Are miRNAs Dynamic Biomarkers in Keratoconus? A Review of the Literature
Abstract
:1. Introduction
2. Materials and Methods
3. Results
3.1. MicroRNAs
3.2. MicroRNA and the Eye
3.3. MicroRNA and the Cornea
miR-184 and miR-205 in the Cornea
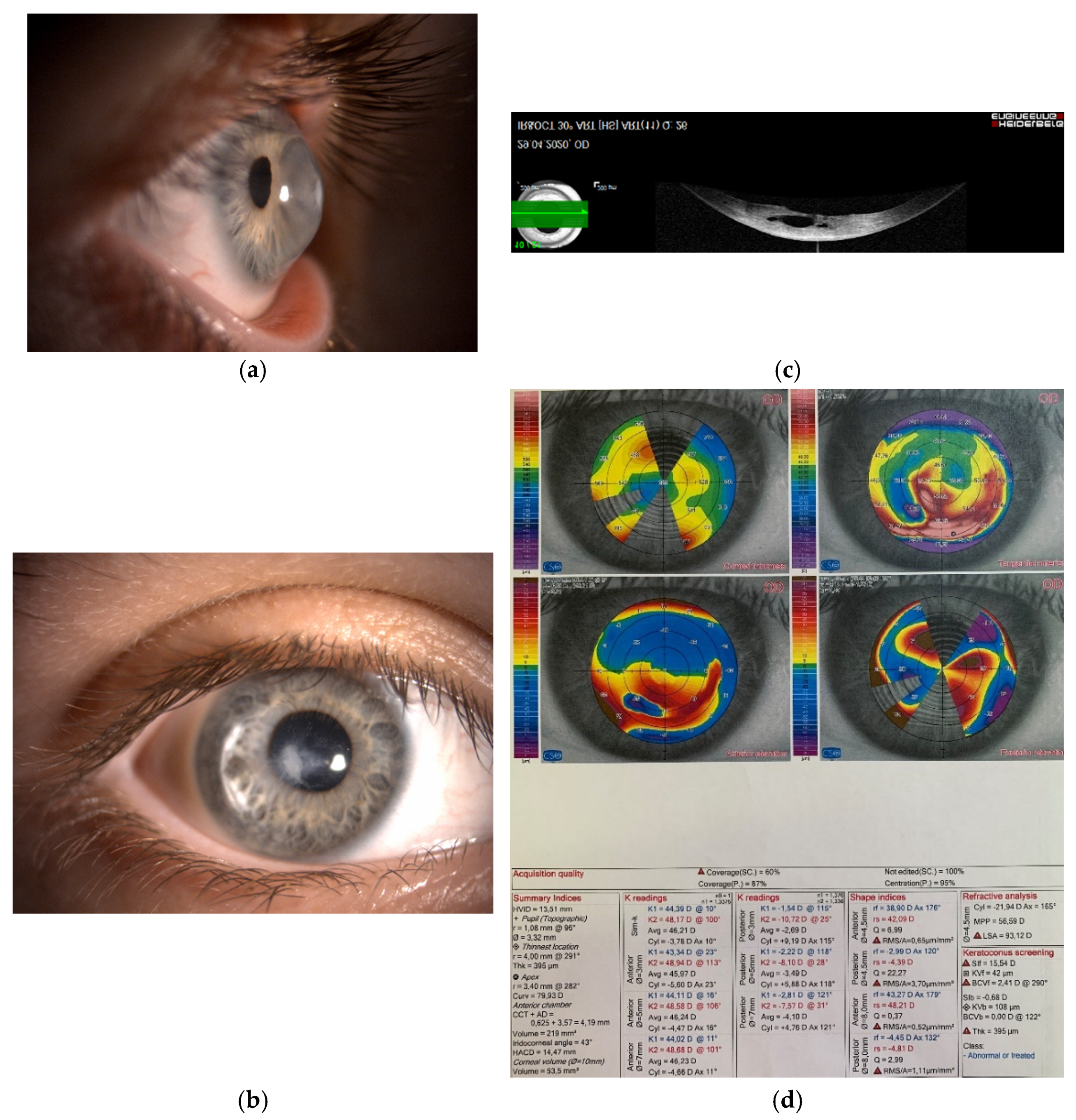
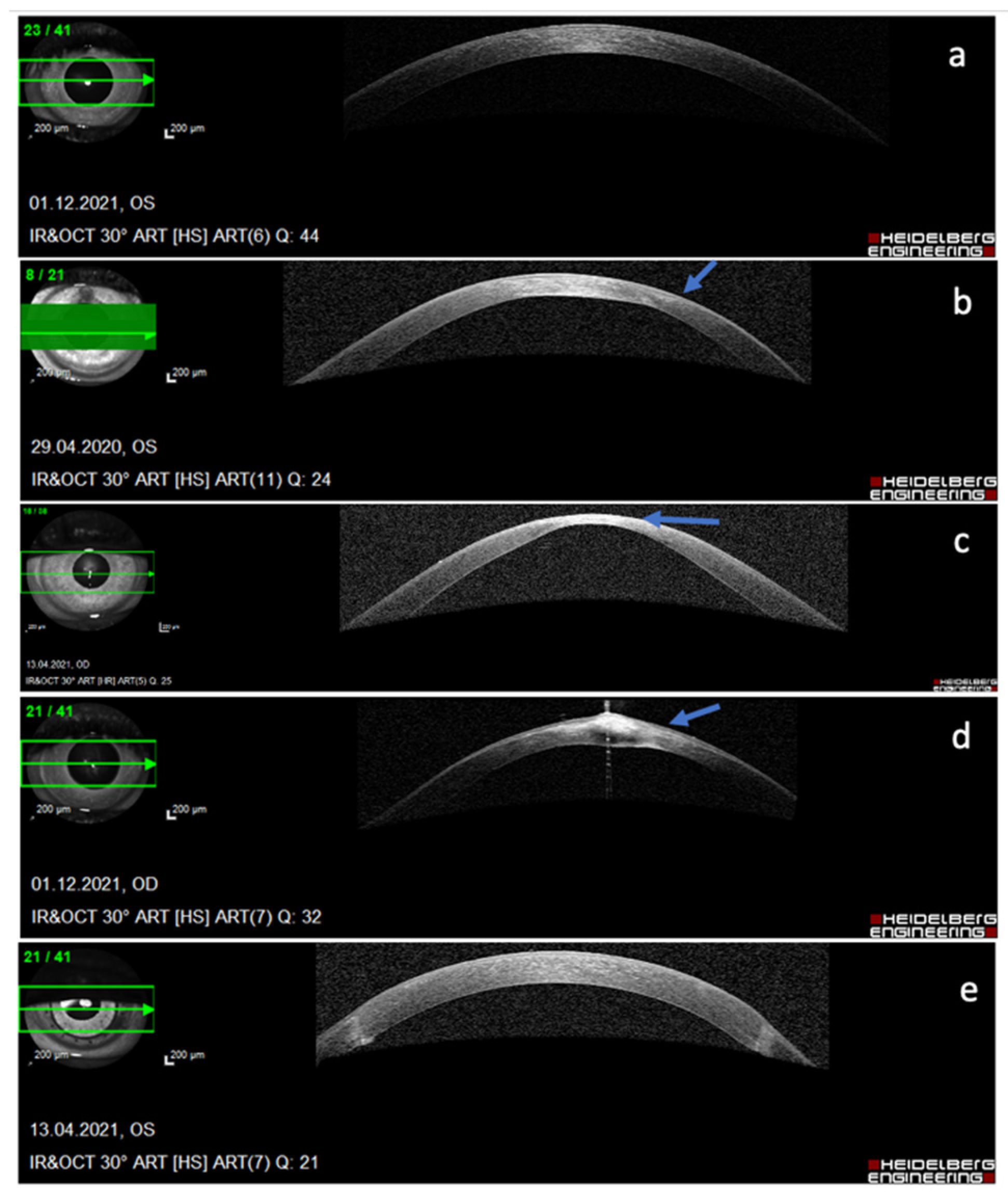
3.4. Keratoconus and MicroRNA
3.4.1. miRNA-184 in Keratoconus
| Type of Mutation Found | Circumstances | Authors | |
|---|---|---|---|
| Mutations in the MIR184 seed region | heterozygous C-to-T transition (r.57C > T) in the MIR184 seed region | Irish family. 18 individuals from three generations were affected. Keratoconus associated with cataract | Hughes et al. [32] |
| Two heterozygous substitutions (+3A > G and +8C > A) in the MIR184 seed region | Two patients. Sporadic keratoconus, no other ocular comorbidities, one had concurrent cataract | Lechner et al. [41] | |
| heterozygous C-to-T transition (c.57C > T) in the MIR184 seed region | Spanish family from Galicia. five generations. Congenital cataracts, varying corneal abnormalities, from severe keratoconus to non-ectatic corneal thinning. | Bykhovsaya et al. [42] | |
| Other miRNAs, altered expression | upregulated, highly expressed: miR-143-3p miR-182-5p miR-92a-3p | Keratoconus corneal tissue. | Drewry M et al. [19] |
| altered expression of six miRNAs: miR-151a-3p miR-138-5p miR-146b-5p miR-194-5p, miR-28-5p miR-181a-2-3p altered expression of four miRNAs: miR-151a-3p miR-195-5p miR-185-5p miR-194-5p | Corneas obtained from keratoconus transplant surgeries. Samples of epithelium obtained by means of impression cytology | Wang YM et al. [37] | |
| 3 miRNAs upregulated: miR-128, miR-32, miR-221 4 miRNAs downregulated: miR-181a, miR-222, miR-98, miR-301a | Keratoconus-associated RNA regulatory network. | Tyan R et al. [45] |
3.4.2. Other miRNAs in Keratoconus
4. Discussion
Funding
Acknowledgments
Conflicts of Interest
References
- Nowak, D.M.; Gajecka, M. The genetics of keratoconus. Middle East Afr. J. Ophthalmol 2011, 18, 2–6. [Google Scholar] [CrossRef] [PubMed]
- Martin, D.I.; Ward, R.; Suter, C.M. Germline epimutation: A basis for epigenetic disease in humans. Ann. N. Y. Acad. Sci. 2005, 1054, 68–77. [Google Scholar] [CrossRef] [PubMed]
