Association of Androgenic Regulation and MicroRNAs in Acinar Adenocarcinoma of Prostate
Abstract
1. Introduction
2. Materials and Methods
2.1. Tissue Samples
2.2. Western Blotting
2.3. Relative Quantification of MicroRNA Expression
2.4. Statistical Analyses
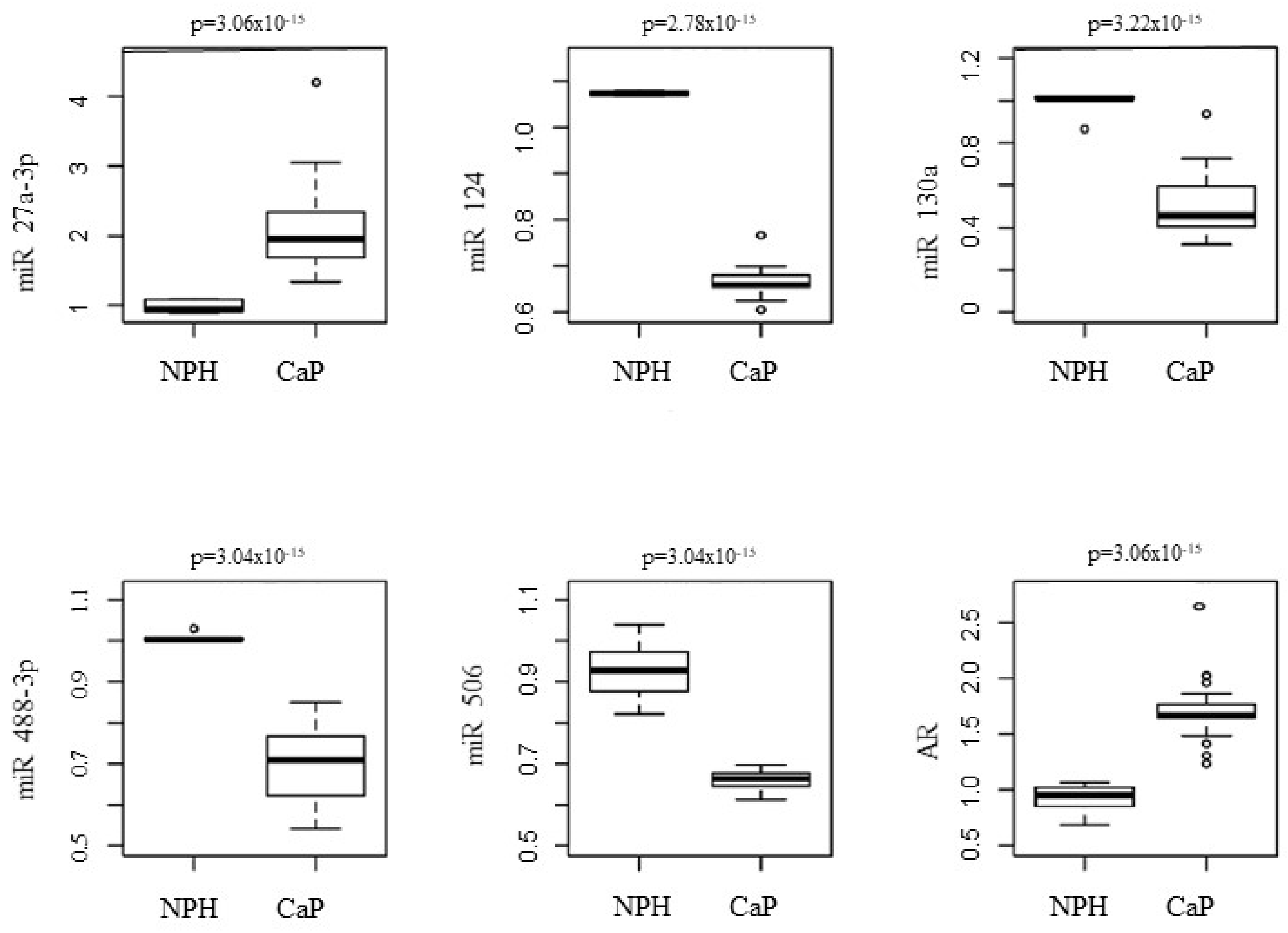
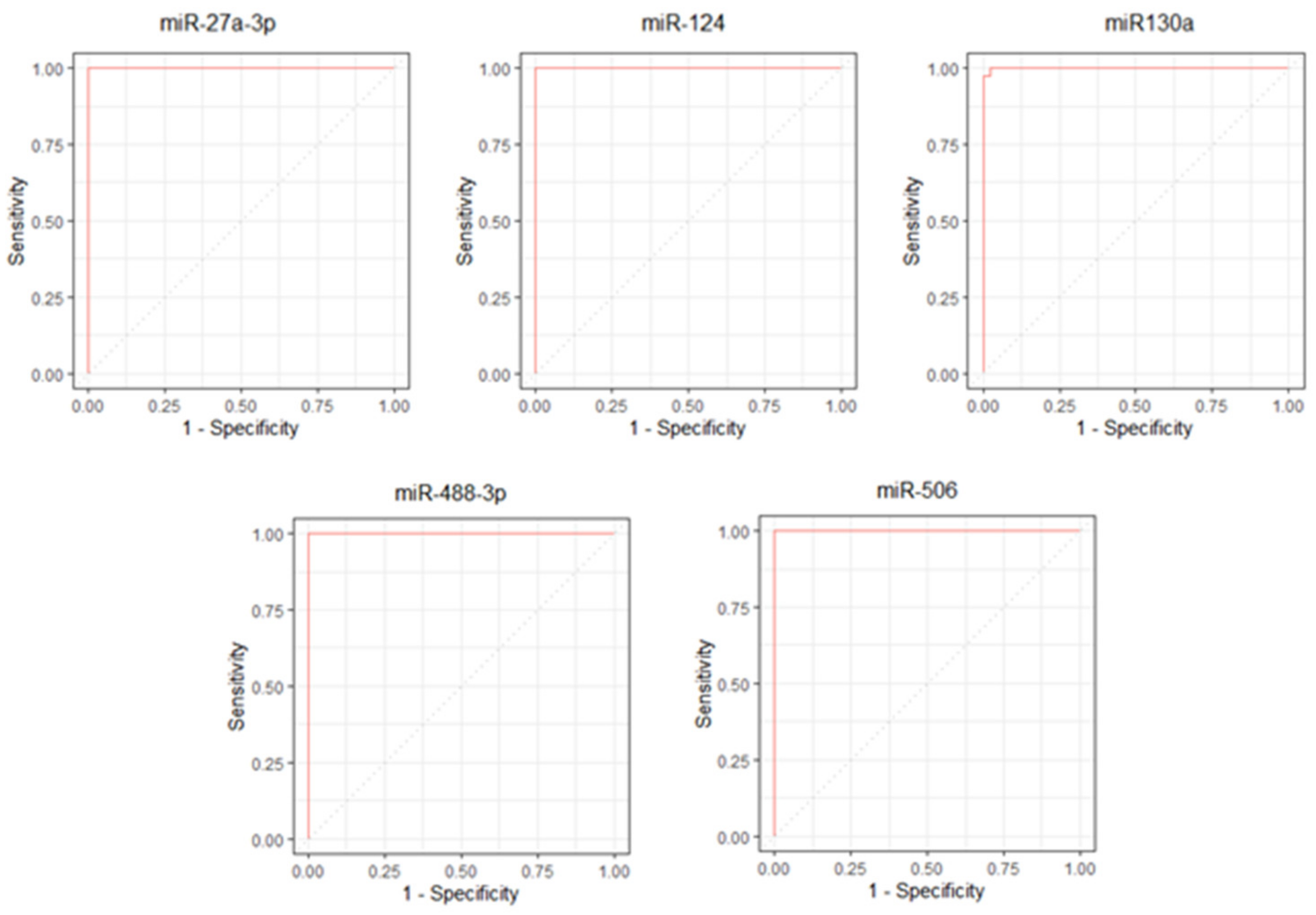
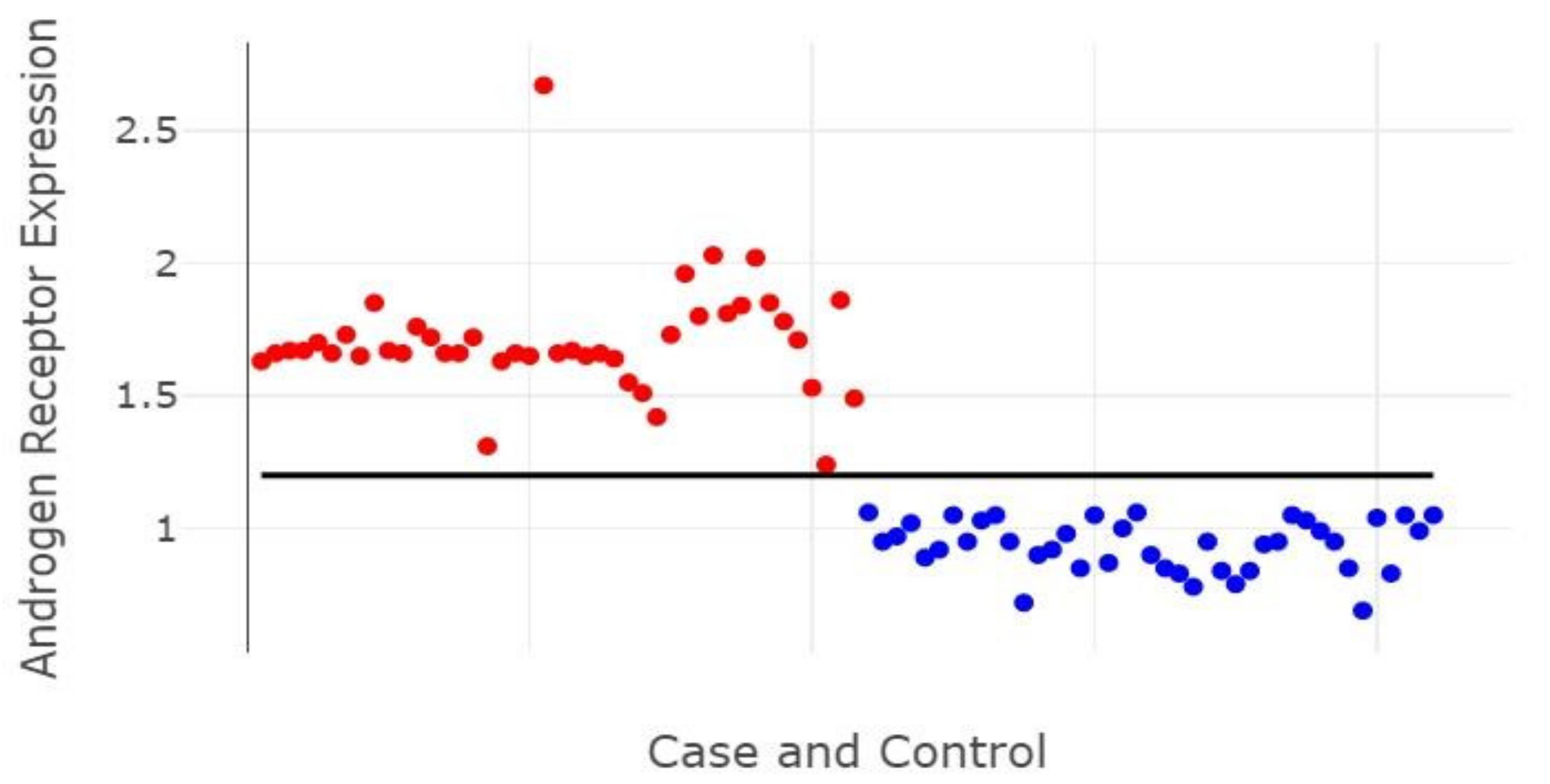
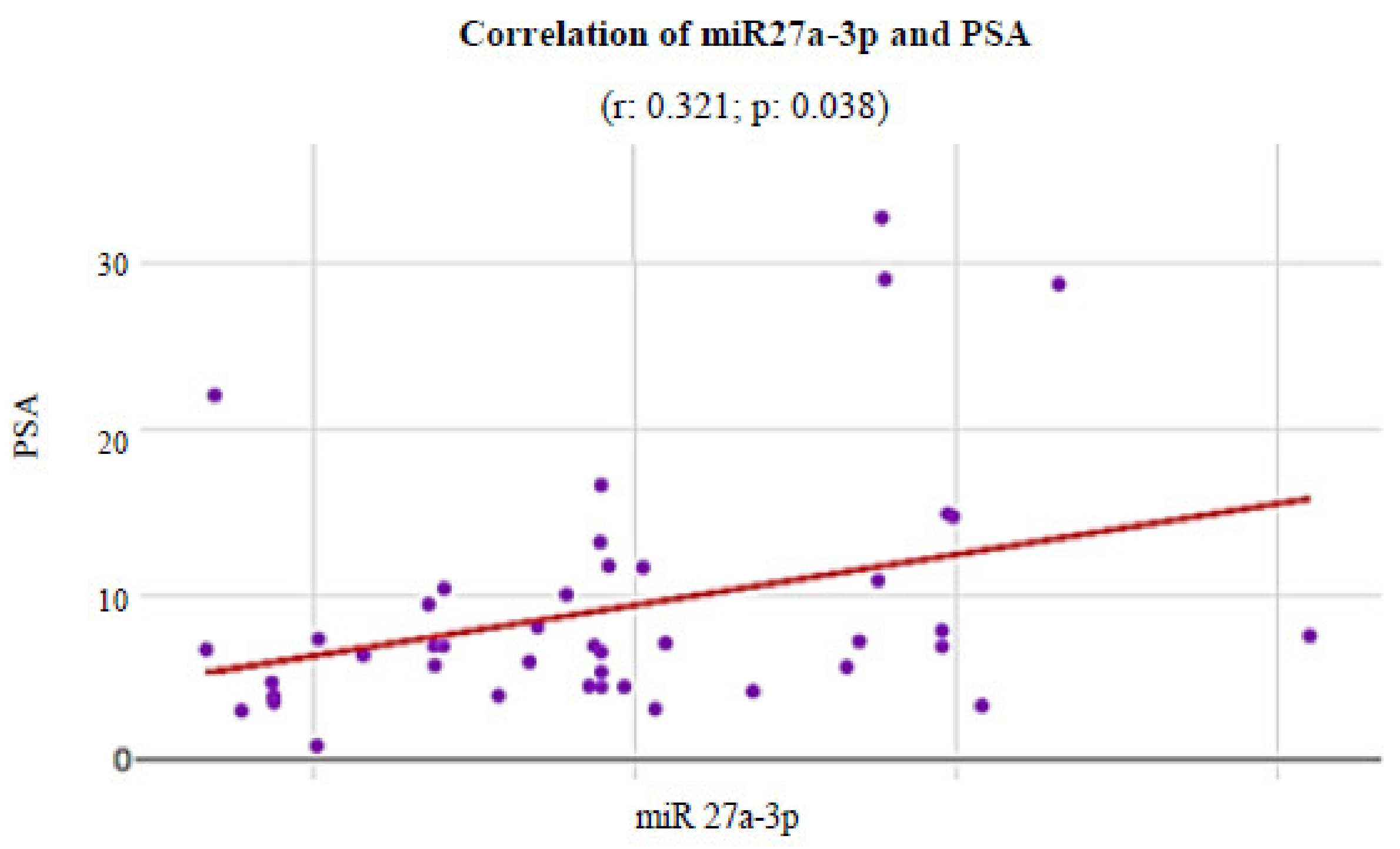
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA. Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef]
- Steffen, R.E.; Trajman, A.; Santos, M.; Caetano, R. Population Screening for Prostate Cancer: More Risks than Benefits. Physis 2018, 28, 1–12. [Google Scholar] [CrossRef]
- Filella, X.; Foj, L. Novel Biomarkers for Prostate Cancer Detection and Prognosis. Adv. Exp. Med. Biol. 2018, 1095, 15–39. [Google Scholar] [CrossRef]
- D’Amico, A.V.; Whittington, R.; Bruce Malkowicz, S.; Schultz, D.; Blank, K.; Broderick, G.A.; Tomaszewski, J.E.; Renshaw, A.A.; Kaplan, I.; Beard, C.J.; et al. Biochemical Outcome after Radical Prostatectomy, External Beam Radiation Therapy, or Interstitial Radiation Therapy for Clinically Localized Prostate Cancer. J. Am. Med. Assoc. 1998, 280, 969–974. [Google Scholar] [CrossRef] [PubMed]
- Cozar, J.M.; Robles-Fernandez, I.; Rodriguez-Martinez, A.; Puche-Sanz, I.; Vazquez-Alonso, F.; Lorente, J.A.; Martinez-Gonzalez, L.J.; Alvarez-Cubero, M.J. The Role of MiRNAs as Biomarkers in Prostate Cancer. Mutat. Res. Mutat. Res. 2019, 781, 165–174. [Google Scholar] [CrossRef] [PubMed]
- Bidarra, D.; Constâncio, V.; Barros-Silva, D.; Ramalho-Carvalho, J.; Moreira-Barbosa, C.; Antunes, L.; Maurício, J.; Oliveira, J.; Henrique, R.; Jerónimo, C. Circulating Micro RNAs as Biomarkers for Prostate Cancer Detection and Metastasis Development Prediction. Front. Oncol. 2019, 9, 900. [Google Scholar] [CrossRef]
- Chua, F.Y.; Adams, B.D. Androgen Receptor and MiR-206 Regulation in Prostate Cancer. Transcription 2017, 8, 313–327. [Google Scholar] [CrossRef] [PubMed]
- Pacult, J.; Rams-Baron, M.; Chmiel, K.; Jurkiewicz, K.; Antosik, A.; Szafraniec, J.; Kurek, M.; Jachowicz, R.; Paluch, M. How Can We Improve the Physical Stability of Co-Amorphous System Containing Flutamide and Bicalutamide? The Case of Ternary Amorphous Solid Dispersions. Eur. J. Pharm. Sci. 2019, 136, 1–9. [Google Scholar] [CrossRef]
