The ABI3 Transcription Factor Interaction and Antagonism with Ubiquitin E3 Ligase ScPRT1 in Syntrichia caninervis
Abstract
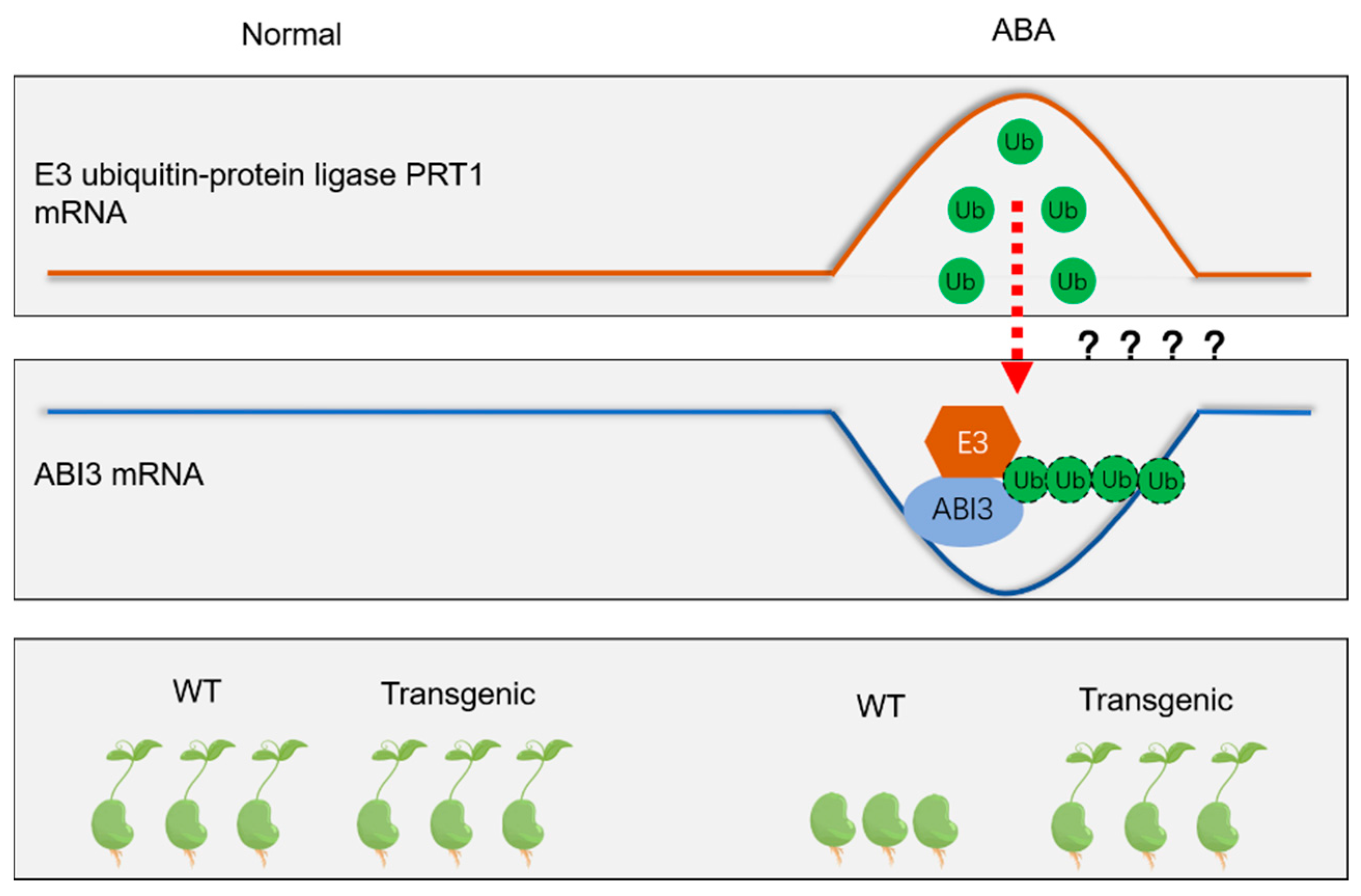
1. Introduction
2. Materials and Methods
2.1. Gene Cloning and Bioinformatics Analysis
2.2. Analysis of ScABI3 Transgenic A. thaliana Germination under ABA Treatment
2.3. Yeast Two-Hybrid Assay
2.4. Pull-Down Assays
2.5. Bimolecular Fluorescence Complementation (BiFC) Assays
2.6. Expression Analysis of ScABI3 and ScPRT1 Genes under Different Abiotic Stress Treatments in S. caninervis
2.7. Statistical Analysis
3. Results
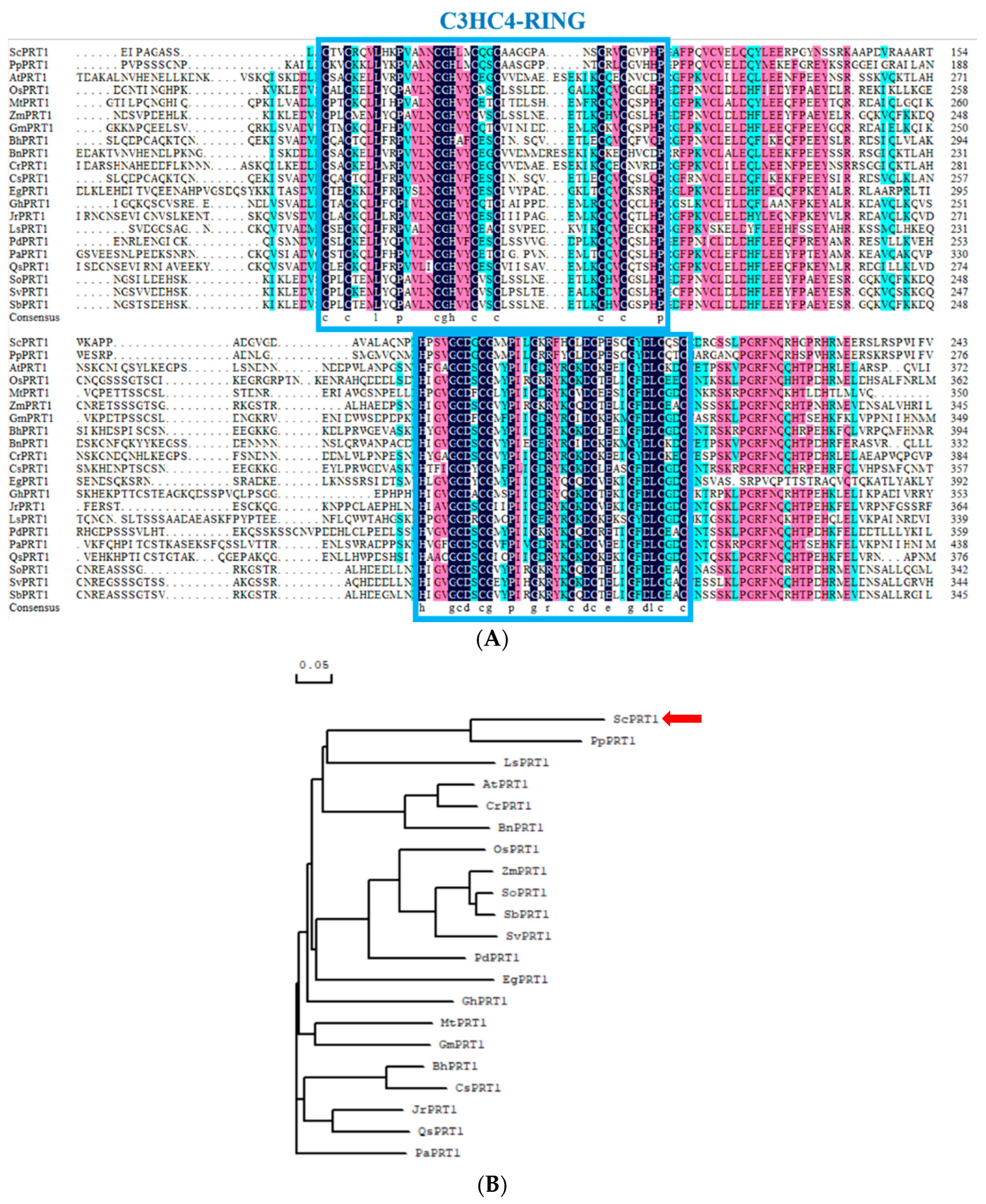
3.1. Multiple Sequence Alignment and Phylogenetic Analysis of ScPRT1
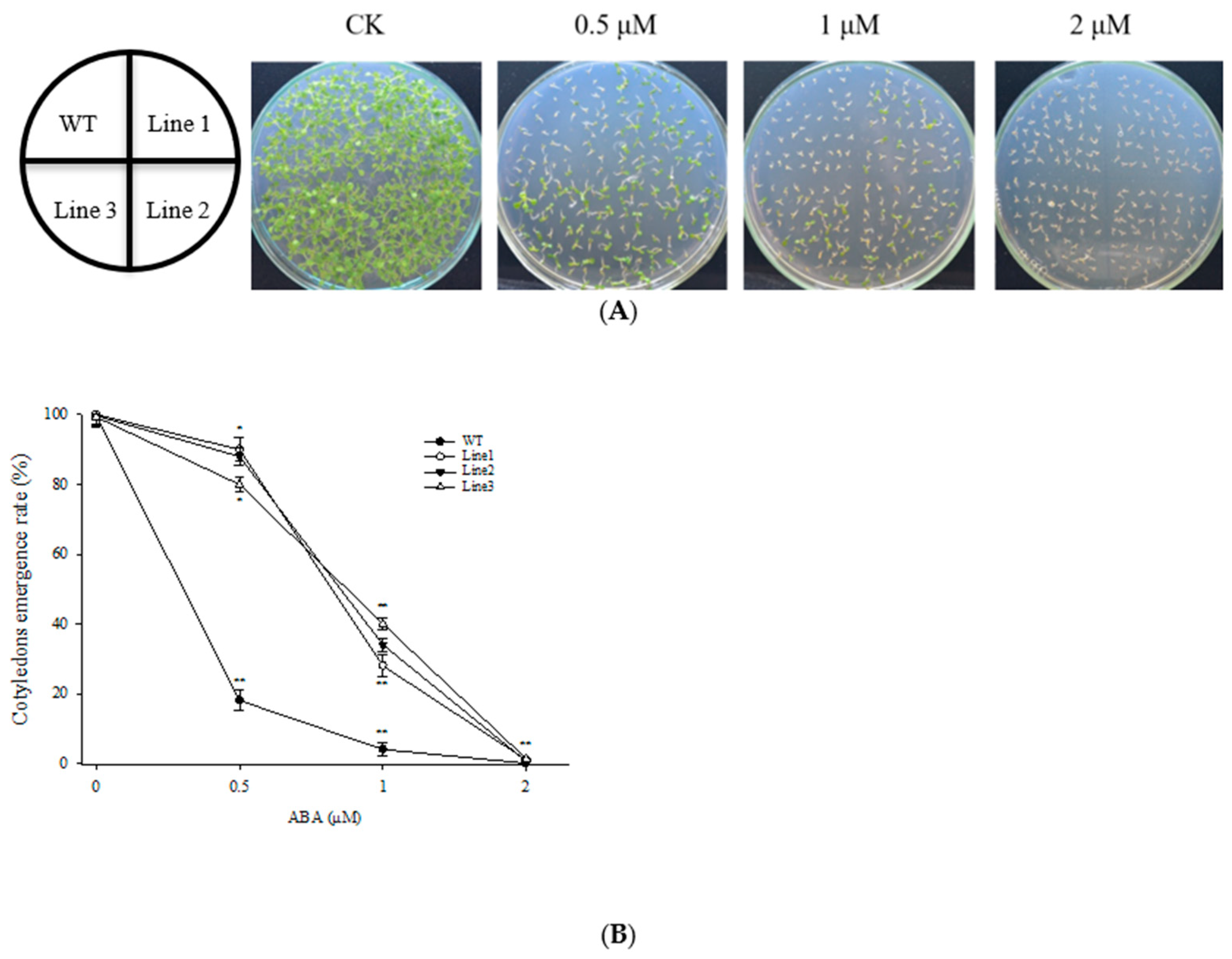
3.2. Overexpression of ScABI3 Reduced the Sensitivity of Seeds under ABA Treatment
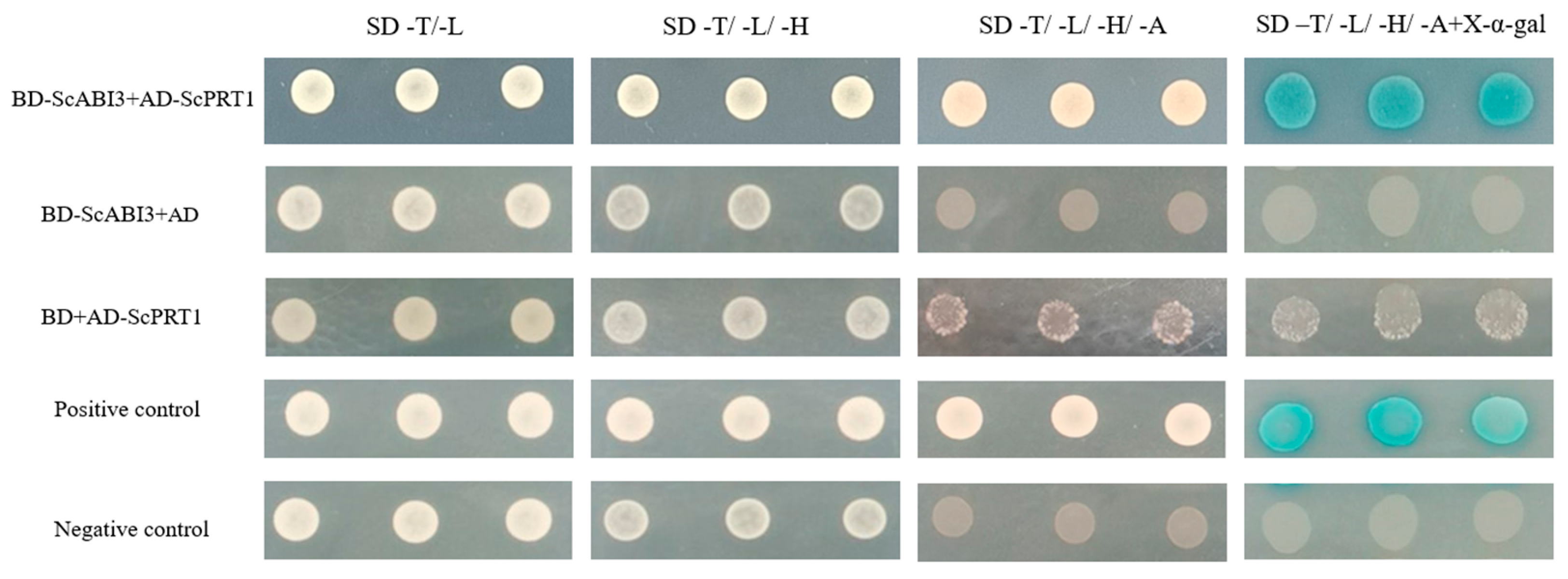
3.3. ScABI3 Interacts with ScPRT1 in Yeast Two-Hybrid Assays
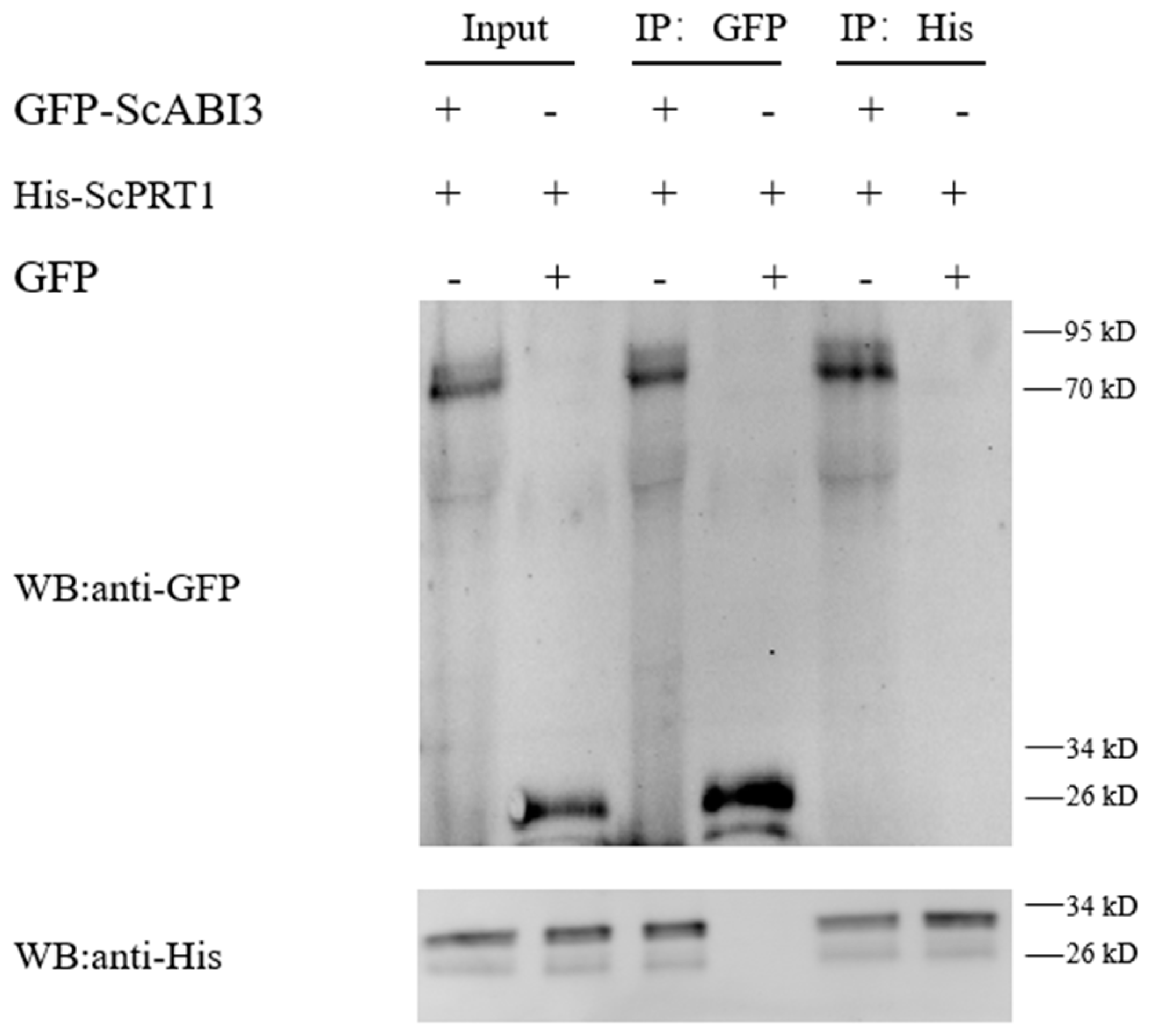
3.4. ScABI3 Interaction with ScPRT1 In Vitro by Pull-Down Assays
3.5. ScABI3 Interaction with ScPRT1 by BiFC in N. benthamiana
3.6. ScABI3 and ScPRT1 Expression Induced by Multiple Abiotic Stresses
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Xu, F.Q.; Xue, H.A.-O. The ubiquitin-proteasome system in plant responses to environments. Plant Cell Environ. 2019, 42, 2931–2944. [Google Scholar] [CrossRef] [PubMed]
