Genome-Wide Identification and Functional Analysis of the Calcineurin B-like Protein and Calcineurin B-like Protein-Interacting Protein Kinase Gene Families in Chinese Cabbage (Brassica rapa ssp. pekinensis)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Identification and Structure Analyses of the CBL and CIPK Gene Family in Chinese Cabbage
2.2. Phylogenetic Analysis and Chromosomal Location
2.3. Plant Material, Growth Condition, and Stress Treatments
2.4. RNA Isolation and Quantitative Real-Time PCR (qRT-PCR) Analysis
2.5. Yeast Two-Hybrid Assay
2.6. Arabidopsis Treatments and Estimation of the Chlorophyll Content
3. Results
3.1. Identification of BraCBL and BraCIPK Genes
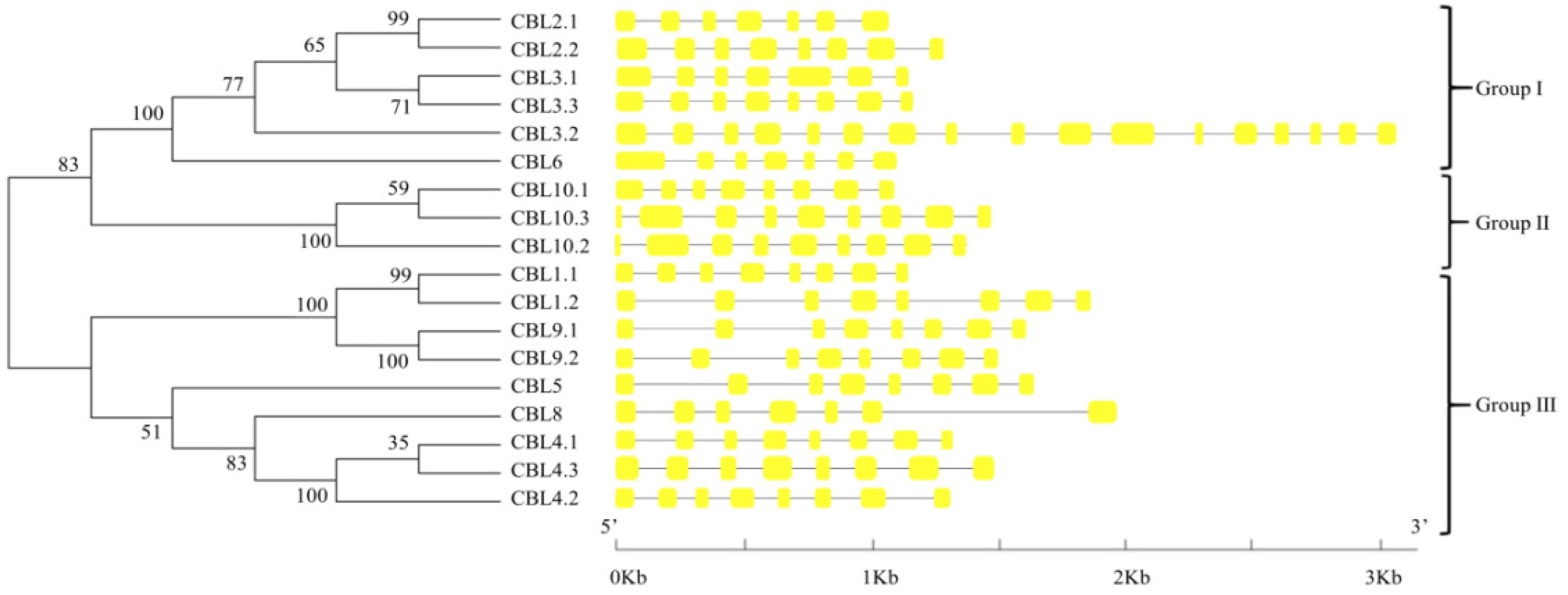
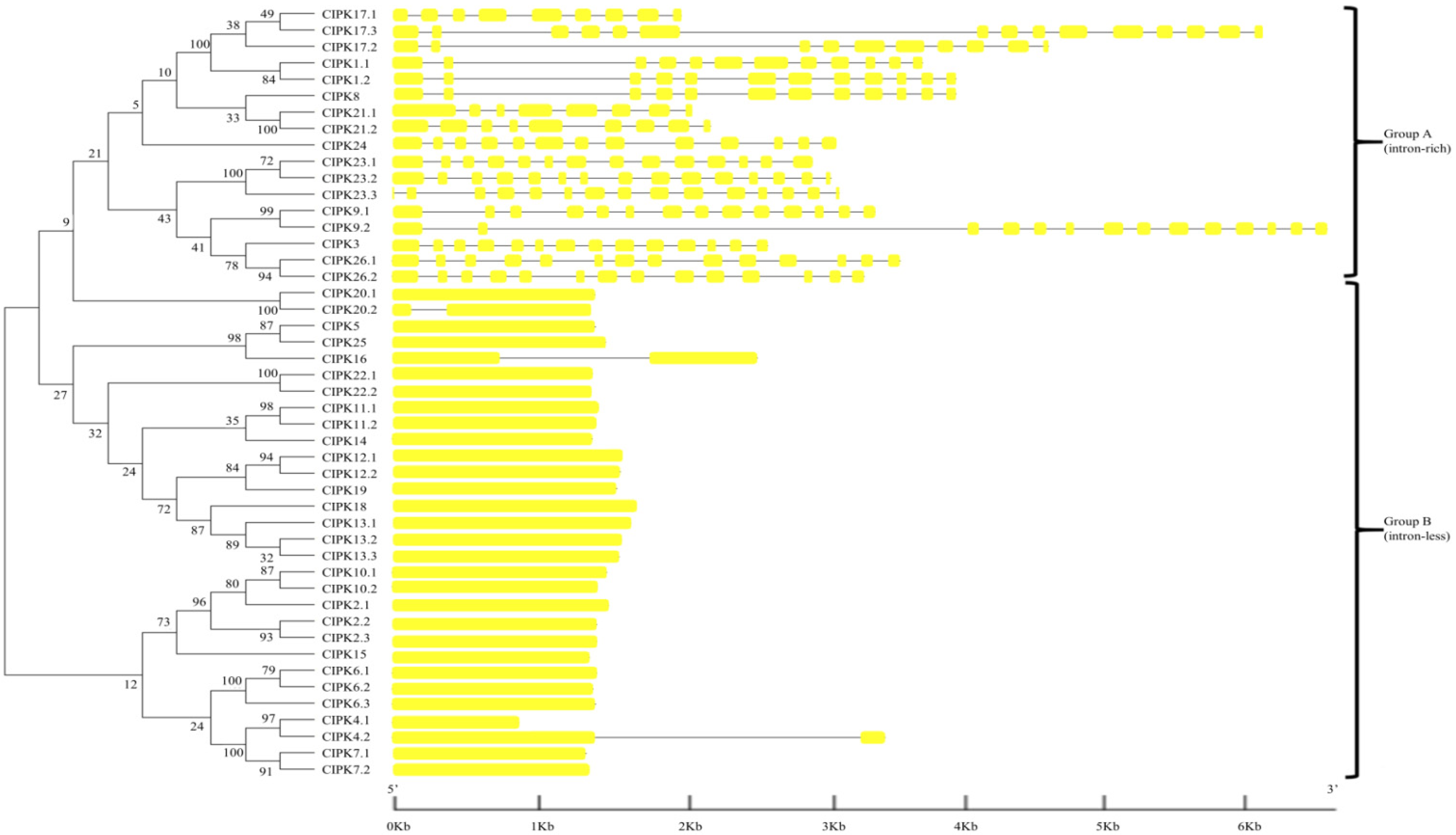
3.2. Phylogenetic Relationships and Gene Structure Analysis of BraCBLs and BraCIPKs
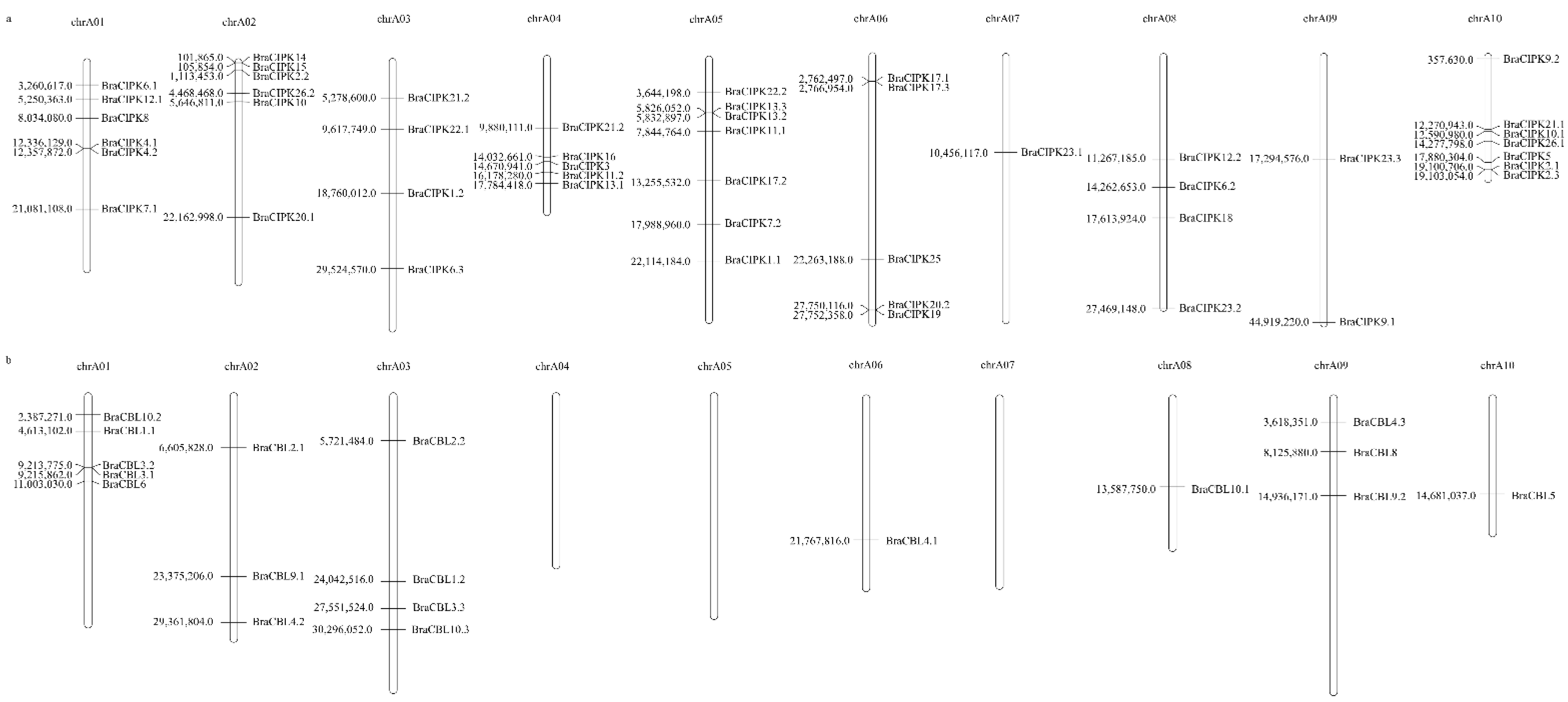
3.3. Chromosomal Localization and Phylogenetic Analysis of the BraCBL and BraCIPK Gene Families in Chinese Cabbage
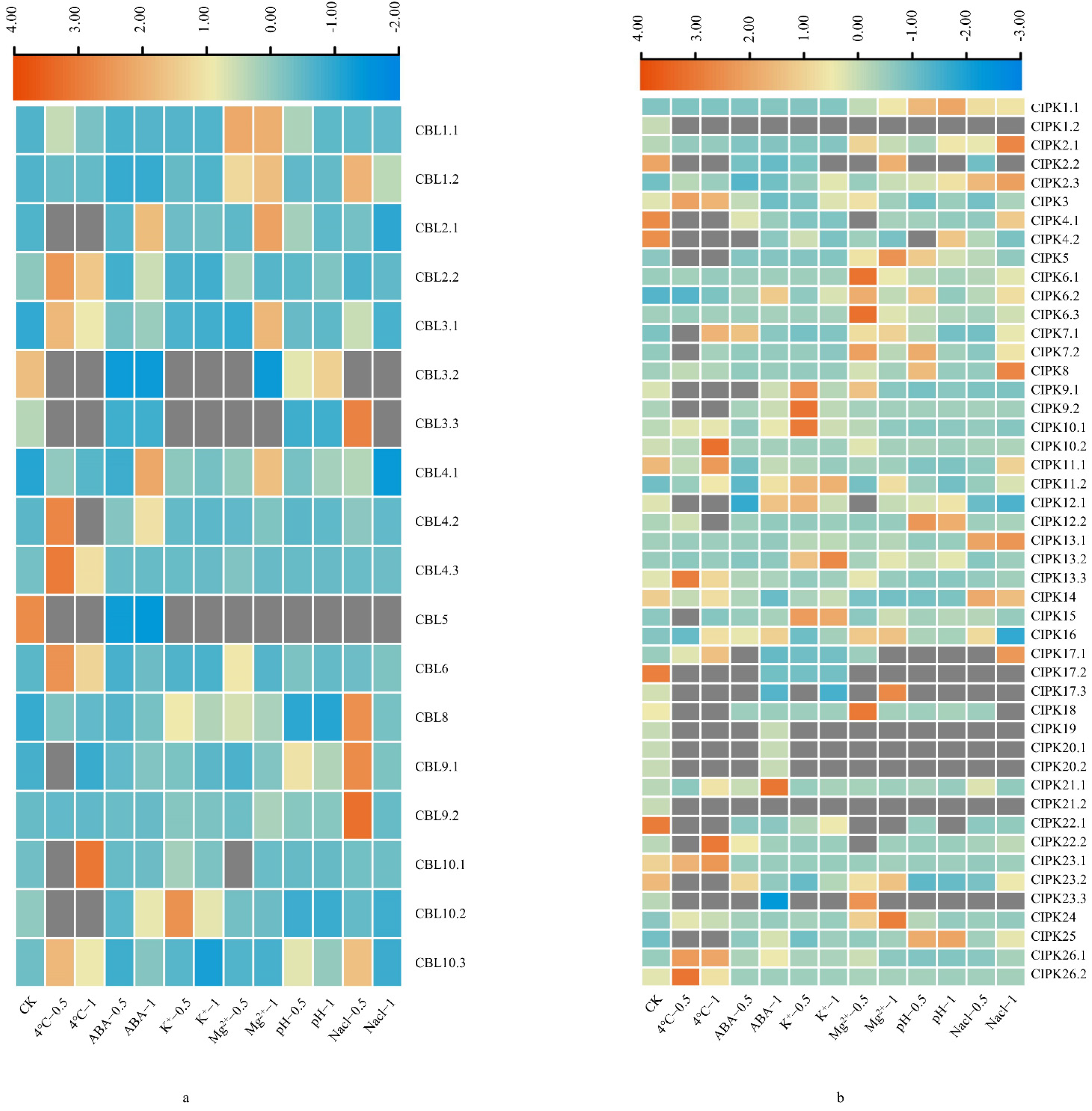
3.4. Expression Profiles of BraCBL and BraCIPK Genes after Stress Treatment
3.5. Analysis of the Interactions between BraCIPK23 Duplicate Genes and BraCBLs Proteins
3.6. Phenotypic Observation and Physiological Indicator Measurements of Trans-BraCIPK23 Repeat Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Peck, S.; Mittler, R. Plant signaling in biotic and abiotic stress. J. Exp. Bot. 2020, 71, 1649–1651. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Batistic, O.; Kudla, J.J.P. Integration and channeling of calcium signaling through the CBL calcium sensor/CIPK protein kinase network. Planta 2004, 219, 915–924. [Google Scholar] [CrossRef] [PubMed]
- Xiong, L.; Schumaker, K.S.; Zhu, J.-K. Cell Signaling during Cold, Drought, and Salt Stress. Plant Cell 2002, 14, S165–S183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McAinsh, M.R.; Pittman, J.K. Shaping the calcium signature. New Phytol. 2009, 181, 275–294. [Google Scholar] [CrossRef]
- Boudsocq, M.; Sheen, J. CDPKs in immune and stress signaling. Trends Plant Sci. 2013, 18, 30–40. [Google Scholar] [CrossRef] [Green Version]
- Sanyal, S.K.; Pandey, A.; Pandey, G.K. The CBL-CIPK signaling module in plants: A mechanistic perspective. Physiol. Plant. 2015, 155, 89–108. [Google Scholar] [CrossRef]
- Lu, T.; Zhang, G.; Sun, L.; Wang, J.; Hao, F. Genome-wide identification of CBL family and expression analysis of CBLs in response to potassium deficiency in cotton. PeerJ 2017, 5, e3653. [Google Scholar] [CrossRef] [Green Version]
- Guo, Y.; Halfter, U.; Ishitani, M.; Zhu, J.K. Molecular characterization of functional domains in the protein kinase SOS2 that is required for plant salt tolerance. Plant Cell 2001, 13, 1383–1399. [Google Scholar] [CrossRef] [Green Version]
- Sánchez-Barrena, M.J.; Fujii, H.; Angulo, I.; Martínez-Ripoll, M.; Zhu, J.-K.; Albert, A. The Structure of the C-Terminal Domain of the Protein Kinase AtSOS2 Bound to the Calcium Sensor AtSOS3. Mol. Cell 2007, 26, 427–435. [Google Scholar] [CrossRef] [Green Version]
- Kolukisaoglu, Ü.; Weinl, S.; Blazevic, D.; Batistic, O.; Kudla, J. Calcium Sensors and Their Interacting Protein Kinases: Genomics of the Arabidopsis and Rice CBL-CIPK Signaling Networks. Plant Physiol. 2004, 134, 43–58. [Google Scholar] [CrossRef] [Green Version]
