Whole-Genome Sequencing Reveals Lignin-Degrading Capacity of a Ligninolytic Bacterium (Bacillus cereus) from Buffalo (Bubalus bubalis) Rumen
Abstract
:1. Introduction
2. Materials and Methods
2.1. Isolation and Identification of Lignin-Degrading Bacteria
2.2. DNA Library Preparation, Whole-Genome Sequencing and Assembly
2.3. Open Reading Frames (ORFs), Evolutionary Genealogy of Genes: Non-Supervised Orthologous Groups (eggNOG), Kyoto Encyclopedia of Genes and Genomes (KEGG) and Carbohydrate-Active Enzymes Analysis
2.4. Guaiacol Colour Reaction
2.5. Biodegradation of Kraft Lignin by Strain B. cereus AH7-7
3. Results
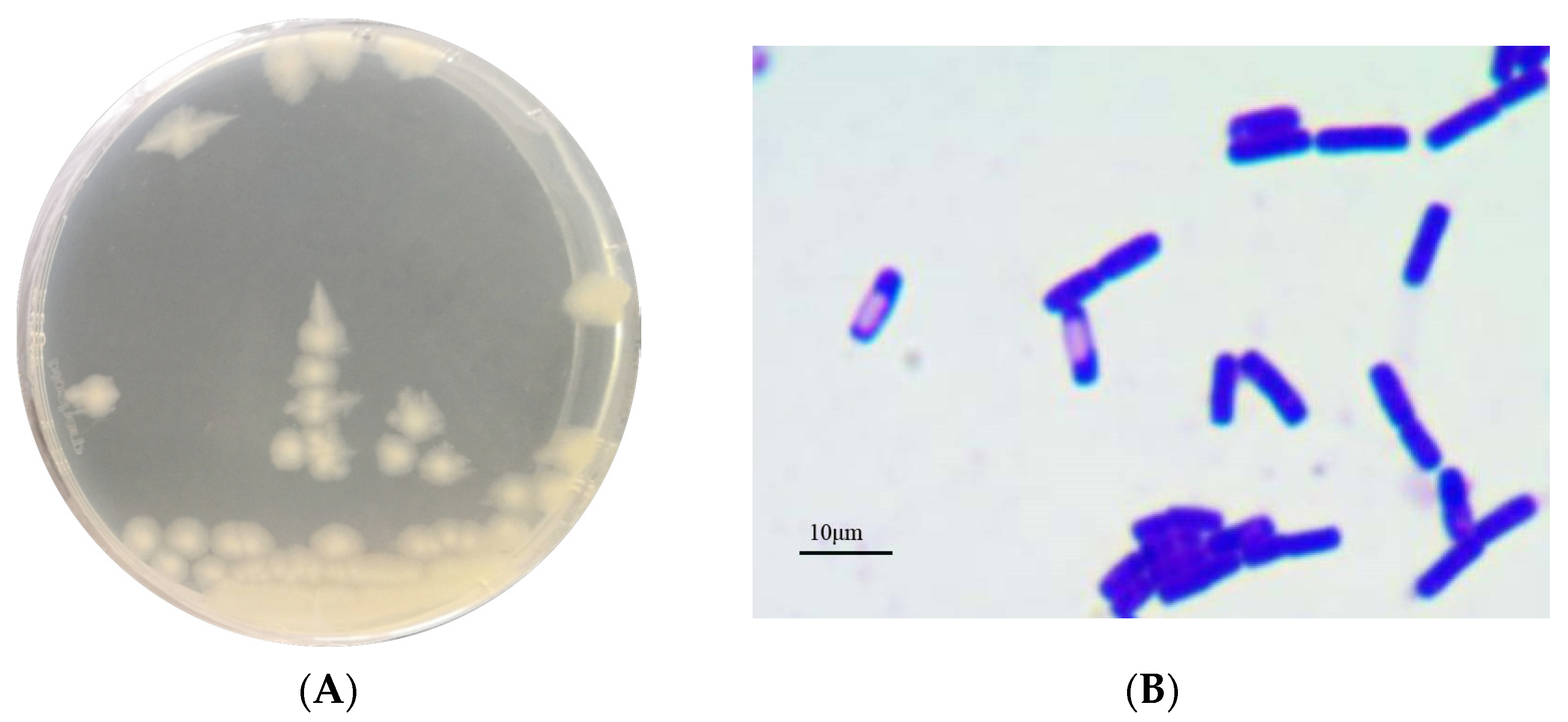
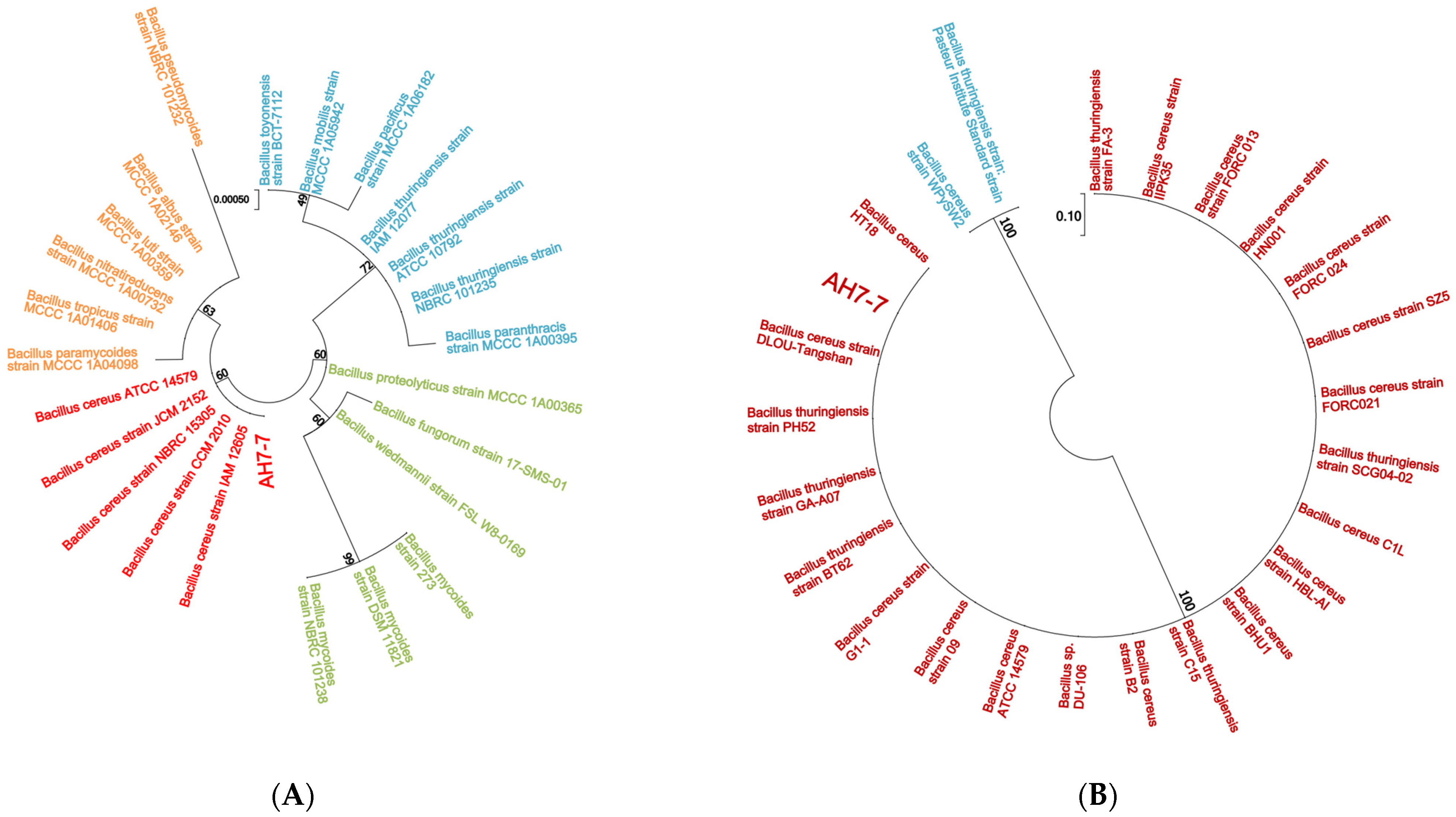
3.1. Identification of Strain AH7-7
3.2. Basic Genomic Characteristics of B. cereus AH7-7
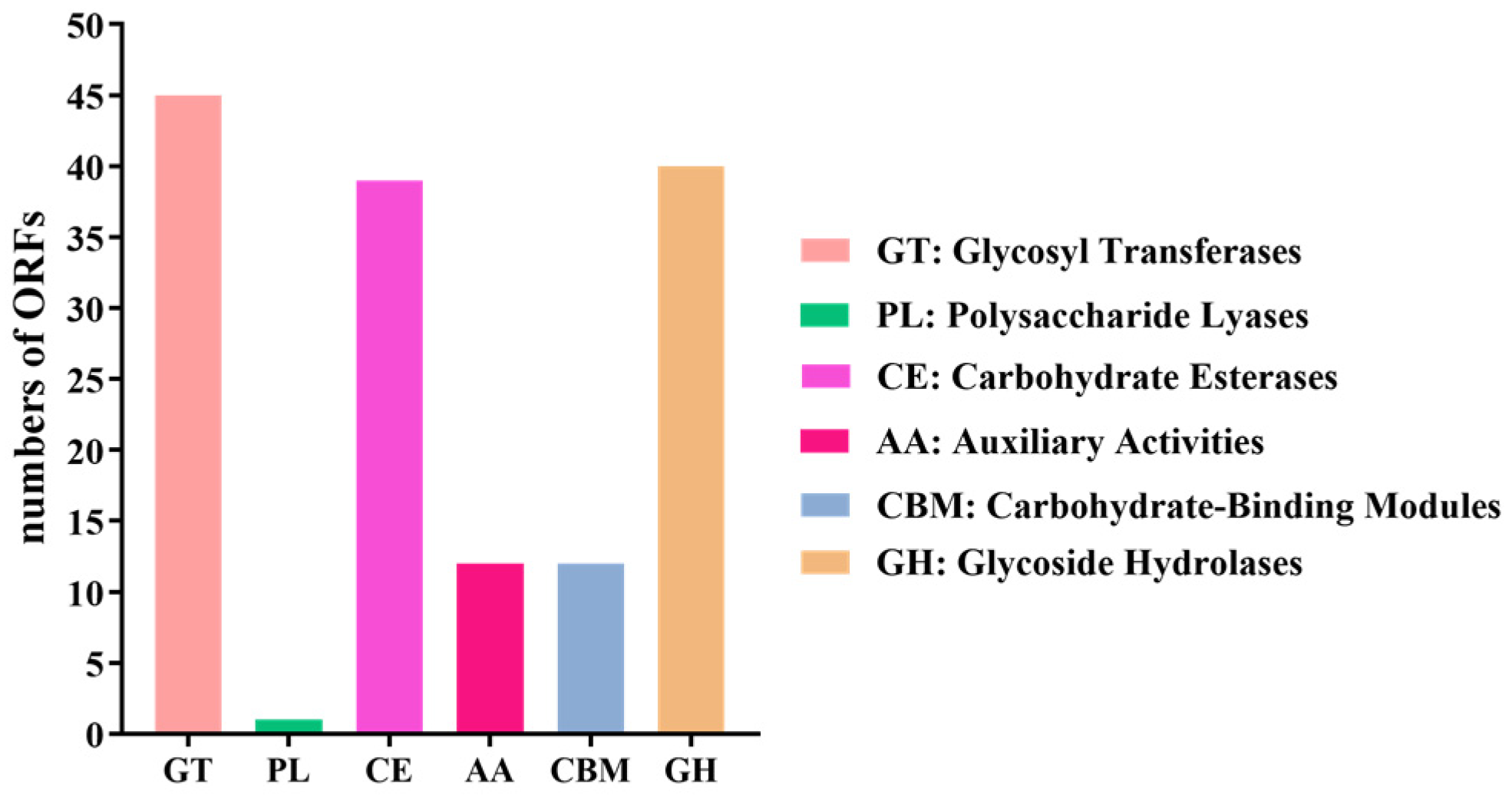
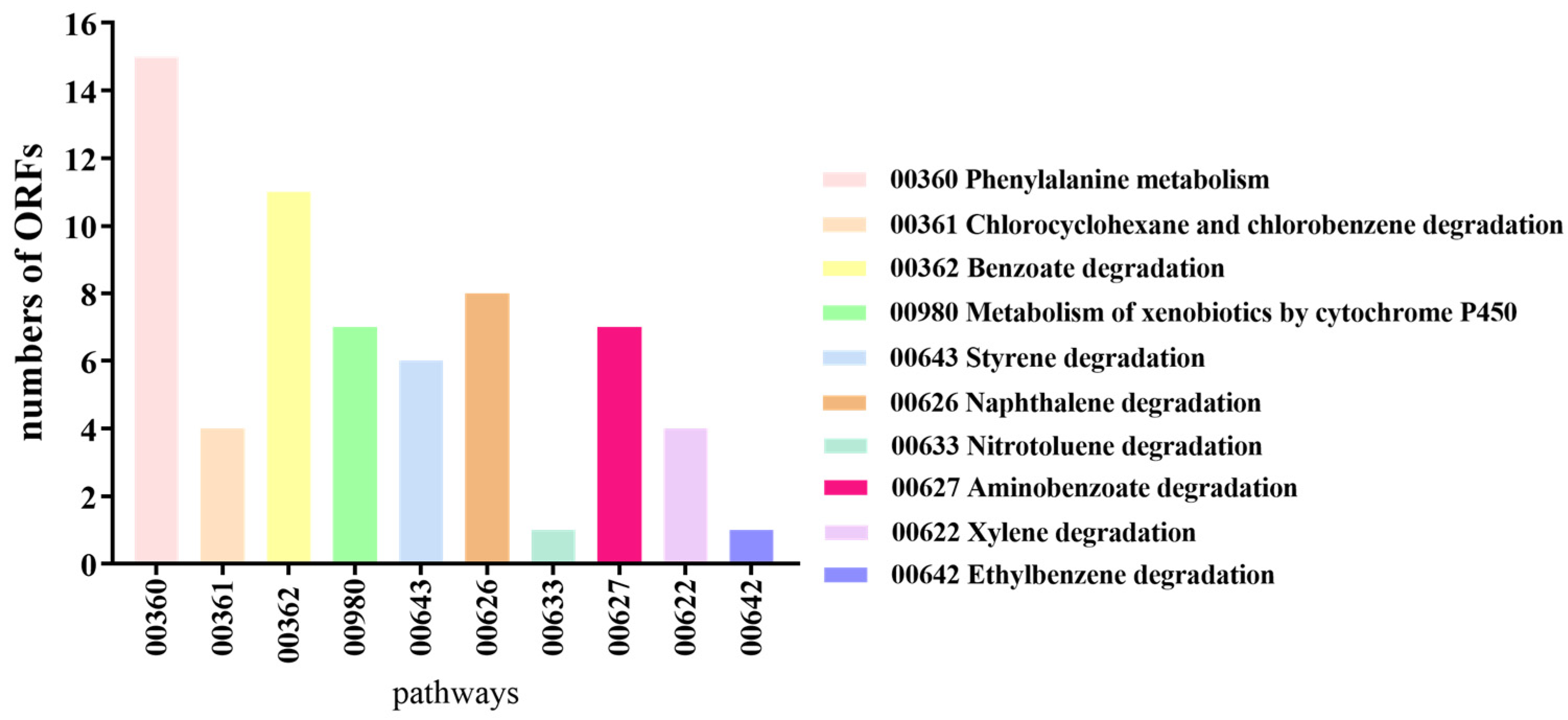
3.3. Annotation Results of B. cereus AH7-7 in eggNOG, KEGG and Carbohydrate-Active Enzymes Database
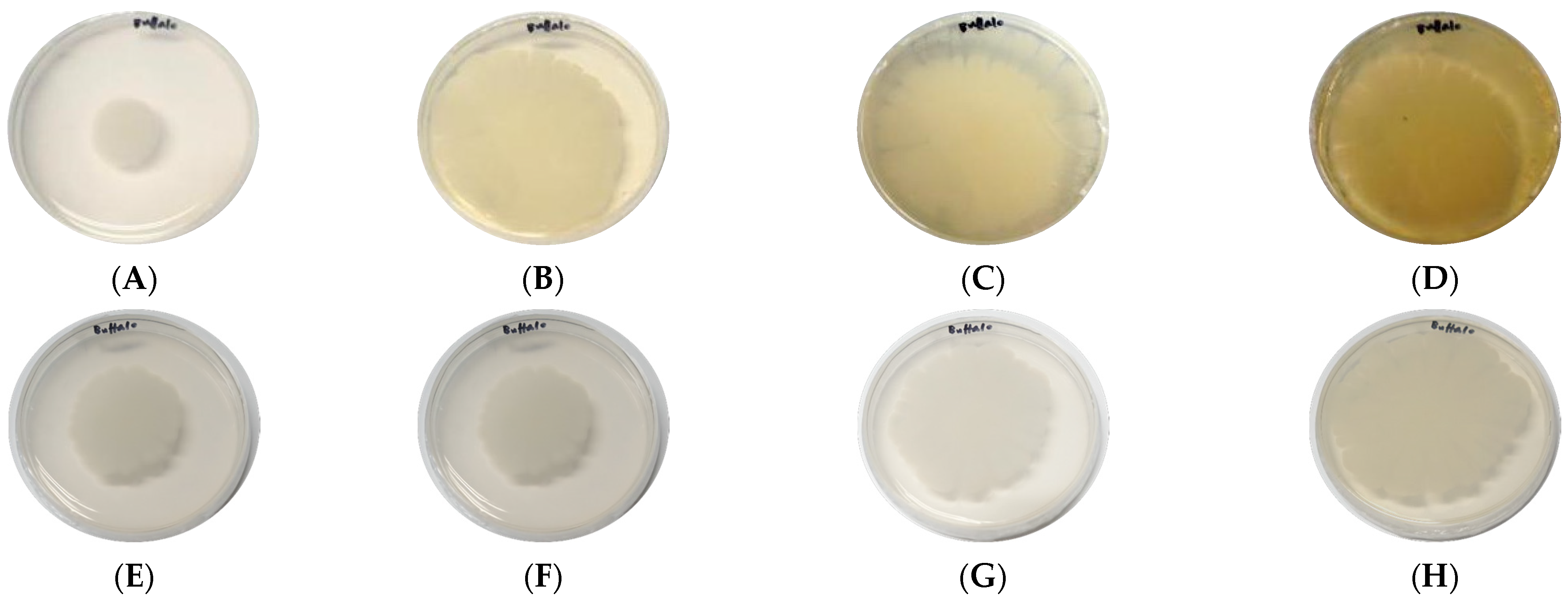
3.4. Laccase Secretion of B. cereus AH7-7
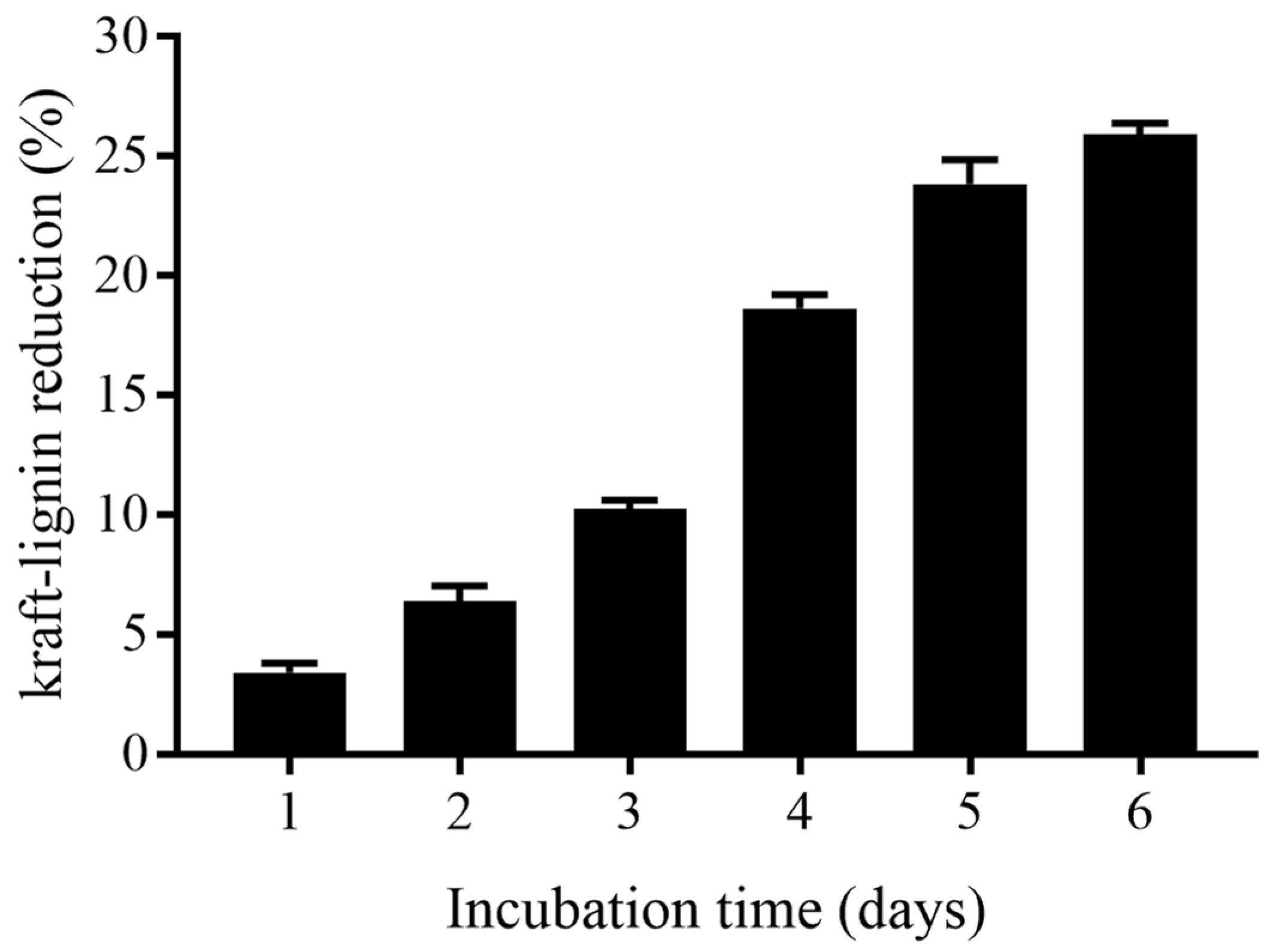
3.5. Kraft-Lignin Degradation of B. cereus AH7-7
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
| Query Acc.ver | Subject Acc.ver | % Identity | Alignment Length | Mismatches | Gap Opens | Q. Start | Q. End | S. Start | S. End | Evalue | Bit Score |
|---|---|---|---|---|---|---|---|---|---|---|---|
| AH7-7 | NR_115526.1 | 99.86 | 1424 | 0 | 2 | 3 | 1424 | 29 | 1452 | 0 | 2617 |
| AH7-7 | NR_115714.1 | 99.86 | 1424 | 0 | 2 | 3 | 1424 | 49 | 1472 | 0 | 2617 |
| AH7-7 | NR_112630.1 | 99.86 | 1424 | 0 | 2 | 3 | 1424 | 28 | 1451 | 0 | 2617 |
| AH7-7 | NR_113266.1 | 99.86 | 1424 | 0 | 2 | 3 | 1424 | 29 | 1452 | 0 | 2617 |
| AH7-7 | NR_114582.1 | 99.86 | 1424 | 0 | 2 | 3 | 1424 | 39 | 1462 | 0 | 2617 |
| AH7-7 | NR_152692.1 | 99.789 | 1424 | 1 | 2 | 3 | 1424 | 49 | 1472 | 0 | 2612 |
| AH7-7 | NR_157736.1 | 99.789 | 1424 | 1 | 2 | 3 | 1424 | 49 | 1472 | 0 | 2612 |
| AH7-7 | NR_157735.1 | 99.789 | 1424 | 1 | 2 | 3 | 1424 | 49 | 1472 | 0 | 2612 |
| AH7-7 | NR_157732.1 | 99.789 | 1424 | 1 | 2 | 3 | 1424 | 49 | 1472 | 0 | 2612 |
| AH7-7 | NR_157730.1 | 99.789 | 1424 | 1 | 2 | 3 | 1424 | 49 | 1472 | 0 | 2612 |
| AH7-7 | NR_157729.1 | 99.789 | 1424 | 1 | 2 | 3 | 1424 | 49 | 1472 | 0 | 2612 |
| AH7-7 | NR_157734.1 | 99.719 | 1424 | 2 | 2 | 3 | 1424 | 49 | 1472 | 0 | 2606 |
| AH7-7 | NR_170494.1 | 99.719 | 1424 | 2 | 2 | 3 | 1424 | 80 | 1503 | 0 | 2606 |
| AH7-7 | NR_157733.1 | 99.649 | 1424 | 3 | 2 | 3 | 1424 | 49 | 1472 | 0 | 2601 |