- Vierstra, R.D. The ubiquitin-26s proteasome system at the nexus of plant biology. Nat. Rev. Mol. Cell Biol. 2009, 10, 385–397. [Google Scholar] [CrossRef] [PubMed]
- Sadanandom, A.; Bailey, M.; Ewan, R.; Lee, J.; Nelis, S. The ubiquitin-proteasome system: Central modifier of plant signalling. New Phytol. 2012, 196, 13–28. [Google Scholar] [CrossRef] [PubMed]
- Smalle, J.; Vierstra, R.D. The ubiquitin 26s proteasome proteolytic pathway. Annu. Rev. Plant Biol. 2004, 55, 555–590. [Google Scholar] [CrossRef] [PubMed]
- Stone, S.L. Chapter three—Role of the ubiquitin proteasome system in plant response to abiotic stress. In International Review of Cell and Molecular Biology; Galluzzi, L., Ed.; Academic Press: Cambridge, MA, USA, 2019; Volume 343, pp. 65–110. [Google Scholar]
- Zhang, H.; Cui, F.; Wu, Y.; Lou, L.; Liu, L.; Tian, M.; Ning, Y.; Shu, K.; Tang, S.; Xie, Q. The ring finger ubiquitin e3 ligase sdir1 targets sdir1-interacting protein1 for degradation to modulate the salt stress response and aba signaling in arabidopsis. Plant Cell 2015, 27, 214–227. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yang, C.; Li, Y.; Zheng, N.; Chen, H.; Zhao, Q.; Gao, T.; Guo, H.; Xie, Q. Sdir1 is a ring finger e3 ligase that positively regulates stress-responsive abscisic acid signaling in arabidopsis. Plant Cell 2007, 19, 1912–1929. [Google Scholar] [CrossRef]
- Lin, J.-H.; Yu, L.; Xiang, C.-B. Arabidopsis nitrate regulated 1 acts as a negative modulator of seed germination by activating abi3 expression. New Phytol. 2019, 225, 835–847. [Google Scholar] [CrossRef]