- Balacescu, O.; Dumitrescu, R.G.; Marian, C. MicroRNAs Role in Prostate Cancer. Methods Mol. Biol. 2018, 1856, 103–117. [Google Scholar] [CrossRef]
- Fabris, L.; Ceder, Y.; Chinnaiyan, A.M.; Jenster, G.W.; Sorensen, K.D.; Tomlins, S.; Visakorpi, T.; Calin, G.A. The Potential of MicroRNAs as Prostate Cancer Biomarkers. Eur. Urol. 2016, 70, 312–322. [Google Scholar] [CrossRef]
- Kanwal, R.; Plaga, A.R.; Liu, X.; Shukla, G.C.; Gupta, S. MicroRNAs in Prostate Cancer: Functional Role as Biomarkers. Cancer Lett. 2017, 407, 9–20. [Google Scholar] [CrossRef] [PubMed]
- Fletcher, C.E.; Sulpice, E.; Combe, S.; Shibakawa, A.; Leach, D.A.; Hamilton, M.P.; Chrysostomou, S.L.; Sharp, A.; Welti, J.; Yuan, W.; et al. Androgen Receptor-Modulatory MicroRNAs Provide Insight into Therapy Resistance and Therapeutic Targets in Advanced Prostate Cancer. Oncogene 2019, 38, 5700–5724. [Google Scholar] [CrossRef] [PubMed]
- Recouvreux, M.V.; Wu, J.B.; Gao, A.C.; Zonis, S.; Chesnokova, V.; Bhowmick, N.; Chung, L.W.; Melmed, S. Androgen Receptor Regulation of Local Growth Hormone in Prostate Cancer Cells. Endocrinology 2017, 158, 2255–2268. [Google Scholar] [CrossRef] [PubMed]
- Gao, W.; Hong, Z.; Huang, H.; Zhu, A.; Lin, S.; Cheng, C.; Zhang, X.; Zou, G.; Shi, Z. MiR-27a in Serum Acts as Biomarker for Prostate Cancer Detection and Promotes Cell Proliferation by Targeting Sprouty2. Oncol. Lett. 2018, 16, 5291–5298. [Google Scholar] [CrossRef] [PubMed]
- Barros-Silva, D.; Costa-Pinheiro, P.; Duarte, H.; Sousa, E.J.; Evangelista, A.F.; Graça, I.; Carneiro, I.; Martins, A.T.; Oliveira, J.; Carvalho, A.L.; et al. MicroRNA-27a-5p Regulation by Promoter Methylation and MYC Signaling in Prostate Carcinogenesis. Cell Death Dis. 2018, 9, 1–15. [Google Scholar] [CrossRef]
- Moustafa, A.A.; Kim, H.; Albeltagy, R.S.; El-Habit, O.H.; Abdel-Mageed, A.B. MicroRNAs in Prostate Cancer: From Function to Biomarker Discovery. Exp. Biol. Med. 2018, 243, 817–825. [Google Scholar] [CrossRef] [PubMed]