| AH7-7 | NR_157728.1 | 99.649 | 1424 | 3 | 2 | 3 | 1424 | 49 | 1472 | 0 | 2601 |
| AH7-7 | NR_043403.1 | 99.579 | 1424 | 4 | 2 | 3 | 1424 | 29 | 1452 | 0 | 2595 |
| AH7-7 | NR_121761.1 | 99.579 | 1424 | 4 | 2 | 3 | 1424 | 46 | 1469 | 0 | 2595 |
| AH7-7 | NR_114581.1 | 99.579 | 1424 | 4 | 2 | 3 | 1424 | 39 | 1462 | 0 | 2595 |
| AH7-7 | NR_112780.1 | 99.508 | 1424 | 5 | 2 | 3 | 1424 | 29 | 1452 | 0 | 2591 |
| AH7-7 | NR_157731.1 | 99.508 | 1424 | 5 | 2 | 3 | 1424 | 49 | 1472 | 0 | 2590 |
| AH7-7 | NR_113991.1 | 99.438 | 1424 | 6 | 2 | 3 | 1424 | 29 | 1452 | 0 | 2584 |
| AH7-7 | NR_036880.1 | 99.368 | 1424 | 7 | 2 | 3 | 1424 | 25 | 1448 | 0 | 2579 |
| AH7-7 | NR_024697.1 | 99.368 | 1424 | 7 | 2 | 3 | 1424 | 43 | 1466 | 0 | 2579 |
| AH7-7 | NR_113996.1 | 99.368 | 1424 | 7 | 2 | 3 | 1424 | 29 | 1452 | 0 | 2579 |
| Query Acc.ver | Subject Acc.ver | % Identity | Alignment Length | Mismatches | Gap Opens | Q. Start | Q. End | S. Start | S. End | Evalue | Bit Score |
|---|---|---|---|---|---|---|---|---|---|---|---|
| AH7-7 | CP053289.1 | 100 | 343 | 0 | 0 | 1 | 343 | 42,757 | 42,415 | 2.08 × 10−177 | 634 |
| AH7-7 | CP040340.1 | 100 | 343 | 0 | 0 | 1 | 343 | 621,370 | 621,712 | 2.08 × 10−177 | 634 |
| AH7-7 | KY283089.1 | 100 | 343 | 0 | 0 | 1 | 343 | 91 | 433 | 2.08 × 10−177 | 634 |
| AH7-7 | CP042270.1 | 100 | 343 | 0 | 0 | 1 | 343 | 1,208,589 | 1,208,931 | 2.08 × 10−177 | 634 |
| AH7-7 | CP044978.1 | 100 | 343 | 0 | 0 | 1 | 343 | 791,108 | 791,450 | 2.08 × 10−177 | 634 |
| AH7-7 | CP042929.1 | 100 | 343 | 0 | 0 | 1 | 343 | 4959 | 5301 | 2.08 × 10−177 | 634 |
| AH7-7 | CP042874.1 | 100 | 343 | 0 | 0 | 1 | 343 | 4959 | 5301 | 2.08 × 10−177 | 634 |
| AH7-7 | CP034551.1 | 100 | 343 | 0 | 0 | 1 | 343 | 4677 | 5019 | 2.08 × 10−177 | 634 |
| AH7-7 | CP026607.1 | 100 | 343 | 0 | 0 | 1 | 343 | 4781 | 5123 | 2.08 × 10−177 | 634 |
| AH7-7 | MH427046.1 | 100 | 343 | 0 | 0 | 1 | 343 | 107 | 449 | 2.08 × 10−177 | 634 |
| AH7-7 | CP021436.1 | 100 | 343 | 0 | 0 | 1 | 343 | 5096 | 5438 | 2.08 × 10−177 | 634 |
| AH7-7 | CP023727.1 | 100 | 343 | 0 | 0 | 1 | 343 | 5,200,294 | 5,200,636 | 2.08 × 10−177 | 634 |
| AH7-7 | CP023245.1 | 100 | 343 | 0 | 0 | 1 | 343 | 4677 | 5019 | 2.08 × 10−177 | 634 |
| AH7-7 | CP022445.1 | 100 | 343 | 0 | 0 | 1 | 343 | 4677 | 5019 | 2.08 × 10−177 | 634 |
| AH7-7 | CP017577.1 | 100 | 343 | 0 | 0 | 1 | 343 | 4,203,994 | 4,204,336 | 2.08 × 10−177 | 634 |
| AH7-7 | CP014486.1 | 100 | 343 | 0 | 0 | 1 | 343 | 4967 | 5309 | 2.08 × 10−177 | 634 |
| AH7-7 | KT923660.1 | 100 | 343 | 0 | 0 | 1 | 343 | 107 | 449 | 2.08 × 10−177 | 634 |