- Khandelwal, A.; Cho, S.H.; Marella, H.; Sakata, Y.; Perroud, P.F.; Pan, A.; Quatrano, R.S. Role of aba and abi3 in desiccation tolerance. Science 2010, 327, 546. [Google Scholar] [CrossRef]
- Tomoi, T.; Kawade, K.; Kitagawa, M.; Sakata, Y.; Tsukaya, H.; Fujita, T. Quantitative imaging reveals distinct contributions of snrk2 and abi3 in plasmodesmatal permeability in physcomitrella patens. Plant Cell Physiol. 2020, 61, 942–956. [Google Scholar] [CrossRef]
- Yotsui, I.; Saruhashi, M.; Kawato, T.; Taji, T.; Hayashi, T.; Quatrano, R.; Sakata, Y. Abscisic acid insensitive3 regulates abscissic acid-responsive gene expression with the nuclear factor y complex through the actt-core element in physcomitrella patens. New Phytol. 2013, 199, 101–109. [Google Scholar] [CrossRef]
- Bedi, S.; Nag Chaudhuri, R. Transcription factor abi3 auto-activates its own expression during dehydration stress response. FEBS Lett. 2018, 592, 2594–2611. [Google Scholar] [CrossRef] [PubMed]
- Sengupta, S.; Ray, A.; Mandal, D.; Nag Chaudhuri, R. Abi3 mediated repression of rav1 gene expression promotes efficient dehydration stress response in arabidopsis thaliana. BBA-Gene Regul. Mech. 2020, 1863, 194582. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Garreton, V.F.; Chua, N.-H.; Chua, N.H. The aip2 e3 ligase acts as a novel negative regulator of aba signaling by promoting abi3 degradation. Genes Dev. 2005, 19, 1532–1543. [Google Scholar] [CrossRef] [PubMed]