- Xiang, Y.; Huang, Y.; Xiong, L. Characterization of Stress-Responsive CIPK Genes in Rice for Stress Tolerance Improvement. Plant Physiol. 2007, 144, 1416–1428. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, J.; Jing, L.; Hui, L.; Li, X.; Yang, Q.; Cheng, Z.M.; Chang, Y. Characterization of CIPK Family in Asian Pear (Pyrus bretschneideri Rehd) and Co-expression Analysis Related to Salt and Osmotic Stress Responses. Front. Plant Sci. 2016, 7, 1361. [Google Scholar] [CrossRef] [Green Version]
- Weinl, S.; Kudla, J. The CBL-CIPK Ca(2+)-decoding signaling network: Function and perspectives. New Phytol. 2009, 184, 517–528. [Google Scholar] [CrossRef] [PubMed]
- Mohanta, T.K.; Mohanta, N.; Mohanta, Y.K.; Parida, P.; Bae, H. Genome-wide identification of Calcineurin B-Like (CBL) gene family of plants reveals novel conserved motifs and evolutionary aspects in calcium signaling events. BMC Plant Biol. 2015, 15, 189. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Yang, B.; Liu, W.-Z.; Li, H.; Wang, L.; Wang, B.; Deng, M.; Liang, W.; Deyholos, M.K.; Jiang, Y.-Q. Identification and characterization of CBL and CIPK gene families in canola (Brassica napus L.). BMC Plant Biol. 2014, 14, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xin, Y.; Wang, Q.; Qian, C.; Xiang, N.; Yang, Y.; Yang, Y. Genome-Wide Identification and Functional Analysis of the Calcineurin B-like Protein and Calcineurin B-like Protein-Interacting Protein Kinase Gene Families in Turnip (Brassica rapa var. rapa). Front. Plant Sci. 2017, 8, 1191–1209. [Google Scholar]
- Chen, X.; Gu, Z.; Xin, D.; Hao, L.; Liu, C.; Huang, J.; Ma, B.; Zhang, H. Identification and characterization of putative CIPK genes in maize. J. Genet. Genom. 2011, 38, 77–87. [Google Scholar] [CrossRef]
- Zhu, J.K. Regulation of ion homeostasis under salt stress—ScienceDirect. Curr. Opin. Plant Biol. 2003, 6, 441–445. [Google Scholar] [CrossRef]
- Quan, R.; Lin, H.; Mendoza, I.; Zhang, Y.; Cao, W.; Yang, Y.; Shang, M.; Chen, S.; Pardo, J.M.; Guo, Y. SCABP8/CBL10, a putative calcium sensor, interacts with the protein kinase SOS2 to protect Arabidopsis shoots from salt stress. Plant Cell 2007, 19, 1415–1431. [Google Scholar] [CrossRef] [Green Version]
- Deng, X.; Hu, W.; Wei, S.; Zhou, S.; Zhang, F.; Han, J.; Chen, L.; Li, Y.; Feng, J.; Fang, B.; et al. TaCIPK29, a CBL-Interacting Protein Kinase Gene from Wheat, Confers Salt Stress Tolerance in Transgenic Tobacco. PLoS ONE 2013, 8, e69881. [Google Scholar] [CrossRef] [Green Version]
- Xu, J.; Li, H.D.; Chen, L.Q.; Wang, Y.; Liu, L.L.; He, L.; Wu, W.H. A Protein Kinase, Interacting with Two Calcineurin B-like Proteins, Regulates K+ Transporter AKT1 in Arabidopsis—ScienceDirect. Cell 2006, 125, 1347–1360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Long, Y.; Qi, G.-N.; Li, J.; Xu, Z.-J.; Wu, W.-H.; Wang, Y. The Os-AKT1 Channel Is Critical for K+ Uptake in Rice Roots and Is Modulated by the Rice CBL1-CIPK23 Complex. Plant Cell 2014, 26, 3387–3402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, R.-J.; Zhao, F.-G.; Yang, Y.; Wang, C.; Li, K.; Kleist, T.J.; Lemaux, P.G.; Luan, S. A calcium signalling network activates vacuolar K+ remobilization to enable plant adaptation to low-K environments. Nat. Plants 2020, 6, 384–393. [Google Scholar] [CrossRef] [PubMed]