- Boll, K.; Reiche, K.; Kasack, K.; Mörbt, N.; Kretzschmar, A.K.; Tomm, J.M.; Verhaegh, G.; Schalken, J.; Von Bergen, M.; Horn, F.; et al. MiR-130a, MiR-203 and MiR-205 Jointly Repress Key Oncogenic Pathways and Are Downregulated in Prostate Carcinoma. Oncogene 2013, 32, 277–285. [Google Scholar] [CrossRef]
- Arora, H.; Qureshi, R.; Park, W. miR-506 regulates epithelial mesenchymal transition in breast cancer cell lines. PLoS ONE 2013, 8, e64273. [Google Scholar] [CrossRef]
- Das, V.; Bhattacharya, S.; Chikkaputtaiah, C.; Hazra, S.; Pal, M. The basics of epithelial–mesenchymal transition (EMT): A study from a structure, dynamics, and functional perspective. J. Cell. Physiol. 2019, 234, 14535–14555. [Google Scholar] [CrossRef] [PubMed]
- Pena, S.D.J.; di Pietro, G.; Fuchshuber-Moraes, M.; Genro, J.P.; Hutz, M.H.; Kehdy, F.D.S.G.; Kohlrausch, F.; Magno, L.A.V.; Montenegro, R.C.; Moraes, M.O.; et al. The Genomic Ancestry of Individuals from Different Geographical Regions of Brazil Is More Uniform than Expected. PLoS ONE 2011, 6, 1–9. [Google Scholar] [CrossRef]
- Suarez-Kurtz, G.; Paula, D.P.; Struchiner, C.J. Pharmacogenomic Implications of Population Admixture: Brazil as a Model Case. Pharmacogenomics 2014, 15, 209–219. [Google Scholar] [CrossRef] [PubMed]
- Epstein, J.I.; Egevad, L.; Amin, M.B.; Delahunt, B.; Srigley, J.R.; Humphrey, P.A. The 2014 International Society of Urological Pathology (ISUP) Consensus Conference on Gleason Grading of Prostatic Carcinoma Definition of Grading Patterns and Proposal for a New Grading System. Am. J. Surg. Pathol. 2016, 40, 244–252. [Google Scholar] [CrossRef]
- D’Amico, A.V. Prostate-Specific Antigen (PSA) and PSA Velocity: Competitors or Collaborators in the Prediction of Curable and Clinically Significant Prostate Cancer. J. Clin. Oncol. 2008, 26, 823–824. [Google Scholar] [CrossRef]
- Leal, M.F.; Mazzotti, T.K.F.; Calcagno, D.Q.; Cirilo, P.D.R.; Martinez, M.C.; Demachki, S.; Assumpção, P.P.; Chammas, R.; Burbano, R.R.; Smith, M.C. Deregulated Expression of Nucleophosmin 1 in Gastric Cancer and Its Clinicopathological Implications. BMC Gastroenterol. 2014, 14, 1–10. [Google Scholar] [CrossRef]