- Gao, D.-Y.; Xu, Z.-S.; He, Y.; Sun, Y.-W.; Ma, Y.-Z.; Xia, L.-Q. Functional analyses of an e3 ligase gene aip2 from wheat in arabidopsis revealed its roles in seed germination and pre-harvest sprouting. J. Integr. Plant Biol. 2014, 56, 480–491. [Google Scholar] [CrossRef]
- Park, G.-G.; Park, J.-J.; Yoon, J.; Yu, S.-N.; An, G. A ring finger e3 ligase gene, oryza sativa delayed seed germination 1 (osdsg1), controls seed germination and stress responses in rice. Plant Mol. Biol. 2010, 74, 467–478. [Google Scholar] [CrossRef] [PubMed]
- Qin, F.; Sakuma, Y.; Tran, L.-S.; Maruyama, K.; Kidokoro, S.; Fujita, Y.; Fujita, M.; Umezawa, T.; Sawano, Y.; Miyazono, K.-I.; et al. Arabidopsis dreb2a-interacting proteins function as ring e3 ligases and negatively regulate plant drought stress-responsive gene expression. Plant Cell 2008, 20, 1693–1707. [Google Scholar] [CrossRef] [PubMed]
- Duong, S.; Vonapartis, E.; Li, C.-Y.; Patel, S.; Gazzarrini, S. The e3 ligase abi3-interacting protein2 negatively regulates fusca3 and plays a role in cotyledon development in arabidopsis thaliana. J. Exp. Bot. 2017, 68, 1555–1567. [Google Scholar] [CrossRef][Green Version]
- Pan, Z.; Pitt, W.G.; Zhang, Y.; Wu, N.; Tao, Y.; Truscott, T.T. The upside-down water collection system of Syntrichia caninervis. Nat. Plants 2016, 2, 16076. [Google Scholar] [CrossRef]