- Cuéllar, T.; Azeem, F.; Andrianteranagna, M.; Pascaud, F.O.; Verdeil, J.L.; Sentenac, H.; Zimmermann, S.; Gaillard, I. Potassium transport in developing fleshy fruits: The grapevine inward K(+) channel VvK1.2 is activated by CIPK-CBL complexes and induced in ripening berry flesh cells. Plant J. 2013, 73, 1006–1018. [Google Scholar] [CrossRef] [PubMed]
- Ho, C.-H.; Lin, S.-H.; Hu, H.-C.; Tsay, Y.-F. CHL1 Functions as a Nitrate Sensor in Plants. Cell 2009, 138, 1184–1194. [Google Scholar] [CrossRef] [Green Version]
- Tang, R.-J.; Liu, H.; Yang, Y.; Yang, L.; Gao, X.-S.; Garcia, V.J.; Luan, S.; Zhang, H.-X. Tonoplast calcium sensors CBL2 and CBL3 control plant growth and ion homeostasis through regulating V-ATPase activity in Arabidopsis. Cell Res. 2012, 22, 1650–1665. [Google Scholar] [CrossRef]
- Tang, R.-J.; Zhao, F.-G.; Garcia, V.J.; Kleist, T.J.; Yang, L.; Zhang, H.-X.; Luan, S. Tonoplast CBL–CIPK calcium signaling network regulates magnesium homeostasis in Arabidopsis. Proc. Natl. Acad. Sci. USA 2015, 112, 3134–3139. [Google Scholar] [CrossRef] [Green Version]
- Huang, C.; Ding, S.; Zhang, H.; Du, H.; An, L. CIPK7 is involved in cold response by interacting with CBL1 in Arabidopsis thaliana. Plant Sci. 2011, 181, 57–64. [Google Scholar] [CrossRef]
- Nieves-Cordones, M.; Caballero, F.; Martínez, V.; Rubio, F. Disruption of the Arabidopsis thaliana inward-rectifier K+ channel AKT1 improves plant responses to water stress. Plant Cell Physiol. 2012, 53, 423–432. [Google Scholar] [CrossRef]
- Fuglsang, A.T.; Yan, G.; Cuin, T.A.; Qiu, Q.; Song, C.; Kristiansen, K.A.; Bych, K.; Schulz, A.; Shabala, S.; Schumaker, K. Arabidopsis Protein Kinase PKS5 Inhibits the Plasma Membrane H1-ATPase by Preventing Interaction. Plant Cell 2007, 19, 1617–1634. [Google Scholar] [CrossRef] [Green Version]
- Sanyal, S.K.; Kanwar, P.; Yadav, A.K.; Sharma, C.; Kumar, A.; Pandey, G.K. Arabidopsis CBL interacting protein kinase 3 interacts with ABR1, an APETALA2 domain transcription factor, to regulate ABA responses. Plant Sci. 2017, 254, 48–59. [Google Scholar] [CrossRef] [PubMed]
- D’Angelo, C.; Weinl, S.; Batistic, O.; Pandey, G.K.; Cheong, Y.H.; Schültke, S.; Albrecht, V.; Ehlert, B.; Schulz, B.; Harter, K.; et al. Alternative complex formation of the Ca2+-regulated protein kinase CIPK1 controls abscisic acid-dependent and independent stress responses in Arabidopsis. Plant J. 2006, 48, 857–872. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y. Current Situation and Treatment of Heavy Metal Pollution in Soil. Compr. Util. Resour. China 2019, 37, 114–116. [Google Scholar]
- Tao, S.; Yan, W.; Meng, W.; Li, T.; Yi, Z.; Wang, X.; Wei, S.; He, G.; Yang, G. Identification and comprehensive analyses of the CBL and CIPK gene families in wheat (Triticum aestivum L.). BMC Plant Biol. 2015, 15, 269–285. [Google Scholar]
- Yuanchun, M.; Qunkang, C.; Zongming, C.; Hui, L.; Youhong, C.; Jing, L. Identification of Important Physiological Traits and Moderators That Are Associated with Improved Salt Tolerance in CBL and CIPK Overexpressors through a Meta-Analysis. Front. Plant Sci. 2017, 8, 856–868. [Google Scholar]