| AH7-7 | CP012691.1 | 100 | 343 | 0 | 0 | 1 | 343 | 4677 | 5019 | 2.08 × 10−177 | 634 |
| AH7-7 | CP011155.1 | 100 | 343 | 0 | 0 | 1 | 343 | 5083 | 5425 | 2.08 × 10−177 | 634 |
| AH7-7 | CP011145.1 | 100 | 343 | 0 | 0 | 1 | 343 | 4677 | 5019 | 2.08 × 10−177 | 634 |
| AH7-7 | KJ813006.1 | 100 | 343 | 0 | 0 | 1 | 343 | 88 | 430 | 2.08 × 10−177 | 634 |
| AH7-7 | AP014864.1 | 100 | 343 | 0 | 0 | 1 | 343 | 5,891,887 | 5,891,545 | 2.08 × 10−177 | 634 |
| AH7-7 | CP068984.1 | 100 | 343 | 0 | 0 | 1 | 343 | 5470 | 5812 | 2.08 × 10−177 | 634 |
| AH7-7 | AP024504.2 | 100 | 343 | 0 | 0 | 1 | 343 | 4677 | 5019 | 2.08 × 10−177 | 634 |
| Substrate | Fermentation/Oxidation | Enzyme Activity | Results | Substrate Utilisation | Results |
|---|---|---|---|---|---|
| Glucose | − | β-Galactosidase | − | Citrate sodium | + |
| Mannitol | − | Arginine bishydrolase | + | Sodium thiosulfate (production of H2S) | − |
| Inose | − | Lysine decarboxylase | − | Tryptophan (production of indole) | − |
| Sorbic alcohol | − | Ornithine decarboxylase | − | Pyruvate (production of acetylmethyl methanol) | w |
| Rhamnose | − | Urease | − | ||
| Saccharose | − | Tryptophan deaminase | − | ||
| Melibiose | − | Gelatinase | + | ||
| Amygdalin | − | ||||
| Arabinose | − |
| Substrate | Results | Substrate | Results | Substrate | Results |
|---|---|---|---|---|---|
| Control | − | Urosside | + | Glucose | + |
| Glycerol | − | Aesculin | + | Fructose | + |
| Erythritol | − | Salicin | + | Seminose | − |
| D-Arabinose | − | Cellobiose | − | Sorbose | − |
| L-Arabinose | − | Maltose | + | Rhamnose | − |
| Ribose | w | Lactose | − | Melampyrin | − |
| D-Xylose | − | Melibiose | - | Increositol | − |
| L-Xylose | − | Saccharose | + | Mannitol | − |
| Adonite | − | Trehalose | + | Sorbic alcohol | − |
| Galactose | − | Inulin | − | α-Methyl-D-mannoside | − |
| D-Arabinol | − | Melezitose | − | α-Methyl-d-glucoside | − |
| L-Arabinol | − | Raffinose | − | N-Acetyl-glucosamine | − |
| Amygdalin | − | Starch | − | 2-Keto-gluconate | − |
| Gluconate | − | Glycogen | + | 5-keto-gluconate | − |
| D-Turanose | − | Xylosic alcohol | − | β-Methyl-D-xyloside | − |
| D-Tagatose | − | D-Lyxose | − |
References
- Zakzeski, J.; Bruijnincx, P.C.A.; Jongerius, A.L.; Weckhuysen, B.M. The catalytic valorization of lignin for the production of renewable chemicals. Chem. Rev. 2010, 110, 3552–3599. [Google Scholar] [CrossRef] [PubMed]