- Zhang, Y.; Mutailifu, A.; Zhang, Y.; Yang, H.; Zhang, D. Detection of abscisic acid and relative transcript abundance in syntrichia caninervis mitt. J. Bryol. 2021, 43, 376–383. [Google Scholar] [CrossRef]
- Silva, A.T.; Gao, B.; Fisher, K.; Mishler, B.; Ekwealor, J.; Stark, L.; Li, X.-S.; Zhang, D.; Bowker, M.; Brinda, J.; et al. To dry perchance to live: Insights from the genome of the desiccation-tolerant biocrust moss syntrichia caninervis. Plant J. 2020, 105, 1339–1356. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, X.; Zhang, K.; Zhang, D.; Guan, K. An abscisic acid insensitive3-like gene from the desert moss syntrichia caninervis confers abiotic stress tolerance and reduces aba sensitivity. Plant Cell Tissue Organ Cult. 2018, 133, 417–435. [Google Scholar] [CrossRef]
- Yu, F.; Wu, Y.; Xie, Q. Ubiquitin-proteasome system in aba signaling: From perception to action. Mol. Plant 2016, 9, 21–33. [Google Scholar] [CrossRef]
- Bueso, E.; Rodriguez, L.; Lorenzo-Orts, L.; Gonzalez-Guzman, M.; Sayas, E.; Munoz-Bertomeu, J.; Ibanez, C.; Serrano, R.; Rodriguez, P.L. The single-subunit ring-type e3 ubiquitin ligase rsl1 targets pyl4 and pyr1 aba receptors in plasma membrane to modulate abscisic acid signaling. Plant J. 2014, 80, 1057–1071. [Google Scholar] [CrossRef] [PubMed]
- Pan, W.; Lin, B.; Yang, X.; Liu, L.; Xia, R.; Li, J.; Wu, Y.; Xie, Q. The ubc27–airp3 ubiquitination complex modulates aba signaling by promoting the degradation of abi1 in arabidopsis. Proc. Natl. Acad. Sci. USA 2020, 117, 27694–27702. [Google Scholar] [CrossRef] [PubMed]