- Yue, X.; Liu, J.; Dong, C.; Cheng, Z. The CBL and CIPK Gene Family in Grapevine (Vitis vinifera): Genome-Wide Analysis and Expression Profiles in Response to Various Abiotic Stresses. Front. Plant Sci. 2017, 8, 978–992. [Google Scholar]
- Wilkins, M.R.; Gasteiger, E.; Bairoch, A.; Sanchez, J.C.; Williams, K.L.; Appel, R.D.; Hochstrasser, D.F. Protein Identification and Analysis Tools in the ExPASy Server. Methods Mol. Biol. 1999, 112, 531–552. [Google Scholar]
- Ren, J.; Wen, L.; Gao, X.; Jin, C.; Xue, Y.; Yao, X. CSS-Palm 2.0: An updated software for palmitoylation sites prediction. Protein Eng. Des. Sel. 2008, 21, 639–644. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Bailey, T.L.; Nadya, W.; Chris, M.; Li, W.W. MEME: Discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006, 34, W369–W373. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Voorrips, R.E. MapChart: Software for the graphical presentation of linkage maps and QTLs. J. Hered. 2002, 93, 77–78. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Z. PAML 4: Phylogenetic Analysis by Maximum Likelihood. Mol. Biol. Evol. 2007, 24, 1586–1591. [Google Scholar] [CrossRef] [Green Version]
- Cao, J.; Huang, J.; Yang, Y.; Hu, X. Analyses of the oligopeptide transporter gene family in poplar and grape. BMC Genom. 2011, 12, 465. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mogami, J.; Fujita, Y.; Yoshida, T.; Tsukiori, Y.; Nakagami, H.; Nomura, Y.; Fujiwara, T.; Nishida, S.; Yanagisawa, S.; Ishida, T.; et al. Two Distinct Families of Protein Kinases Are Required for Plant Growth under High External Mg2+ Concentrations in Arabidopsis. Plant Physiol. 2015, 167, 1039–1057. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, Y.; Xiong, L.; Song, C.-P.; Gong, D.; Halfter, U.; Zhu, J.-K. A Calcium Sensor and Its Interacting Protein Kinase Are Global Regulators of Abscisic Acid Signaling in Arabidopsis. Dev. Cell 2002, 3, 233–244. [Google Scholar] [CrossRef] [Green Version]
- IBM Corp. IBM SPSS Statistics for Windows, Version 24.0; IBM Corp.: Armonk, NY, USA, 2016. [Google Scholar]
- Arnon, D. Copper induced enzyme in isolated chloroplasts: Polyphenoloxidase in Beta vulgaris. Plant Physiol. 1949, 24, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Xu, C.; Wu, J.; Min, L.; Stefan, G.; Feng, C.; Liang, J.; Cai, C.; Liu, Z.; Bo, L. Improved Brassica rapa reference genome by single-molecule sequencing and chromosome conformation capture technologies. Hortic. Res. 2018, 5, 50–60. [Google Scholar] [CrossRef] [Green Version]
- Batistic, O.; Sorek, N.; Schültke, S.; Yalovsky, S.; Kudla, J. Dual Fatty Acyl Modification Determines the Localization and Plasma Membrane Targeting of CBL/CIPK Ca2+ Signaling Complexes in Arabidopsis. Plant Cell 2008, 20, 1346–1362. [Google Scholar] [CrossRef] [Green Version]
- Albrecht, V.; Ritz, O.; Linder, S.; Harter, K.; Kudla, J. The NAF domain defines a novel protein-protein interaction module conserved in Ca2+-regulated kinases. EMBO J. 2001, 20, 1051–1063. [Google Scholar] [CrossRef] [Green Version]