- Pradervand, S.; Weber, J.; Thomas, J.; Bueno, M.; Wirapati, P.; Lefort, K.; Dotto, G.P.; Harshman, K. Impact of Normalization on MiRNA Microarray Expression Profiling. RNA 2009, 15, 493–501. [Google Scholar] [CrossRef] [PubMed]
- Schmittgen, T.D.; Livak, K.J. Analyzing Real-Time PCR Data by the Comparative CT Method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
- Leal, M.F.; Ribeiro, H.F.; Rey, J.A.; Pinto, G.R.; Smith, M.C.; Moreira-Nunes, C.A.; Assumpção, P.P.; Lamarão, L.M.; Calcagno, D.Q.; Montenegro, R.C.; et al. YWHAE Silencing Induces Cell Proliferation, Invasion and Migration through the up-Regulation of CDC25B and MYC in Gastric Cancer Cells: New Insights about YWHAE Role in the Tumor Development and Metastasis Process. Oncotarget 2016, 7, 85393–85410. [Google Scholar] [CrossRef]
- Logozzi, M.; Angelini, D.F.; Giuliani, A.; Mizzoni, D.; Raimo, R.D.; Maggi, M.; Gentilucci, A.; Marzio, V.; Salciccia, S.; Borsellino, G.; et al. Increased Plasmatic Levels of Psa-Expressing Exosomes Distinguish Prostate Cancer Patients from Benign Prostatic Hyperplasia: A Prospective Study. Cancers 2019, 11, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Strand, S.H.; Bavafaye-Haghighi, E.; Kristensen, H.; Rasmussen, A.K.; Hoyer, S.; Borre, M.; Mouritzen, P.; Besenbacher, S.; Orntoft, T.F.; Sorensen, K.D. A Novel Combined MiRNA and Methylation Marker Panel (MiMe) for Prediction of Prostate Cancer Outcome after Radical Prostatectomy. Int. J. Cancer 2019, 145, 3445–3452. [Google Scholar] [CrossRef]
- Lyu, J.; Zhao, L.; Wang, F.; Ji, J.; Cao, Z.; Xu, H.; Shi, X.; Zhu, Y.; Zhang, C.; Guo, F.; et al. Discovery and Validation of Serum MicroRNAs as Early Diagnostic Biomarkers for Prostate Cancer in Chinese Population. Biomed Res. Int. 2019, 2019, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Karsh, L.I.; Nissenblatt, M.J.; Canfield, S.E. Androgen Receptor Splice Variant, AR-V7, as a Biomarker of Resistance to Androgen Axis-Targeted Therapies in Advanced Prostate Cancer. Clin. Genitourin. Cancer 2020, 18, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Altschuler, J.; Stockert, J.A.; Kyprianou, N. Non-Coding Rnas Set a New Phenotypic Frontier in Prostate Cancer Metastasis and Resistance. Int. J. Mol. Sci. 2021, 22, 1–27. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.S.; Liu, L.X.; Li, G.P.; Chen, Y.; Li, C.Y.; Jin, D.Y.; Wang, X.L. Combined Serum CA19-9 and MiR-27a-3p in Peripheral Blood Mononuclear Cells to Diagnose Pancreatic Cancer. Cancer Prev. Res. 2013, 6, 331–338. [Google Scholar] [CrossRef] [PubMed]