- Kudla, J.; Bock, R. Lighting the way to protein-protein interactions: Recommendations on best practices for bimolecular fluorescence complementation analyses. Plant Cell 2016, 28, 1002–1008. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhang, D.; Li, H.; Gao, B.; Yang, H.; Zhang, Y.; Wood, A.J. Characterization of reference genes for rt-qpcr in the desert moss syntrichia caninervis in response to abiotic stress and desiccation/rehydration. Front. Plant Sci. 2015, 6, 38. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative pcr and the 2−δδct method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Raghavendra, A.S.; Gonugunta, V.K.; Christmann, A.; Grill, E. Aba perception and signalling. Trends Plant Sci. 2010, 15, 395–401. [Google Scholar] [CrossRef]
- Stone, S.; Williams, L.; Farmer, L.; Vierstra, R.; Callis, J. Keep on going, a ring e3 ligase essential for arabidopsis growth and development, is involved in abscisic acid signaling. Plant Cell 2007, 18, 3415–3428. [Google Scholar] [CrossRef]
- Wu, Q.; Zhang, X.; Peirats-Llobet, M.; Belda-Palazon, B.; Wang, X.; Cui, S.; Yu, X.A.-O.; Rodriguez, P.A.-O.; An, C. Ubiquitin ligases rglg1 and rglg5 regulate abscisic acid signaling by controlling the turnover of phosphatase pp2ca. Plant Cell 2016, 28, 2178–2196. [Google Scholar] [CrossRef]
- Cheng, C.; Wang, Z.; Ren, Z.; Zhi, L.; Yao, B.; Chao, S.; Liu, L.; Li, X. Scfatpp2-b11 modulates aba signaling by facilitating snrk2.3 degradation in arabidopsis thaliana. PLoS Genet. 2017, 13, e1006947. [Google Scholar] [CrossRef] [PubMed]