- Lanza, M.; Benincasa, G.; Costa, D.; Napoli, C. Clinical Role of Epigenetics and Network Analysis in Eye Diseases: A Translational Science Review. J. Ophthalmol. 2019, 2019, 2424956. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bartel, D.P.; Chen, C.Z. Micromanagers of gene expression: The potentially widespread influence of metazoan microRNAs. Nat. Rev. Genet. 2004, 5, 396–400. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol 2018, 9, 402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, M.; Marin-Muller, C.; Bharadwaj, U.; Chow, K.H.; Yao, Q.; Chen, C. MicroRNAs: Control and loss of control in human physiology and disease. World J. Surg. 2009, 33, 667–684. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McCall, M.N.; Kim, M.S.; Adil, M.; Patil, A.H.; Lu, Y.; Mitchell, C.J.; Leal-Rojas, P.; Xu, J.; Kumar, M.; Dawson, L.V.; et al. Toward the human cellular microRNAome. Genome Res. 2017, 27, 1769–1781. [Google Scholar] [CrossRef] [Green Version]
- Available online: http://www.mirbase.org (accessed on 4 February 2022).
- Van Rooij, E. The art of microRNA research. Circ. Res. 2011, 108, 219–234. [Google Scholar] [CrossRef]
- Amin, M.M.J.; Trevelyan, C.J.; Turner, N.A. MicroRNA-214 in Health and Disease. Cells 2021, 10, 3274. [Google Scholar] [CrossRef]
- Schmiedel, J.M.; Klemm, S.L.; Zheng, Y.; Sahay, A.; Blüthgen, N.; Marks, D.S.; van Oudenaarden, A. Gene expression. MicroRNA control of protein expression noise. Science 2015, 348, 128–132. [Google Scholar] [CrossRef]
- Halvorsen, A.R.; Helland, Å.; Gromov, P.; Wielenga, V.T.; Talman, M.M.; Brunner, N.; Sandhu, V.; Børresen-Dale, A.; Gromova, I.D.V. HaakensenProfiling of microRNAs in tumor interstitial fluid of breast tumors—A novel resource to identify biomarkers for prognostic classification and detection of cancer. Mol. Oncol. 2017, 11, 220–234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Daniel, R.; Wu, Q.; Williams, V.; Clark, G.; Guruli, G.; Zehner, Z. A Panel of MicroRNAs as Diagnostic Biomarkers for the Identification of Prostate Cancer. Int. J. Mol. Sci. 2017, 18, 1281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laffont, B.; Rayner, K.J. MicroRNAs in the Pathobiology and Therapy of Atherosclerosis. Can. J. Cardiol. 2017, 33, 313–324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gomez, J.L.; Chen, A.; Diaz, M.P.; Zirn, N.; Gupta, A.; Britto, C.; Sauler, M.; Yan, X.; Stewart, E.; Santerian, K. A Network of Sputum MicroRNAs Is Associated with Neutrophilic Airway Inflammation in Asthma. Am. J. Respir. Crit. Care Med. 2020, 202, 51–64. [Google Scholar] [CrossRef] [PubMed]