- Bowers, J.; Chapman, B.; Rong, J.; Paterson, A.H. Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 2003, 422, 433–438. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Barrena, M.J.; Martínez-Ripoll, M.; Zhu, J.K.; Albert, A. The Structure of the Arabidopsis thaliana SOS3: Molecular Mechanism of Sensing Calcium for Salt Stress Response. J. Mol. Biol. 2005, 345, 1253–1264. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Li, Z.; Fu, Q.; Liang, C.; Liu, Z.; Liu, Q.; Pu, G.; Li, J. Genome-Wide Identification of CBL-CIPK Gene Family in Honeysuckle (Lonicera japonica Thunb.) and Their Regulated Expression Under Salt Stress. Front. Genet. 2021, 12, 751040. [Google Scholar] [CrossRef] [PubMed]
- Du, W.; Yang, J.; Ma, L.; Su, Q.; Pang, Y. Identification and Characterization of Abiotic Stress Responsive CBL-CIPK Family Genes in Medicago. Int. J. Mol. Sci. 2021, 22, 4634. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Zhang, B.; Deng, J.; Chen, L.; Ullah, A.; Yang, X. Genome-wide analysis of CBL and CIPK family genes in cotton: Conserved structures with divergent interactions and expression. Physiol. Mol. Biol. Plants 2021, 27, 359–368. [Google Scholar] [CrossRef] [PubMed]
- Gigolashvili, T.; Berger, B.; Flügge, U.I. Specific and coordinated control of indolic and aliphatic glucosinolate biosynthesis by R2R3-MYB transcription factors in Arabidopsis thaliana. Phytochem. Rev. 2009, 8, 3–13. [Google Scholar] [CrossRef]
- Cannon, S.B.; Mitra, A.; Baumgarten, A.; Young, N.D.; May, G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004, 4, 10. [Google Scholar] [CrossRef] [Green Version]
- Zhu, K.; Chen, F.; Liu, J.; Chen, X.; Hewezi, T.; Cheng, Z.-M. Evolution of an intron-poor cluster of the CIPK gene family and expression in response to drought stress in soybean. Sci. Rep. 2016, 6, 28225. [Google Scholar] [CrossRef] [Green Version]
- Roy, S.W.; Penny, D. Patterns of Intron Loss and Gain in Plants: Intron Loss–Dominated Evolution and Genome-Wide Comparison of O. sativa and A. thaliana. Mol. Biol. Evol. 2007, 24, 171–181. [Google Scholar] [CrossRef] [Green Version]
- Mao, J.; Manik, S.M.; Shi, S.; Chao, J.; Jin, Y.; Wang, Q.; Liu, H. Mechanisms and Physiological Roles of the CBL-CIPK Networking System in Arabidopsis thaliana. Genes 2016, 7, 62. [Google Scholar] [CrossRef] [Green Version]
- Li, R.; Zhang, J.; Wei, J.; Wang, H.; Wang, Y.; Ma, R. Functions and mechanisms of the CBL–CIPK signaling system in plant response to abiotic stress. Prog. Nat. Sci. 2009, 19, 667–676. [Google Scholar] [CrossRef]
- Liang, W.H.; Liu, X.F.; Bi, J.-J.; Lv, H.-C.; Peng, W.F. Effect of G15V and T20N Point Mutations of OsRacD from Oryza sativa on Cellular Localization and Interaction with Target Proteins. Chin. J. Biochem. Mol. Biol. 2009, 25, 828–833. [Google Scholar]
- Behera, S.; Long, Y.; Schmitz-Thom, I.; Wang, X.P.; Zhang, C.; Li, H.; Steinhorst, L.; Manishankar, P.; Ren, X.-L.; Wang, Y.; et al. Two spatially and temporally distinct Ca2+ signals convey Arabidopsis thaliana responses to K+ deficiency. New Phytol. 2016, 213, 739–750. [Google Scholar] [CrossRef] [PubMed] [Green Version]


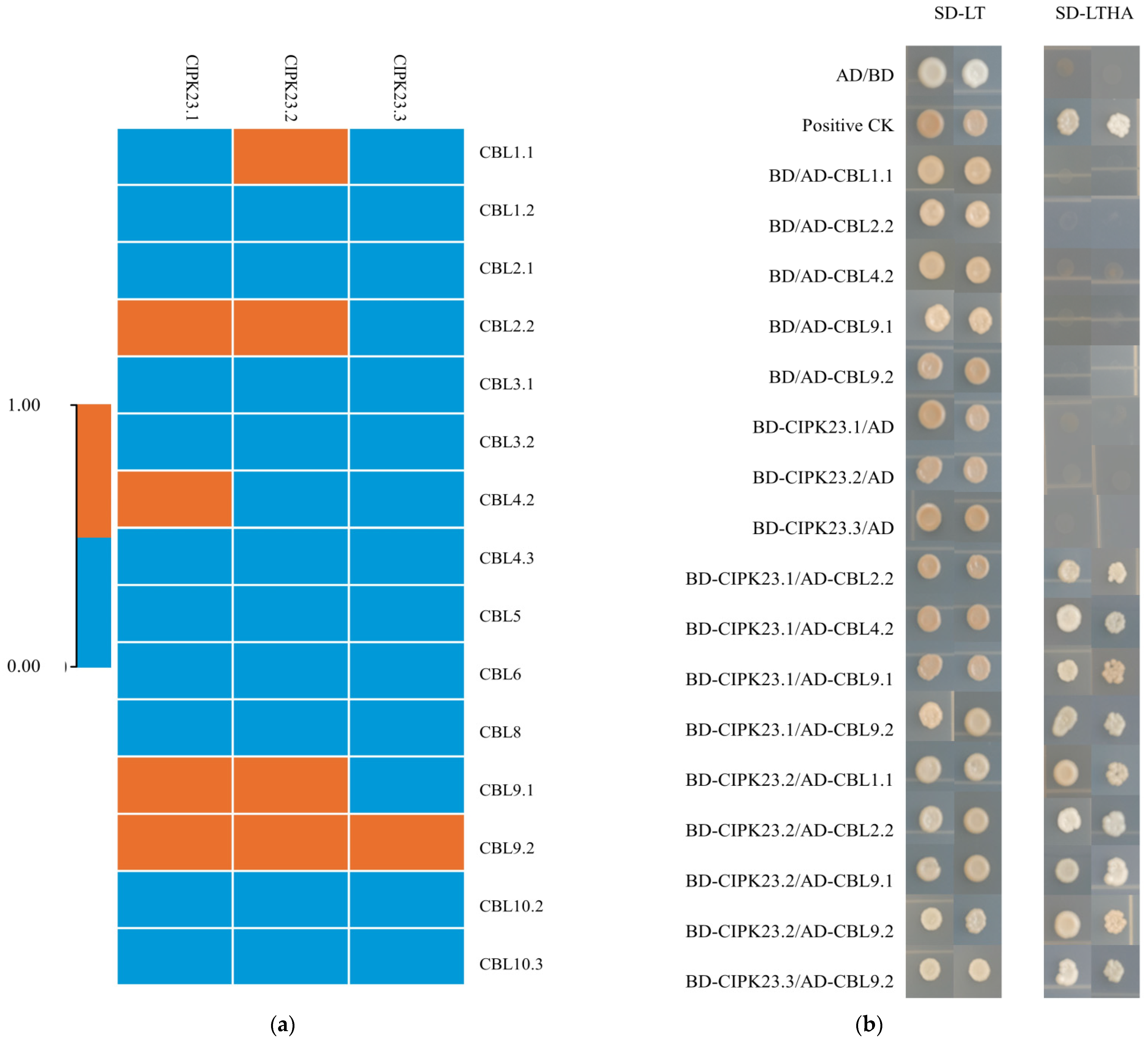
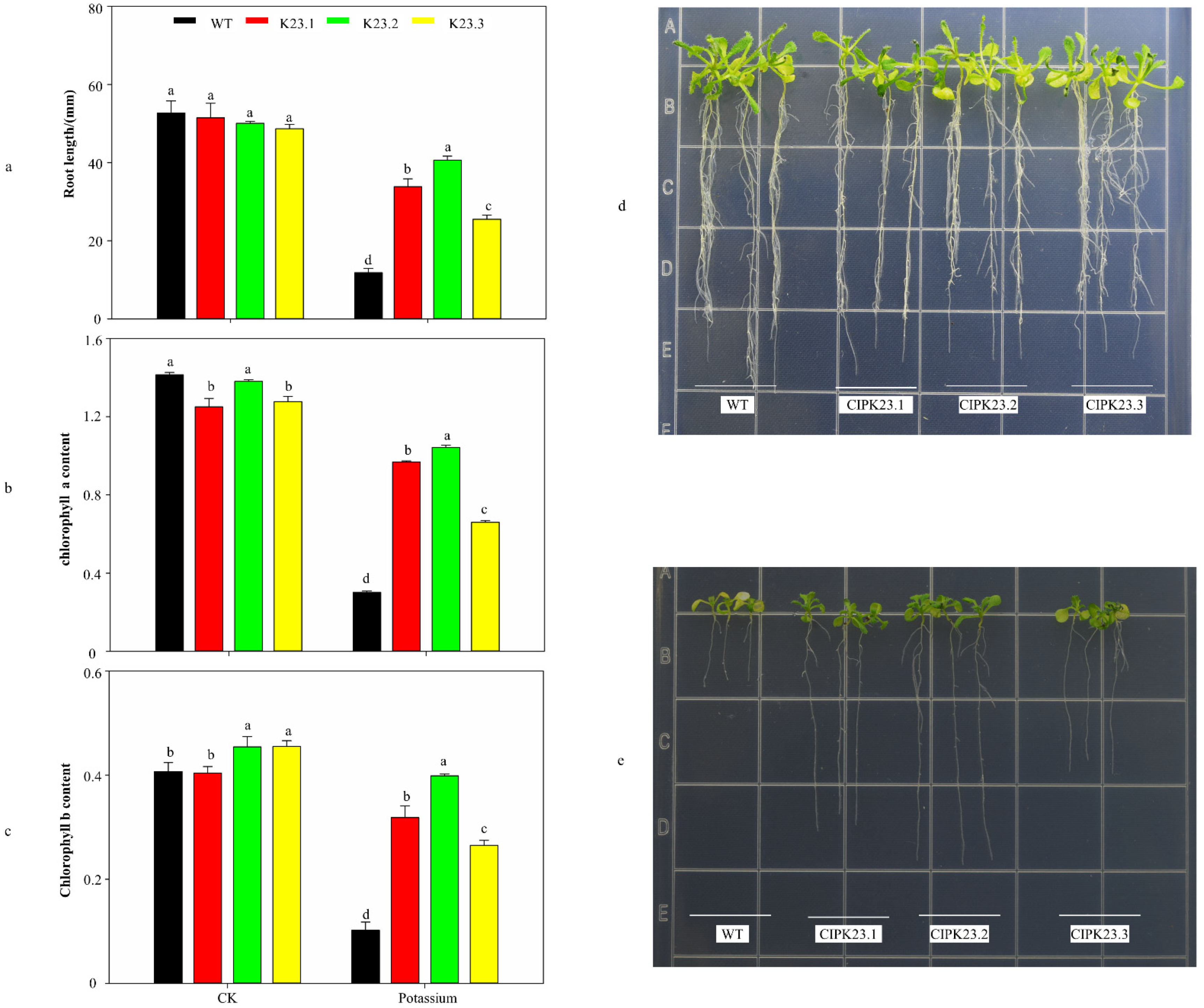
| Gene Name | Arabidopsis Ortholog | Identity/% | Gene Locus | MW (Da) | PI | GRAVY | NO. of Amino Acids | No. of EF-Hand | Myristoylaton Motif | Palmitoylation Prediction |