- Kong, L.; Cheng, J.; Zhu, Y.; Ding, Y.; Meng, J.; Chen, Z.; Xie, Q.; Guo, Y.; Li, J.; Yang, S.; et al. Degradation of the aba co-receptor abi1 by pub12/13 u-box e3 ligases. Nat. Commun. 2015, 6, 8630. [Google Scholar] [CrossRef] [PubMed]
- Seo, J.S.; Zhao, P.; Jung, C.; Chua, N.H. Plant u-box protein 10 negatively regulates abscisic acid response in arabidopsis. Appl. Biol. Chem. 2019, 62, 39. [Google Scholar] [CrossRef]
- Girod, P.-A.; Fu, H.; Zrÿd, J.-P.; Vierstra, R. Multiubiquitin chain binding subunit mcb1 (rpn10) of the 26s proteasome is essential for developmental progression in physcomitrella patens. Plant Cell 1999, 11, 1457–1472. [Google Scholar] [CrossRef] [PubMed]
- Ranjan, A.; Dickopf, S.; Ullrich, K.K.; Rensing, S.A.; Hoecker, U. Functional analysis of cop1 and spa orthologs from physcomitrella and rice during photomorphogenesis of transgenic arabidopsis reveals distinct evolutionary conservation. BMC Plant Biol. 2014, 14, 178. [Google Scholar] [CrossRef]
- Wang, J.; Liu, S.; Liu, H.; Chen, K.; Zhang, P. Pnsag1, an e3 ubiquitin ligase of the antarctic moss pohlia nutans, enhanced sensitivity to salt stress and aba. Plant Physiol. Bioch. 2019, 141, 343–352. [Google Scholar] [CrossRef]
- Guillory, A.; Bonhomme, S. Phytohormone biosynthesis and signaling pathways of mosses. Plant Mol. Biol. 2021, 107, 245–277. [Google Scholar] [CrossRef]
