |
| Stromal | 72 | 19 | 54 | 0 | 1 |
| Club | 55 | 5 | 88 | 0 | 0 |
| Goblet | 13 | 0 | 0 | 0 | 0 |
| Neuroendocrine | 88 | 3 | 16 | 0 | 0 |
| Total | 1773 | 160 | 1703 | 49 (45) | 195 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, T.; Brown, A.P.; Cai, L.P.; Quon, G.; Ji, H. Single-Cell RNA-Seq Analysis Reveals Lung Epithelial Cell Type-Specific Responses to HDM and Regulation by Tet1. Genes 2022, 13, 880. https://doi.org/10.3390/genes13050880
Zhu T, Brown AP, Cai LP, Quon G, Ji H. Single-Cell RNA-Seq Analysis Reveals Lung Epithelial Cell Type-Specific Responses to HDM and Regulation by Tet1. Genes. 2022; 13(5):880. https://doi.org/10.3390/genes13050880
Chicago/Turabian StyleZhu, Tao, Anthony P. Brown, Lucy P. Cai, Gerald Quon, and Hong Ji. 2022. "Single-Cell RNA-Seq Analysis Reveals Lung Epithelial Cell Type-Specific Responses to HDM and Regulation by Tet1" Genes 13, no. 5: 880. https://doi.org/10.3390/genes13050880
APA StyleZhu, T., Brown, A. P., Cai, L. P., Quon, G., & Ji, H. (2022). Single-Cell RNA-Seq Analysis Reveals Lung Epithelial Cell Type-Specific Responses to HDM and Regulation by Tet1. Genes, 13(5), 880. https://doi.org/10.3390/genes13050880