|---|---|---|---|---|---|---|---|---|---|---|
| BraCBL1.1 | AtCBL1 | 92.21 | A01:4613102–4615104 (+strand) | 24,587.01 | 4.69 | −0.19 | 213 | 3 | Y | Y |
| BraCBL1.2 | 91.12 | A03:24042516–24044378 (+strand) | 24,603.09 | 4.64 | −0.173 | 213 | 3 | Y | Y | |
| BraCBL2.1 | AtCBL2 | 94.57 | A02:6605828–6607158 (−strand) | 25,865.49 | 4.89 | −0.22 | 226 | 3 | N | Y |
| BraCBL2.2 | 80.84 | A03:5721464–5722801 (−strand) | 29,846.17 | 5.12 | −0.123 | 260 | 2 | N | Y | |
| BraCBL3.1 | AtCBL3 | 89.61 | A01:9215862–9217195 (−strand) | 25,783.26 | 4.78 | −0.227 | 226 | 3 | N | Y |
| BraCBL3.2 | 41.07 | A03:27551525–27554719 (−strand) | 53,216.02 | 5.92 | −0.249 | 475 | 3 | N | Y | |
| BraCBL3.3 | 60.78 | A01:9213775–9215163 (−strand) | 28,005.91 | 5.27 | −0.296 | 250 | 2 | N | Y | |
| BraCBL4.1 | AtCBL4 | 86.85 | A06:21767816–21769240 (−strand) | 25,583.31 | 4.97 | −0.282 | 221 | 3 | Y | Y |
| BraCBL4.2 | 77.28 | A02:29361806–29362997 (+strand) | 22,451.92 | 5.04 | −0.184 | 196 | 3 | Y | Y | |
| BraCBL4.3 | 88.19 | A09:3618351–3619847 (+strand) | 24,986.75 | 4.95 | −0.218 | 217 | 3 | Y | Y | |
| BraCBL5 | AtCBL5 | 80.58 | A10:14681036–14683065 (−strand) | 22,430.59 | 5.23 | −0.332 | 194 | 3 | Y | Y |
| BraCBL6 | AtCBL6 | 83.19 | A01:11003030–11004304 (+strand) | 26,206.29 | 5.99 | −0.21 | 227 | 3 | N | Y |
| BraCBL8 | AtCBL8 | 89.15 | A09:8125880–8127444 (−strand) | 24,751.32 | 5.23 | −0.308 | 214 | 3 | N | Y |
| BraCBL9.1 | AtCBL9 | 91.9 | A02:23375206–23376913 (−strand) | 24,350.67 | 4.62 | −0.17 | 213 | 3 | Y | Y |
| BraCBL9.2 | 90.34 | A09:14936171–14937957 (−strand) | 24,354.62 | 4.62 | −0.22 | 213 | 3 | Y | Y | |
| BraCBL10.1 | AtCBL10 | 83.33 | A08:13587750–13589239 (−strand) | 28,462.58 | 4.82 | −0.079 | 246 | 3 | N | Y |
| BraCBL10.2 | 72.04 | A01:2387271–2388596 (−strand) | 24,335.99 | 4.67 | −0.075 | 211 | 3 | N | Y | |
| BraCBL10.3 | 82.69 | A03:30296053–30297485 (+strand) | 28,705.94 | 4.49 | −0.001 | 249 | 3 | N | Y |
| Gene Name | Arabidopsis Ortholog | Identity/% | Gene Locus | MW (Da) | PI | GRAVY | NO. of Amino Acids |
|---|---|---|---|---|---|---|---|
| BraCIPK1.1 | AtCIPK1 | 90.83 | A05:22114185–22117606 (+strand) | 50,008.16 | 6.08 | −0.39 | 446 |
| BraCIPK1.2 | 86.29 | A03:18760011–18763637 (−strand) | 47,266.56 | 8.94 | −0.32 | 420 | |
| BraCIPK2.1 | AtCIPK2 | 75 | A10:19100706–19102103 (+strand) | 52,509.11 | 7.2 | −0.425 | 465 |
| BraCIPK2.2 | 74.4 | A02:1113453–1114772 (−strand) | 49,660.32 | 6.78 | −0.394 | 439 | |
| BraCIPK2.3 | 73.45 | A10:19103054–19104376 (+strand) | 49,404.33 | 9.02 | −0.378 | 440 | |
| BraCIPK3 | AtCIPK3 | 88.94 | A04:14670940–14673364 (−strand) | 50,253.49 | 6.82 | −0.534 | 440 |
| BraCIPK4.1 | AtCIPK4 | 50.86 | A01:12336129–12336953 (+strand) | 30,459.14 | 9.74 | −0.31 | 274 |
| BraCIPK4.2 | 66.87 | A01:12357872–12361057 (+strand) | 54,941.5 | 7.31 | −0.171 | 490 | |
| BraCIPK5 | AtCIPK5 | 79.29 | A10:17880305–17881609 (+strand) | 49,526.82 | 6.14 | −0.329 | 434 |
| BraCIPK6.1 | AtCIPK6 | 85.48 | A01:3260617–3261942 (−strand) | 49,199.57 | 8.69 | −0.289 | 441 |
| BraCIPK6.2 | 83.79 | A08:14262653–14263951 (−strand) | 48,461.95 | 9.04 | −0.303 | 432 | |
| BraCIPK6.3 | 80.88 | A03:29524568–29525881 (+strand) | 49,009.42 | 8.9 | −0.316 | 437 | |