- Holdsworth, M.; Vicente, J.; Sharma, G.; Abbas, M.; Zubrycka, A. The plant n-degron pathways of ubiquitin-mediated proteolysis. J. Integr. Plant Biol. 2019, 62, 70–89. [Google Scholar] [CrossRef]


| Gene | Forward Primer | Reverse Primer | Annealing Temperature |
|---|---|---|---|
| ScABI3 | GGTACTTCATCGTTCTGG | GTCACCGTCTAATCTCTG | 60 °C |
| ScPRT1 | GGAGCGCGACCAGAACAACTAC | CATCAGGTGGCCGCAGTTCATC | 59 °C |
| Scα-TUB | CGGTCATTACACCGTGGGAA | CCTCTCCAGCAACAGCGAA | 60 °C |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Zhou, J.; Zhang, Y.; Zhang, D. The ABI3 Transcription Factor Interaction and Antagonism with Ubiquitin E3 Ligase ScPRT1 in Syntrichia caninervis. Genes 2022, 13, 718. https://doi.org/10.3390/genes13050718
Zhang Y, Zhou J, Zhang Y, Zhang D. The ABI3 Transcription Factor Interaction and Antagonism with Ubiquitin E3 Ligase ScPRT1 in Syntrichia caninervis. Genes. 2022; 13(5):718. https://doi.org/10.3390/genes13050718
Chicago/Turabian StyleZhang, Yigong, Jiyang Zhou, Yi Zhang, and Daoyuan Zhang. 2022. "The ABI3 Transcription Factor Interaction and Antagonism with Ubiquitin E3 Ligase ScPRT1 in Syntrichia caninervis" Genes 13, no. 5: 718. https://doi.org/10.3390/genes13050718
APA StyleZhang, Y., Zhou, J., Zhang, Y., & Zhang, D. (2022). The ABI3 Transcription Factor Interaction and Antagonism with Ubiquitin E3 Ligase ScPRT1 in Syntrichia caninervis. Genes, 13(5), 718. https://doi.org/10.3390/genes13050718