- Nam, R.K.; Wallis, C.J.D.; Amemiya, Y.; Benatar, T.; Seth, A. Identification of a Novel MicroRNA Panel Associated with Metastasis Following Radical Prostatectomy for Prostate Cancer. Anticancer Res. 2018, 38, 5027–5034. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Huang, W.; Chen, B.; Bai, P.D.; Wang, X.G.; Xing, J.C. Up-Regulation of MiR-124 Inhibits Invasion and Proliferation of Prostate Cancer Cells through Mediating JAK-STAT3 Signaling Pathway. Eur. Rev. Med. Pharmacol. Sci. 2017, 21, 2338–2345. [Google Scholar] [PubMed]
- Zhang, W.; Mao, Y.Q.; Wang, H.; Yin, W.J.; Zhu, S.X.; Wang, W.C. MiR-124 Suppresses Cell Motility and Adhesion by Targeting Talin 1 in Prostate Cancer Cells. Cancer Cell Int. 2015, 15, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.B.; Xue, L.; Ma, A.H.; Tepper, C.G.; Gandour-Edwards, R.; Kung, H.J.; Devere White, R.W. Tumor Suppressive MiR-124 Targets Androgen Receptor and Inhibits Proliferation of Prostate Cancer Cells. Oncogene 2013, 32, 4130–4138. [Google Scholar] [CrossRef] [PubMed]
- Chu, M.; Chang, Y.; Guo, Y.; Wang, N.; Cui, J.; Gao, W.Q. Regulation and Methylation of Tumor Suppressor MiR-124 by Androgen Receptor in Prostate Cancer Cells. PLoS ONE 2015, 10, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Ramalho-Carvalho, J.; Martins, J.B.; Cekaite, L.; Sveen, A.; Torres-Ferreira, J.; Graça, I.; Costa-Pinheiro, P.; Eilertsen, I.A.; Antunes, L.; Oliveira, J.; et al. Epigenetic Disruption of MiR-130a Promotes Prostate Cancer by Targeting SEC23B and DEPDC1. Cancer Lett. 2017, 385, 150–159. [Google Scholar] [CrossRef]
- Fujita, Y.; Kojima, T.; Kawakami, K.; Mizutani, K.; Kato, T.; Deguchi, T.; Ito, M. MIR-130a Activates Apoptotic Signaling through Activation of Caspase-8 in Taxane-Resistant Prostate Cancer Cells. Prostate 2015, 75, 1568–1578. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.D.; Jiang, L.H.; Sun, D.W.; Li, J.; Ji, Z.L. The Role of MiR-130a in Cancer. Breast Cancer 2017, 24, 521–527. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Lu, G.; Ke, X.; Lu, X.; Wang, X.; Li, H.; Ren, M.; He, S. MiR-488 Acts as a Tumor Suppressor Gene in Gastric Cancer. Tumor Biol. 2016, 37, 8691–8698. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Zou, X.; Zou, J.; Zhang, G. A Review of Recent Research on the Role of Micrornas in Renal Cancer. Med. Sci. Monit. 2021, 27, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Chi, Y.; Wang, D.; Wang, J.; Yu, W.; Yang, J. Long Non-Coding RNA in the Pathogenesis of Cancers. Cells 2019, 8, 1–44. [Google Scholar] [CrossRef]
- Arnold, J.; Engelmann, J.C.; Schneider, N.; Bosserhoff, A.K.; Kuphal, S. MiR-488-5p and Its Role in Melanoma. Exp. Mol. Pathol. 2020, 112, 1–18. [Google Scholar] [CrossRef]
- Wang, J.; Li, X.; Xiao, Z.; Wang, Y.; Han, Y.; Li, J.; Zhu, W.; Leng, Q.; Wen, Y.; Wen, X. MicroRNA-488 Inhibits Proliferation and Glycolysis in Human Prostate Cancer Cells by Regulating PFKFB3. FEBS Open Bio 2019, 9, 1798–1807. [Google Scholar] [CrossRef]
- Hu, C.Y.; You, P.; Zhang, J.; Zhang, H.; Jiang, N. MiR-506-3p Acts as a Novel Tumor Suppressor in Prostate Cancer through Targeting GALNT4. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 5133–5138. [Google Scholar] [CrossRef]
- Nikolić, Z.Z.; Pavićević, D.L.S.; Romac, S.P.; Brajušković, G.N. Genetic Variants within Endothelial Nitric Oxide Synthase Gene and Prostate Cancer: A Meta-Analysis. Clin. Transl. Sci. 2015, 8, 23–31. [Google Scholar] [CrossRef][Green Version]
- Fletcher, C.E.; Dart, D.A.; Sita-lumsden, A.; Cheng, H.; Rennie, P.S.; Bevan, C.L. Androgen-Regulated Processing of the Oncomir MiR-27a, Which Targets Prohibitin in Prostate Cancer. Hum. Mol. Genet. 2012, 21, 3112–3127. [Google Scholar] [CrossRef]
- Park, J.-L.; Kim, M.; Song, K.-S.; Kim, S.-Y.; Kim, Y.S. Cell-Free MiR-27a, a Potential Diagnostic and Prognostic Biomarker for Gastric Cancer. Genom. Inform. 2015, 13, 70. [Google Scholar] [CrossRef]
- Wang, G.; Zhao, D.; Spring, D.J.; Depinho, R.A. Genetics and Biology of Prostate Cancer. Genes Dev. 2018, 32, 1105–1140. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Qian, K.; Wei, X.; Deng, H.; Zhao, B.; Chen, Q.; Zhang, J.; Liu, H. MiR-27a Promotes Proliferation, Migration, and Invasion of Colorectal Cancer by Targeting FAM172A and Acts as a Diagnostic and Prognostic Biomarker. Oncol. Rep. 2017, 37, 3554–3564. [Google Scholar] [CrossRef]