- Konovalova, J.; Gerasymchuk, D.; Parkkinen, I.; Chmielarz, P.; Domanskyi, A. Interplay between MicroRNAs and Oxidative Stress in Neurodegenerative Diseases. Int. J. Mol. Sci. 2019, 20, 6055. [Google Scholar] [CrossRef] [Green Version]
- Xu, S. microRNA expression in the eyes and their significance in relation to functions. Prog. Retin. Eye Res. 2009, 28, 87–116. [Google Scholar] [CrossRef]
- Li, Y.; Piatigorsky, J. Targeted deletion of Dicer disrupts lens morphogenesis, corneal epithelium stratification, and whole eye development. Dev. Dyn. 2009, 238, 2388–2400. [Google Scholar] [CrossRef] [Green Version]
- Drewry, M.; Helwa, I.; Allingham, R.R.; Hauser, M.A.; Liu, Y. miRNA Profile in Three Different Normal Human Ocular Tissues by miRNA-Seq. Invest Ophthalmol. Vis. Sci. 2016, 57, 3731–3739. [Google Scholar] [CrossRef] [Green Version]
- The RNAcentral Consortium; Petrov, A.I.; Kay, S.J.E.; Kalvari, I.; Howe, K.L.; Gray, K.A.; Bruford, E.A.; Paul, J.; Kersey, G.; Cochrane, R.D.F. RNAcentral: A comprehensive database of non-coding RNA sequences. Nucleic. Acids Res. 2017, 45, D128–D134. [Google Scholar] [CrossRef]
- Zhang, Y.; Cai, S.; Jia, Y.; Chen, Q.; Jing, S.; Hong, Z.; Fei, W.; Yunshan, C.; Xiaorong, L. Decoding Noncoding RNAs: Role of MicroRNAs and Long Noncoding RNAs in Ocular Neovascularization. Theranostics 2017, 7, 3155–3167. [Google Scholar] [CrossRef]
- Mukwaya, A.; Jensen, L.; Peebo, B.; Lagali, N. MicroRNAs in the cornea: Role and implications for treatment of corneal neovascularization. Ocul. Surf. 2019, 17, 400–411. [Google Scholar] [CrossRef] [PubMed]
- Naderan, M.; Jahanrad, A.; Balali, S. Histopathologic findings of keratoconus corneas underwent penetrating keratoplasty according to topographic measurements and keratoconus severity. Int. J. Ophthalmol. 2017, 10, 1640–1646. [Google Scholar] [CrossRef] [PubMed]
- Ryan, D.G.; Oliveira-Fernandes, M.; Lavker, R.M. MicroRNAs of the mammalian eye display distinct and overlapping tissue specificity. Mol. Vis. 2006, 12, 1175–1184. [Google Scholar] [PubMed]