- Bandounas, L.; Wierckx, N.J.P.; de Winde, J.H.; Ruijssenaars, H.J. Isolation and characterization of novel bacterial strains exhibiting ligninolytic potential. BMC Biotechnol. 2011, 11, 94. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, S.; Kang, M.; Bae, J.H.; Sohn, J.H.; Sung, B.H. Bacterial Valorization of Lignin: Strains, Enzymes, Conversion Pathways, Biosensors, and Perspectives. Front. Bioeng. Biotechnol. 2019, 7, 1–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Masai, E.; Ichimura, A.; Sato, Y.; Miyauchi, K.; Katayama, Y.; Fukuda, M. Roles of the enantioselective glutathione S-transferases in cleavage of β-aryl ether. J. Bacteriol. 2003, 185, 1768–1775. [Google Scholar] [CrossRef] [Green Version]
- Santos, A.; Mendes, S.; Brissos, V.; Martins, L.O. New dye-decolorizing peroxidases from Bacillus subtilis and Pseudomonas putida MET94: Towards biotechnological applications. Appl. Microbiol. Biotechnol. 2014, 98, 2053–2065. [Google Scholar] [CrossRef]
- Ahmad, M.; Roberts, J.N.; Hardiman, E.M.; Singh, R.; Eltis, L.D.; Bugg, T.D.H. Identification of DypB from rhodococcus jostii RHA1 as a lignin peroxidase. Biochemistry 2011, 50, 5096–5107. [Google Scholar] [CrossRef]
- Trojanowski, J.; Haider, K.; Sundman, V. Decomposition of c-14-labeled lignin and phenols by a nocardia-sp. Arch. Microbiol. 1977, 114, 149–153. [Google Scholar] [CrossRef]
- Sahm, L.E.; Sahm, H. Degradation of Coniferyl Alcohol and Other Lignin-Related Aromatic Compounds by Nocardia sp. DSM 1069. Arch. Microbiol. 1980, 126, 141–148. [Google Scholar]
- Min, K.; Gong, G.; Woo, H.M.; Kim, Y.; Um, Y. A dye-decolorizing peroxidase from Bacillus subtilis exhibiting substrate-dependent optimum temperature for dyes and b-ether lignin dimer. Sci. Rep. 2015, 5, 1–8. [Google Scholar] [CrossRef]
- Chandra, R.; Chowdhary, P. Properties of bacterial laccases and their application in bioremediation of industrial wastes. Environ. Sci. Process. Impacts 2015, 17, 326–342. [Google Scholar] [CrossRef]
- De Gonzalo, G.; Colpa, D.I.; Habib, M.H.M.; Fraaije, M.W. Bacterial enzymes involved in lignin degradation. J. Biotechnol. 2016, 236, 110–119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brown, M.E.; Chang, M.C.Y. Exploring bacterial lignin degradation. Curr. Opin. Chem. Biol. 2014, 19, 1–7. [Google Scholar] [CrossRef]