| BraCIPK7.1 | AtCIPK7 | 80.47 | A01:21081109–21082353 (+strand) | 46,300.42 | 9.21 | −0.234 | 414 |
| BraCIPK7.2 | 79.84 | A05:17988961–17990229 (−strand) | 47,280.64 | 8.73 | −0.214 | 422 | |
| BraCIPK8 | AtCIPK8 | 91.57 | A01:8034080–8036979 (+strand) | 50,459.22 | 8.63 | −0.215 | 446 |
| BraCIPK9.1 | AtCIPK9 | 90.33 | A09:44919220–44922333 (−strand) | 49,822.14 | 8.33 | −0.393 | 445 |
| BraCIPK9.2 | 91.59 | A10:357630–363660 (+strand) | 50,384.78 | 8.03 | −0.402 | 447 | |
| BraCIPK10.1 | AtCIPK10 | 84.36 | A10:12590980–12592368 (−strand) | 52,751.48 | 8.02 | −0.539 | 462 |
| BraCIPK10.2 | 75.76 | A02:5646811–5648142 (+strand) | 50,887.75 | 8.91 | −0.455 | 443 | |
| BraCIPK11.1 | AtCIPK11 | 80.27 | A05:7844764–7846092 (+strand) | 49,700.13 | 8.29 | −0.331 | 442 |
| BraCIPK11.2 | 79.24 | A04:16178280–16179593 (−strand) | 49,403.9 | 8.62 | −0.283 | 437 | |
| BraCIPK12.1 | AtCIPK12 | 80.98 | A01:5250363–5251847 (−strand) | 55,311.48 | 6.75 | −0.259 | 494 |
| BraCIPK12.2 | 83.33 | A08:11267185–11268651 (−strand) | 55,031.29 | 6.75 | −0.261 | 488 | |
| BraCIPK13.1 | AtCIPK13 | 83.72 | A04:17784418–17785956 (−strand) | 57,880.06 | 8.17 | −0.267 | 512 |
| BraCIPK13.2 | 80.98 | A05:5832897–5834375 (+strand) | 55,244.17 | 8.99 | −0.207 | 492 | |
| BraCIPK13.3 | 81.92 | A05:5826052–5827512 (+strand) | 54,789.57 | 9.13 | −0.221 | 486 | |
| BraCIPK14 | AtCIPK14 | 75.05 | A02:101865–103160 (+strand) | 48,561.02 | 9.11 | −0.241 | 431 |
| BraCIPK15 | AtCIPK15 | 81.41 | A02:105854–107125 (−strand) | 47,963.47 | 8.66 | −0.37 | 423 |
| BraCIPK16 | AtCIPK16 | 81.99 | A04:14032661–14035013 (−strand) | 52,367 | 5.49 | −0.399 | 461 |
| BraCIPK17.1 | AtCIPK17 | 70.08 | A06:2762497–2764360 (+strand) | 39,815.64 | 5.36 | −0.131 | 357 |
| BraCIPK17.3 | 78.34 | A05:13255532–13259898 (+strand) | 44,843.86 | 7.59 | −0.211 | 399 | |
| BraCIPK17.4 | 52.18 | A06:2766954–2772570 (+strand) | 68,620.32 | 9.24 | −0.049 | 620 | |
| BraCIPK18 | AtCIPK18 | 83.11 | A08:17613924–17615498 (+strand) | 58,872 | 7.54 | −0.287 | 524 |
| BraCIPK19 | AtCIPK19 | 81.82 | A06:27752359–27753804 (−strand) | 54,462.93 | 9 | −0.276 | 481 |
| BraCIPK20.1 | AtCIPK20 | 86.59 | A02:22162998–22164305 (−strand) | 49,902.91 | 9.21 | −0.45 | 435 |
| BraCIPK20.2 | 64.27 | A06:27750115–27751398 (+strand) | 40,057.09 | 8.87 | −0.437 | 352 | |
| BraCIPK21.1 | AtCIPK21 | 92.89 | A10:12270943–12272878 (−strand) | 46,403.52 | 8.28 | −0.166 | 416 |
| BraCIPK21.2 | 85.13 | A03:5278600–5280652 (+strand) | 42,800.57 | 8.59 | −0.116 | 386 | |
| BraCIPK22.1 | AtCIPK22 | 78.95 | A03:9617749–9619041 (−strand) | 48,701.31 | 9.11 | −0.362 | 430 |
| BraCIPK22.2 | 76.68 | A05:3644198–3645481 (+strand) | 48,342.69 | 9.15 | −0.354 | 427 | |
| BraCIPK23.1 | AtCIPK23 | 84.89 | A07:10456117–10458829 (−strand) | 52,147.85 | 9.2 | −0.398 | 470 |
| BraCIPK23.2 | 81.02 | A09:27469147–27471976 (−strand) | 48,712.89 | 9.14 | −0.411 | 440 | |
| BraCIPK23.3 | 73.78 | A08:17294577–17297463 (−strand) | 44,789.61 | 8.68 | −0.28 | 400 | |
| BraCIPK24 | AtCIPK24 | 88.18 | A04:9880111–9882974 (−strand) | 51,600.66 | 9.19 | −0.267 | 453 |
| BraCIPK25 | AtCIPK25 | 70.14 | A06:22263189–22264565 (−strand) | 51,839.79 | 8.68 | −0.306 | 458 |