- Yan, X.; Yu, H.; Liu, Y.; Hou, J.; Yang, Q.; Zhao, Y. MiR-27a-3p Functions as a Tumor Suppressor and Regulates Non-Small Cell Lung Cancer Cell Proliferation via Targeting HOXB8. Technol. Cancer Res. Treat. 2019, 18, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Wan, X.; Huang, W.; Yang, S.; Zhang, Y.; Zhang, P.; Kong, Z.; Li, T.; Wu, H.; Jing, F.; Li, Y. Androgen-Induced MiR-27A Acted as a Tumor Suppressor by Targeting MAP2K4 and Mediated Prostate Cancer Progression. Int. J. Biochem. Cell Biol. 2016, 79, 249–260. [Google Scholar] [CrossRef]
- Waltering, K.K.; Porkka, K.P.; Jalava, S.E.; Urbanucci, A.; Kohonen, P.J.; Latonen, L.M.; Kallioniemi, O.P.; Jenster, G.; Visakorpi, T. Androgen Regulation of Micro-RNAs in Prostate Cancer. Prostate 2011, 71, 604–614. [Google Scholar] [CrossRef]
- Conteduca, V.; Gurioli, G.; Brighi, N.; Lolli, C.; Schepisi, G.; Casadei, C.; Burgio, S.L.; Gargiulo, S.; Ravaglia, G.; Rossi, L.; et al. Plasma Androgen Receptor in Prostate Cancer. Cancers 2019, 11, 1–15. [Google Scholar] [CrossRef]
- ShafiAA, S.; AE, Y.; NL, W. Androgen Receptors in Hormone-Dependent and Castration-Resistant Prostate Cancer. Pharmacol. Ther. 2013, 140, 223–238. [Google Scholar] [CrossRef]
- Davies, A.; Conteduca, V.; Zoubeidi, A.; Beltran, H. Biological Evolution of Castration-Resistant Prostate Cancer. Eur. Urol. Focus 2019, 5, 147–154. [Google Scholar] [CrossRef] [PubMed]
- Billis, A.; Magna, L.A.; Watanabe, I.C.; Costa, M.V.; Telles, G.H.; Ferreira, U. Are prostate carcinoma clinical stages T1c and T2 similar? Int. Braz. 2006, 32, 165–171. [Google Scholar] [CrossRef]
- Cambruzzi, E.; Zettler, C.G.; Pegas, K.L.; Teixeira, S.L. Relação entre escore de Gleason e fatoresprognósticos no adenocarcinoma acinar de próstata. J. Bras. Patol. Med. Lab. 2010, 46, 61–68. [Google Scholar] [CrossRef]
| miR | Score D’Amico μ (h) | p Value * | ||||
|---|---|---|---|---|---|---|
| High (n = 5) | Intermediate (n = 24) | Low (n = 14) | H-L a | H-I b | L-I c | |