- Bianchetti, C.M.; Harmann, C.H.; Takasuka, T.E.; Hura, G.L.; Dyer, K.; Fox, B.G. Fusion of dioxygenase and lignin-binding domains in a novel secreted enzyme from cellulolytic streptomyces sp. SIRexaa-e. J. Biol. Chem. 2013, 288, 18574–18587. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perestelo, F.; Falcon, M.A.; De La Fuente, G. Production of vanillic acid from vanillin by resting cells of Serratia marcescens. Appl. Environ. Microbiol. 1989, 55, 1660–1662. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allocati, N.; Federici, L.; Masulli, M.; Di Ilio, C. Glutathione transferases in bacteria. FEBS J. 2009, 276, 58–75. [Google Scholar] [CrossRef] [PubMed]
- Reiter, J.; Strittmatter, H.; Wiemann, L.O.; Schieder, D.; Sieber, V. Enzymatic cleavage of lignin β-O-4 aryl ether bonds via net internal hydrogen transfer. Green Chem. 2013, 15, 1373–1381. [Google Scholar] [CrossRef]
- Masai, E.; Nishikawa, S.; Morohoshi, N.; Haraguchi, T. Detection and localization of a new enzyme catalyzing the fl-aryl ether cleavage in the soil bacterium (Pseudomonas paucimobilis SYK-6). FEBS Lett. 1989, 249, 348–352. [Google Scholar] [CrossRef] [Green Version]
- Picart, P.; Sevenich, M.; Domínguez De María, P.; Schallmey, A. Exploring glutathione lyases as biocatalysts: Paving the way for enzymatic lignin depolymerization and future stereoselective applications. Green Chem. 2015, 17, 4931–4940. [Google Scholar] [CrossRef] [Green Version]
- Rashid, G.M.M.; Taylor, C.R.; Liu, Y.; Zhang, X.; Rea, D.; Fü, V.; Bugg, T.D.H. Identification of Manganese Superoxide Dismutase from Sphingobacterium sp. T2 as a Novel Bacterial Enzyme for Lignin Oxidation. ACS Chem. Biol. 2015, 10, 2286–2294. [Google Scholar] [CrossRef]
- Koren, S.; Walenz, B.P.; Berlin, K.; Miller, J.R.; Bergman, N.H.; Phillippy, A.M. Canu: Scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017, 27, 722–736. [Google Scholar] [CrossRef] [Green Version]
- Arimoto, M.; Yamagishi, K.; Wang, J.; Tanaka, K.; Miyoshi, T.; Kamei, I.; Kondo, R.; Mori, T.; Kawagishi, H.; Hirai, H. Molecular breeding of lignin-degrading brown-rot fungus Gloeophyllum trabeum by homologous expression of laccase gene. AMB Express 2015, 5, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Zhong, H.; Zhou, J.; Wu, Y.; Ma, Z.; Yang, L.; Wang, Z.; Ling, C.; Li, X. Lignin degradation by water buffalo. Trop. Anim. Health Prod. 2021, 53, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Terramoccia, S.; Bartocci, S.; Amici, A.; Martillotti, F. Protein and protein-free dry matter rumen degradability in buffalo, cattle and sheep fed diets with different forage to concentrate ratios. Livest. Prod. Sci. 2000, 65, 185–195. [Google Scholar] [CrossRef]
- Chanthakhoun, V.; Wanapat, M.; Kongmun, P.; Cherdthong, A. Comparison of ruminal fermentation characteristics and microbial population in swamp buffalo and cattle. Livest. Sci. 2012, 143, 172–176. [Google Scholar] [CrossRef]