| BraCIPK26.1 | AtCIPK26 | 90.35 | A10:14277798–14280847 (−strand) | 49,950.04 | 7.63 | −0.502 | 441 |
| BraCIPK26.2 | 88.08 | A02:4468468–4471749 (+strand) | 49,780.72 | 7.65 | −0.516 | 441 |
| Seq 1. | Seq 2 | Identity/% | Ks | Ka | ω | T(MYA) |
|---|---|---|---|---|---|---|
| CIPK1.1 | CIPK1.2 | 85.87 | 0.2033 | 0.0215 | 0.105755042 | 6.7767 |
| CIPK2.2 | CIPK2.3 | 79.05 | 0.5405 | 0.0785 | 0.145235893 | 18.0167 |
| CIPK4.1 | CIPK4.2 | 44.49 | 0.5557 | 0.1255 | 0.225841281 | 18.5233 |
| CIPK5 | CIPK25 | 67.54 | 0.9199 | 0.0712 | 0.077399717 | 30.6633 |
| CIPK6.1 | CIPK6.2 | 90.02 | 0.5716 | 0.018 | 0.031490553 | 19.0533 |
| CIPK7.1 | CIPK7.2 | 86.49 | 0.3183 | 0.0619 | 0.194470625 | 10.6100 |
| CIPK9.1 | CIPK9.2 | 89.49 | 0.3889 | 0.0302 | 0.077654924 | 12.9633 |
| CIPK10.1 | CIPK10.2 | 84.42 | 0.667 | 0.0565 | 0.084707646 | 22.2333 |
| CIPK11.1 | CIPK11.2 | 82.06 | 0.6492 | 0.0437 | 0.067313617 | 21.6400 |
| CIPK12.1 | CIPK12.2 | 90.49 | 0.4008 | 0.0484 | 0.120758483 | 13.3600 |
| CIPK13.2 | CIPK13.3 | 85.69 | 0.6879 | 0.0625 | 0.090856229 | 22.9300 |
| CIPK17.1 | CIPK17.3 | 54.03 | 0.1041 | 0.0042 | 0.040345821 | 3.4700 |
| CIPK20.1 | CIPK20.2 | 66.06 | 0.4802 | 0.0676 | 0.140774677 | 16.0067 |
| CIPK21.1 | CIPK21.2 | 87.74 | 0.2286 | 0.046 | 0.201224847 | 7.6200 |
| CIPK22.1 | CIPK22.2 | 90.23 | 0.6802 | 0.0693 | 0.101881799 | 22.6733 |
| CIPK23.1 | CIPK23.2 | 80.38 | 0.3699 | 0.0085 | 0.022979184 | 12.3300 |
| CIPK26.1 | CIPK26.2 | 86.43 | 0.1264 | 0.0681 | 0.538765823 | 4.2133 |
| CBL1.1 | CBL1.2 | 96.71 | 0.4019 | 0.0092 | 0.022891266 | 13.3967 |
| CBL2.1 | CBL2.2 | 84.62 | 0.4885 | 0.0093 | 0.019038 | 16.2833 |
| CBL3.1 | CBL3.3 | 52.61 | 0.3695 | 0.2117 | 0.572936 | 12.3167 |
| CBL4.1 | CBL4.3 | 92.31 | 0.4213 | 0.0237 | 0.056254 | 14.0433 |
| CBL9.1 | CBL9.2 | 98.59 | 0.3269 | 0.007 | 0.021413 | 10.8967 |
| CBL10.1 | CBL10.3 | 77.11 | 0.3583 | 0.0661 | 0.184482 | 11.9433 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Q.; Zhao, K.; Gong, Y.; Yang, Y.; Yue, Y. Genome-Wide Identification and Functional Analysis of the Calcineurin B-like Protein and Calcineurin B-like Protein-Interacting Protein Kinase Gene Families in Chinese Cabbage (Brassica rapa ssp. pekinensis). Genes 2022, 13, 795. https://doi.org/10.3390/genes13050795
Wang Q, Zhao K, Gong Y, Yang Y, Yue Y. Genome-Wide Identification and Functional Analysis of the Calcineurin B-like Protein and Calcineurin B-like Protein-Interacting Protein Kinase Gene Families in Chinese Cabbage (Brassica rapa ssp. pekinensis). Genes. 2022; 13(5):795. https://doi.org/10.3390/genes13050795
Chicago/Turabian StyleWang, Qianwen, Kai Zhao, Yuqiang Gong, Yunqiang Yang, and Yanling Yue. 2022. "Genome-Wide Identification and Functional Analysis of the Calcineurin B-like Protein and Calcineurin B-like Protein-Interacting Protein Kinase Gene Families in Chinese Cabbage (Brassica rapa ssp. pekinensis)" Genes 13, no. 5: 795. https://doi.org/10.3390/genes13050795
APA StyleWang, Q., Zhao, K., Gong, Y., Yang, Y., & Yue, Y. (2022). Genome-Wide Identification and Functional Analysis of the Calcineurin B-like Protein and Calcineurin B-like Protein-Interacting Protein Kinase Gene Families in Chinese Cabbage (Brassica rapa ssp. pekinensis). Genes, 13(5), 795. https://doi.org/10.3390/genes13050795