| 27a-3p | 2.48 (1.08–3.87) | 1.99 (1.87–2.10) | 1.89 (1.51–2.27) | 0.395 | 0.326 | 0.241 |
| 124 | 0.65 (0.57–0.74) | 0.67 (0.65–0.68) | 0.67 (0.63–0.69) | 0.272 | 0.319 | 0.540 |
| 130a | 0.45 (0.28–0.61) | 0.52 (0.45–0.58) | 0.45 (0.41–0.49) | 0.864 | 0.386 | 0.455 |
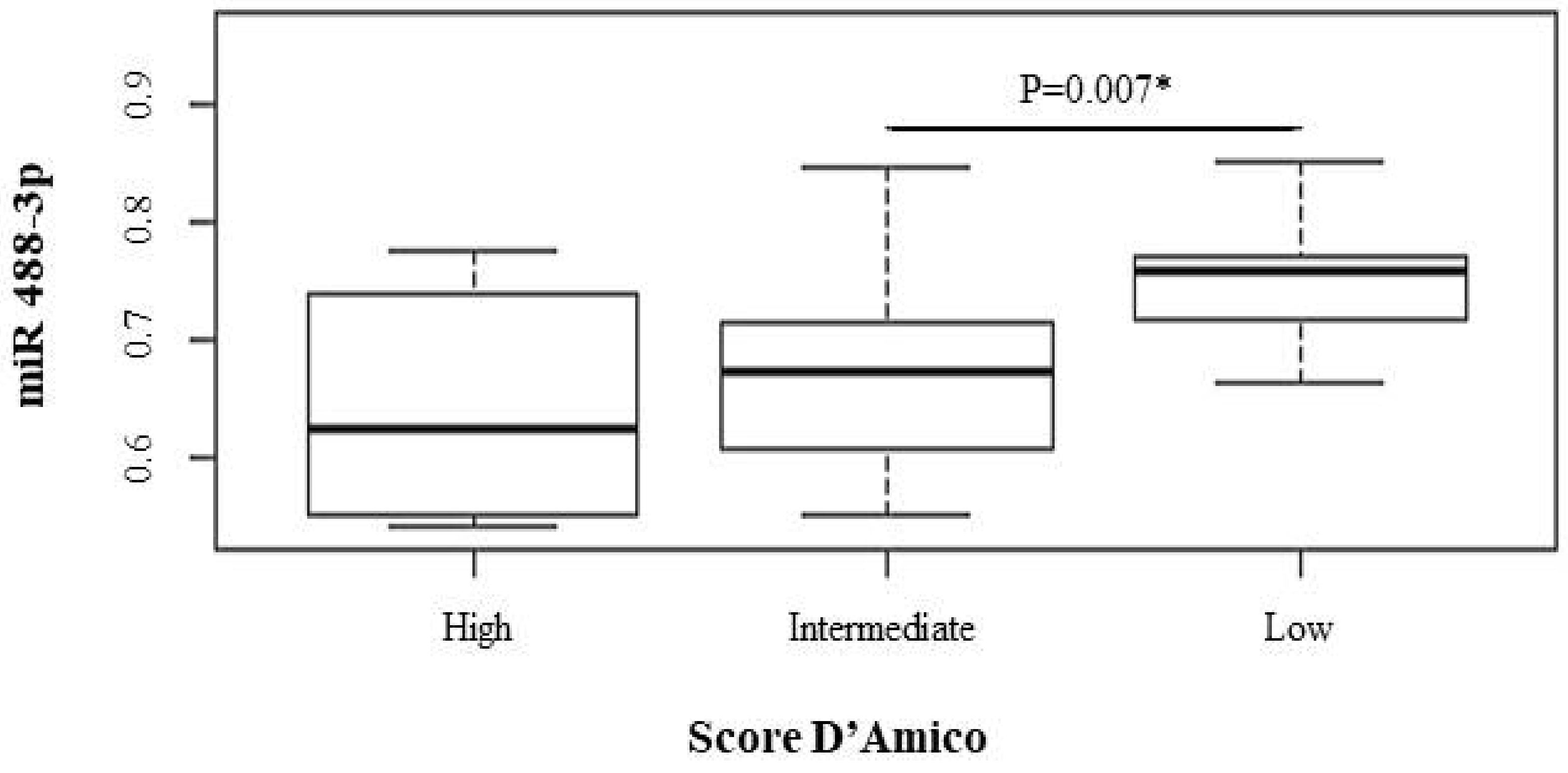
| 488-3p | 0.65 (0.51–0.77) | 0.67 (0.63–0.70) | 0.75 (0.71–0.78) | 0.079 | 0.665 | 0.007 |
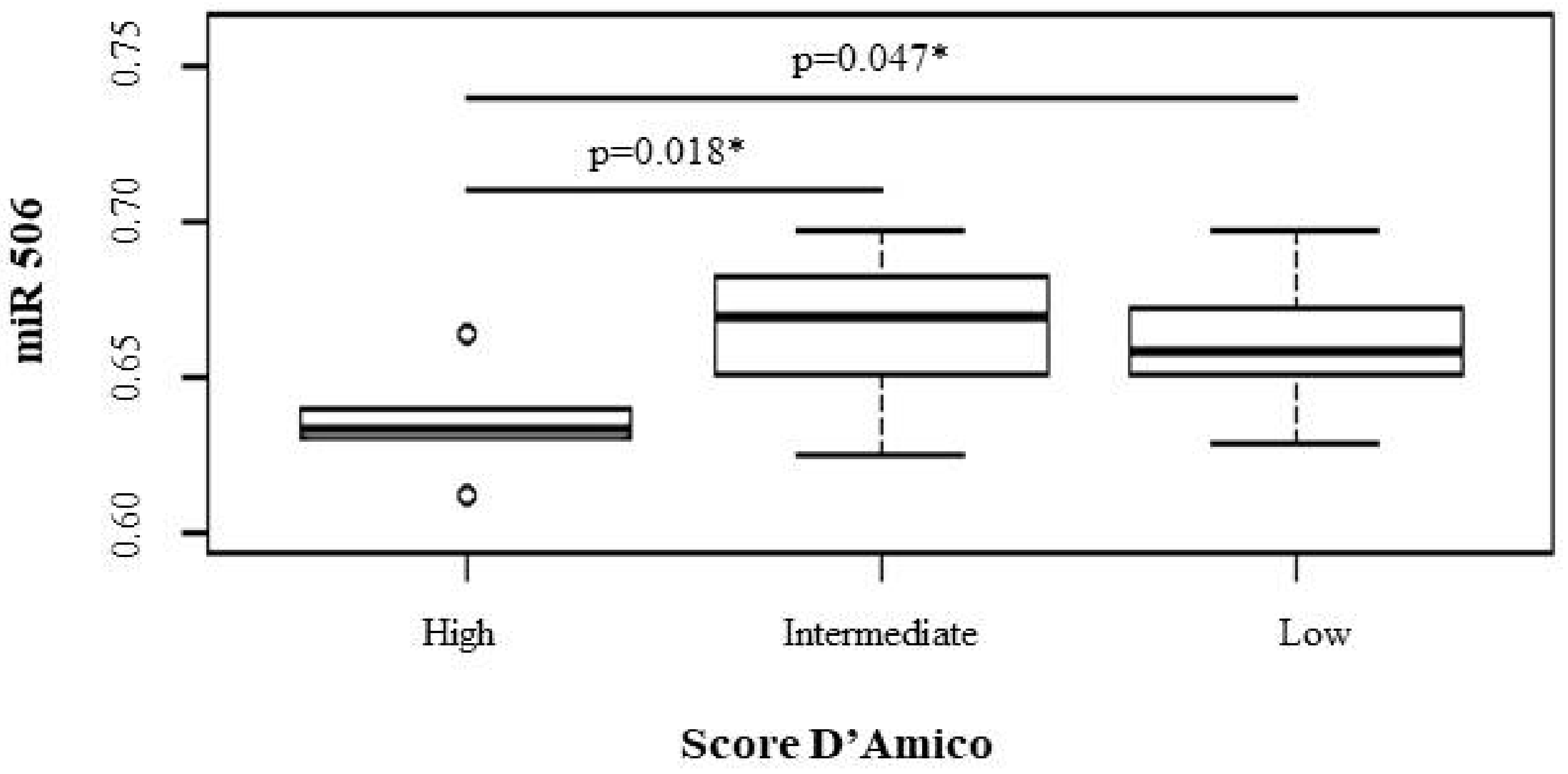
| 506 | 0.64 (0.61–0.66) | 0.67 (0.65–0.67) | 0.66 (0.64–0.67) | 0.047 | 0.018 | 0.581 |
| AR | 1.81 (1.15–2.46) | 1.69 (1.62–1.76) | 1.66 (1.62–1.69) | 0.777 | 0.862 | 0.558 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bernardes, J.G.B.; Fernandes, M.R.; Rodrigues, J.C.G.; Vinagre, L.W.M.S.; Pastana, L.F.; Dobbin, E.A.F.; Medeiros, J.A.G.; Dias Junior, L.B.; Bernardes, G.M.; Bernardes, I.M.M.; et al. Association of Androgenic Regulation and MicroRNAs in Acinar Adenocarcinoma of Prostate. Genes 2022, 13, 622. https://doi.org/10.3390/genes13040622
Bernardes JGB, Fernandes MR, Rodrigues JCG, Vinagre LWMS, Pastana LF, Dobbin EAF, Medeiros JAG, Dias Junior LB, Bernardes GM, Bernardes IMM, et al. Association of Androgenic Regulation and MicroRNAs in Acinar Adenocarcinoma of Prostate. Genes. 2022; 13(4):622. https://doi.org/10.3390/genes13040622
Chicago/Turabian StyleBernardes, Julio Guilherme Balieiro, Marianne Rodrigues Fernandes, Juliana Carla Gomes Rodrigues, Lui Wallacy Morikawa Souza Vinagre, Lucas Favacho Pastana, Elizabeth Ayres Fragoso Dobbin, Jéssyca Amanda Gomes Medeiros, Leonidas Braga Dias Junior, Gabriel Monteiro Bernardes, Izabel Maria Monteiro Bernardes, and et al. 2022. "Association of Androgenic Regulation and MicroRNAs in Acinar Adenocarcinoma of Prostate" Genes 13, no. 4: 622. https://doi.org/10.3390/genes13040622
APA StyleBernardes, J. G. B., Fernandes, M. R., Rodrigues, J. C. G., Vinagre, L. W. M. S., Pastana, L. F., Dobbin, E. A. F., Medeiros, J. A. G., Dias Junior, L. B., Bernardes, G. M., Bernardes, I. M. M., Santos, N. P. C. D., Demachki, S., & Burbano, R. M. R. (2022). Association of Androgenic Regulation and MicroRNAs in Acinar Adenocarcinoma of Prostate. Genes, 13(4), 622. https://doi.org/10.3390/genes13040622