- Wang, Z.; Wu, W.; Cui, L.; Li, X.; Kulyar, M.F.E.A.; Xiong, H.; Zhou, N.; Yin, H.; Li, J.; Li, X. Isolation, characterization, and interaction of lignin-degrading bacteria from rumen of buffalo (Bubalus bubalis). J. Basic Microbiol. 2021, 61, 757–768. [Google Scholar] [CrossRef]
- Zhong, H.; Zhou, J.; Abdelrahman, M.; Xu, H.; Wu, Z.; Cui, L. The Effect of Lignin Composition on Ruminal Fiber Fractions Degradation from Different Roughage Sources in Water Buffalo ( Bubalus bubalis ). Agriculture 2021, 11, 1015. [Google Scholar] [CrossRef]
- Garrity, G.M.; Bell, J.A.; Lilburn, T.G. Taxonomic Outline of the Prokaryotes Bergey’s Manual of Systematic Bacteriology, 2nd ed.; Springer: Berlin/Heidelberg, Germany, 2004. [Google Scholar]
- Patel, R.K.; Jain, M. NGS QC toolkit: A toolkit for quality control of next generation sequencing data. PLoS ONE 2012, 7, e003019. [Google Scholar] [CrossRef]
- Schubert, M.; Lindgreen, S.; Orlando, L. AdapterRemoval v2: Rapid adapter trimming, identification, and read merging. BMC Res. Notes 2016, 9, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Luo, R.; Lam, T.-W.; Liu, B.; Xie, Y.; Li, Z.; Huang, W.; Yuan, J.; He, G.; Chen, Y.; Pan, Q.; et al. SOAPdenovo2: An empirically improved memory-efficient short-read de novo assembler. Gigascience 2012, 1, 1–6. [Google Scholar] [CrossRef]
- Chin, C.S.; Peluso, P.; Sedlazeck, F.J.; Nattestad, M.; Concepcion, G.T.; Clum, A.; Dunn, C.; O’Malley, R.; Figueroa-Balderas, R.; Morales-Cruz, A.; et al. Phased diploid genome assembly with single-molecule real-time sequencing. Nat. Methods 2016, 13, 1050–1054. [Google Scholar] [CrossRef] [Green Version]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K.; et al. Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 2014, 9, e0112963. [Google Scholar] [CrossRef] [PubMed]
- Besemer, J.; Lomsadze, A.; Borodovsky, M. GeneMarkS: A self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res. 2001, 29, 2607–2618. [Google Scholar] [CrossRef] [Green Version]
- Huerta-Cepas, J.; Forslund, K.; Coelho, L.P.; Szklarczyk, D.; Jensen, L.J.; Von Mering, C.; Bork, P. Fast genome-wide functional annotation through orthology assignment by eggNOG-mapper. Mol. Biol. Evol. 2017, 34, 2115–2122. [Google Scholar] [CrossRef] [Green Version]
- Moriya, Y.; Itoh, M.; Okuda, S.; Yoshizawa, A.C.; Kanehisa, M. KAAS: An automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 2007, 35, 182–185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lombard, V.; Golaconda Ramulu, H.; Drula, E.; Coutinho, P.M.; Henrissat, B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014, 42, D490–D495. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raj, A.; Reddy, M.M.K.; Chandra, R.; Purohit, H.J.; Kapley, A. Biodegradation of kraft-lignin by Bacillus sp. isolated from sludge of pulp and paper mill. Biodegradation 2007, 18, 783–792. [Google Scholar] [CrossRef] [PubMed]
- Wanapat, M.; Rowlinson, P. Nutrition and feeding of swamp buffalo: Feed resources and rumen approach. Ital. J. Anim. Sci. 2007, 6, 67–73. [Google Scholar] [CrossRef]
- Giardina, P.; Faraco, V.; Pezzella, C.; Piscitelli, A.; Vanhulle, S.; Sannia, G. Laccases: A never-ending story. Cell. Mol. Life Sci. 2010, 67, 369–385. [Google Scholar] [CrossRef]
- Givaudan, A.; Effosse, A.; Faure, D.; Potier, P.; Bouillant, M.L.; Bally, R. Polyphenol oxidase in Azospirillum lipoferum isolated from rice rhizosphere: Evidence for laccase activity in non-motile strains of Azospirillum lipoferum. FEMS Microbiol. Lett. 1993, 108, 205–210. [Google Scholar] [CrossRef]
- Alexandre, G.; Zhulin, I.B. Laccases are widespread in bacteria. Trends Biotechnol. 2000, 18, 41–42. [Google Scholar] [CrossRef]
- Santhanam, N.; Vivanco, J.M.; Decker, S.R.; Reardon, K.F. Expression of industrially relevant laccases: Prokaryotic style. Trends Biotechnol. 2011, 29, 480–489. [Google Scholar] [CrossRef] [PubMed]
- Martins, L.O.; Durão, P.; Brissos, V.; Lindley, P.F. Laccases of prokaryotic origin: Enzymes at the interface of protein science and protein technology. Cell. Mol. Life Sci. 2015, 72, 911–922. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.; Zhang, K.; Liu, P.; Han, H.; Zhao, S.; Kakade, A.; Khan, A.; Du, D.; Li, X. Lignin depolymerization and utilization by bacteria. Bioresour. Technol. 2018, 269, 557–566. [Google Scholar] [CrossRef]
- Ruijssenaars, H.J.; Hartmans, S. A cloned Bacillus halodurans multicopper oxidase exhibiting alkaline laccase activity. Appl. Microbiol. Biotechnol. 2004, 65, 177–182. [Google Scholar] [CrossRef] [PubMed]
- Masai, E.; Katayama, Y.; Fukuda, M. Genetic and biochemical investigations on bacterial catabolic pathways for lignin-derived aromatic compounds. Biosci. Biotechnol. Biochem. 2007, 71, 1–15. [Google Scholar] [CrossRef] [PubMed]
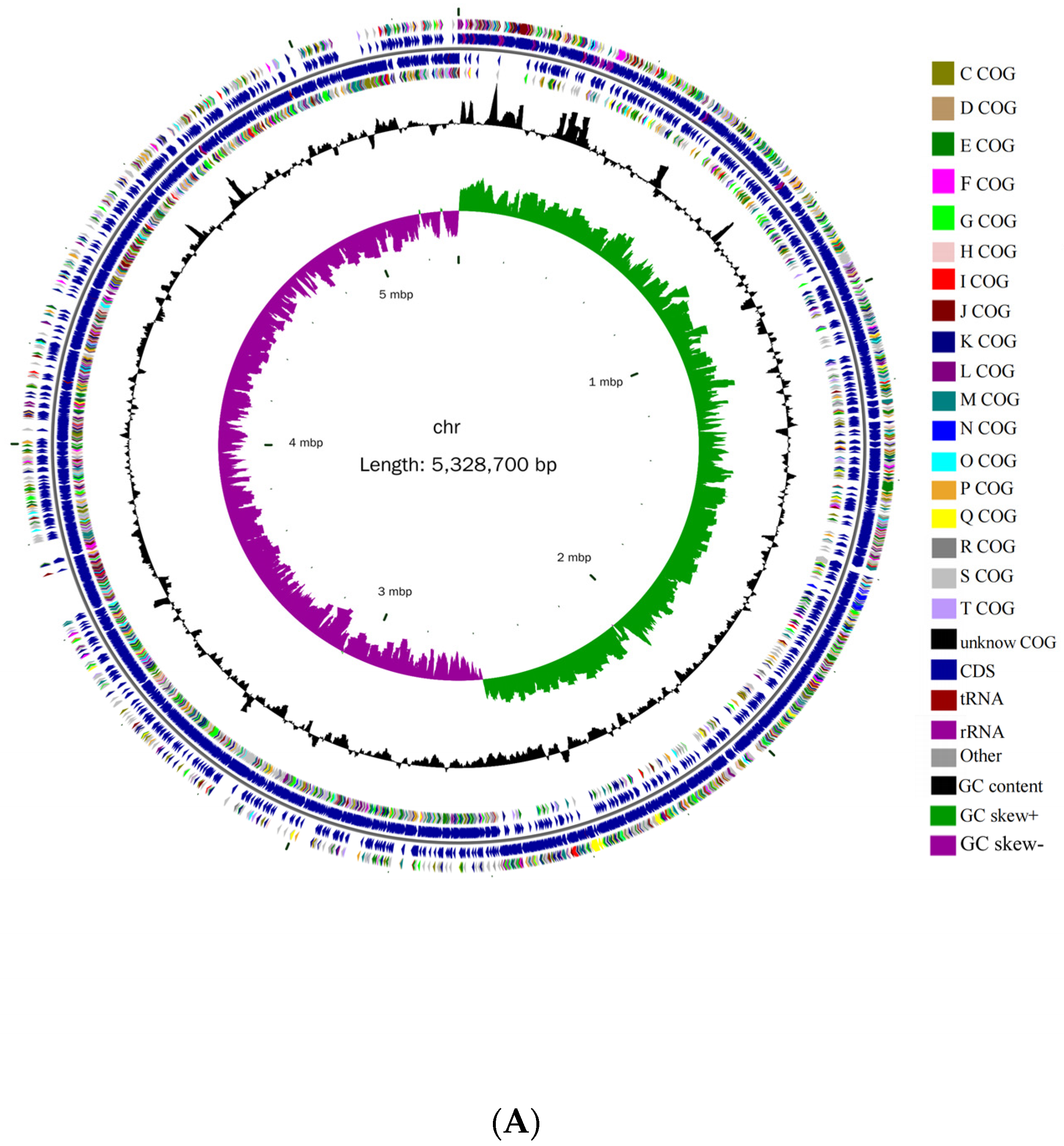
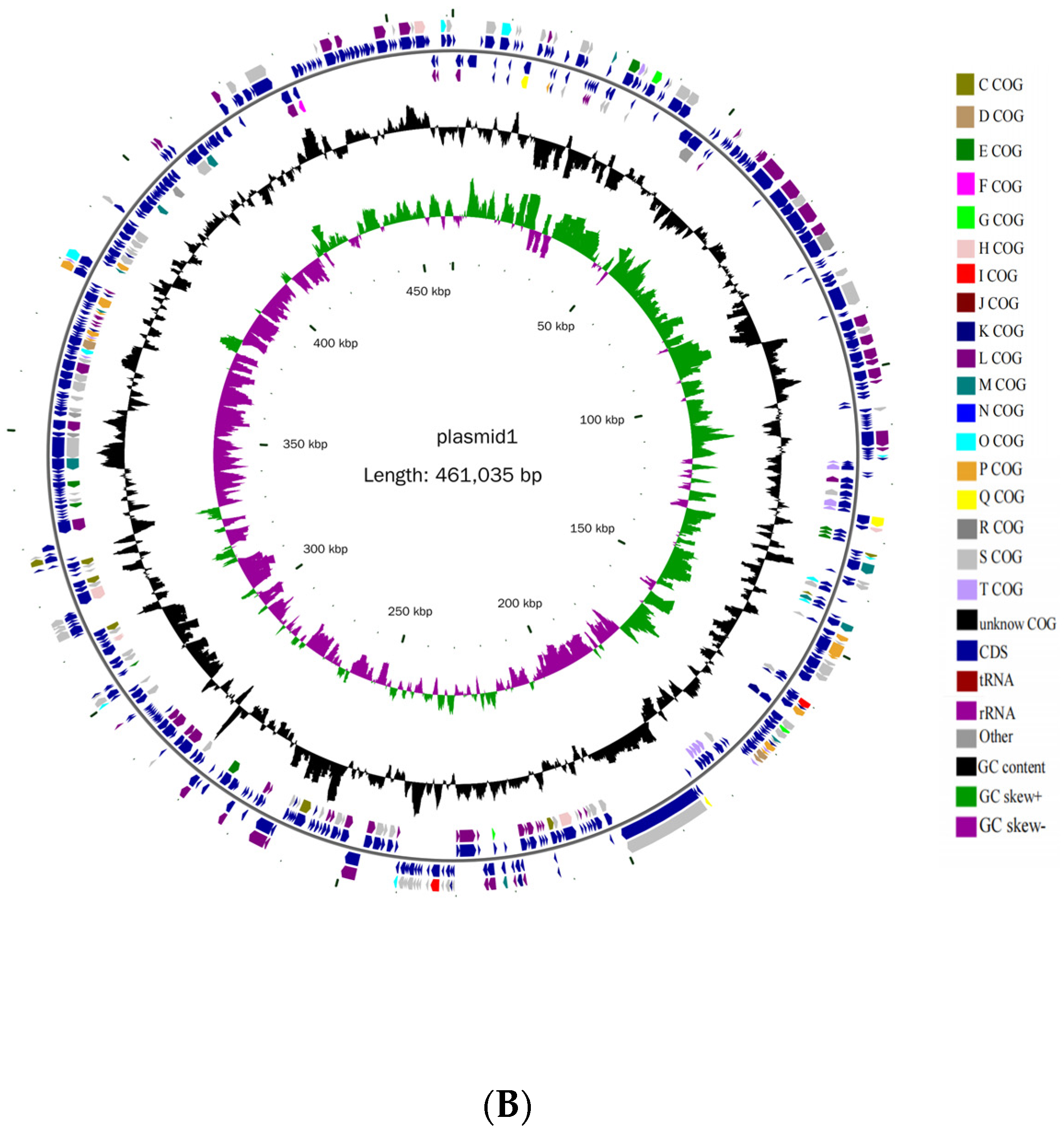
| Genomic Contents | Chromosome | Plasmid |
|---|---|---|
| Sequence length (bp) | 5,328,700 | 461,035 |
| GC content (%) | 35.36 | 33.67 |
| Open reading frames | 5440 | 455 |
| Number of tRNAs | 107 | 0 |
| Number of 5rRNAs | 14 | 0 |
| Number of 6rRNAs | 14 | 0 |
| Number of 23rRNAs | 14 | 0 |
| Number of ncRNAs | 321 | 12 |
| Enzymes | Chromosome | Gene IDs | Plasmid | Gene IDs |
|---|---|---|---|---|
| Laccase | 1 | chr_3833 | 0 | - |
| Multicopper oxidase | 1 | chr_1866 | 1 | plasmid1_118 |
| Cytochrome P450 | 5 | chr_2531, chr_2535 | 1 | plasmid1_13 |
| Monooxygenase | 8 | chr_1061, chr_1702 | 0 | - |
| Dioxygenase | 30 | chr_243, chr_244 | 0 | - |
| Catalase | 7 | chr_782, chr_823 | 0 | - |
| Oxidase | 36 | chr_185, chr_397 | 0 | - |
| Oxidoreductase | 28 | chr_280, chr_384 | 1 | plasmid1_377 |
| Dehydrogenase | 109 | chr_183, chr_325 | 6 | plasmid1_193, plasmid1_241 |
| Auxiliary Activities | Number of ORFs Annotated | Genes IDs | Enzymes |
|---|---|---|---|
| AA1_3 | 2 | chr_1866, plasmid1_118 | laccase (EC 1.10.3.2); dihydrogeodin oxidase (EC 1.10.3) |
| AA4 | 2 | chr_1325, chr_3451 | vanillin alcohol oxidase (EC 1.1.3.38) |
| AA6 | 2 | chr_1403, chr_5117 | 1,4-benzoquinone reductase (EC. 1.6.5.6) |
| AA7 | 3 | chr_185, chr_397, chr_649 | glucooligosaccharide oxidase (EC 1.1.3); chitooligosaccharide oxidase (EC 1.1.3); cellooligosaccharide dehydrogenase (EC 1.1.99) |
| AA10 | 3 | chr_2698, chr_2731, plasmid1_37 | monooxygenase (EC 1.14.99.54; EC 1.14.99.56; EC 1.14.99.53; EC 1.14.99) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhong, H.; Zhou, J.; Wang, F.; Wu, W.; Abdelrahman, M.; Li, X. Whole-Genome Sequencing Reveals Lignin-Degrading Capacity of a Ligninolytic Bacterium (Bacillus cereus) from Buffalo (Bubalus bubalis) Rumen. Genes 2022, 13, 842. https://doi.org/10.3390/genes13050842
Zhong H, Zhou J, Wang F, Wu W, Abdelrahman M, Li X. Whole-Genome Sequencing Reveals Lignin-Degrading Capacity of a Ligninolytic Bacterium (Bacillus cereus) from Buffalo (Bubalus bubalis) Rumen. Genes. 2022; 13(5):842. https://doi.org/10.3390/genes13050842
Chicago/Turabian StyleZhong, Huimin, Jiayan Zhou, Fan Wang, Wenqing Wu, Mohamed Abdelrahman, and Xiang Li. 2022. "Whole-Genome Sequencing Reveals Lignin-Degrading Capacity of a Ligninolytic Bacterium (Bacillus cereus) from Buffalo (Bubalus bubalis) Rumen" Genes 13, no. 5: 842. https://doi.org/10.3390/genes13050842
APA StyleZhong, H., Zhou, J., Wang, F., Wu, W., Abdelrahman, M., & Li, X. (2022). Whole-Genome Sequencing Reveals Lignin-Degrading Capacity of a Ligninolytic Bacterium (Bacillus cereus) from Buffalo (Bubalus bubalis) Rumen. Genes, 13(5), 842. https://doi.org/10.3390/genes13050842