- Kalaimani, L.; Devarajan, B.; Subramanian, U.; Ayyasamy, V.; Namperumalsamy, V.P.; Veerappan, M.; Chidambaranathan, G.P. MicroRNA Profiling of Highly Enriched Human Corneal Epithelial Stem Cells by Small RNA Sequencing. Sci. Rep. 2020, 10, 7418. [Google Scholar] [CrossRef] [PubMed]
- Abu-Amero, K.K.; Helwa, I.; Al-Muammar, A.; Strickland, S.; Hauser, M.A.; Allingham, R.R.; Liu, Y. Screening of the Seed Region of MIR184 in Keratoconus Patients from Saudi Arabia. Biomed Res. Int. 2015, 2015, 604508. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cotsarelis, G.; Cheng, S.Z.; Dong, G.; Sun, T.T.; Lavker, R.M. Existence of slow-cycling limbal epithelial basal cells that can be preferentially stimulated to proliferate: Implications on epithelial stem cells. Cell 1989, 57, 201–209. [Google Scholar] [CrossRef]
- Yi, R.; O’Carroll, D.; Pasolli, H.A.; Zhang, Z.; Dietrich, F.S.; Tarakhovsky, A.; Fuchs, E. Morphogenesis in skin is governed by discrete sets of differentially expressed microRNAs. Nat. Genet. 2006, 38, 356–362. [Google Scholar] [CrossRef]
- Karali, M.; Peluso, I.; Gennarino, V.A.; Bilio, M.; Verde, R.; Lago, G.; Dollé, P.; Banfi, S. miRNeye: A microRNA expression atlas of the mouse eye. BMC Genom. 2010, 11, 715. [Google Scholar] [CrossRef] [Green Version]
- Yu, J.; Ryan, D.G.; Getsios, S.; Oliveira-Fernandes, M.; Fatima, A.; Lavker, R.M. MicroRNA-184 antagonizes microRNA-205 to maintain SHIP2 levels in epithelia. Proc. Natl. Acad. Sci. USA 2008, 105, 19300–19305. [Google Scholar] [CrossRef] [Green Version]
- Cao, Q.; Xu, W.; Chen, W.; Peng, D.; Liu, Q.; Dong, J.; Reinach, P.S.; Yan, D. MicroRNA-184 negatively regulates corneal epithelial wound healing via targeting CDC25A, CARM1, and LASP1. Eye Vis. 2020, 7, 35. [Google Scholar] [CrossRef]
- Hughes, A.E.; Bradley, D.T.; Campbell, M.; Lechner, J.; Dash, D.P.; Simpson, D.A.; Willoughby, C.E. Mutation altering the miR-184 seed region causes familial keratoconus with cataract. Am. J. Hum. Genet 2011, 89, 628–633. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, H.; Park, J.K.; Katsnelson, J.; Kaplan, N.; Yang, W.; Getsios, S.; Lavker, R.M. microRNA-103/107 Family Regulates Multiple Epithelial Stem Cell Characteristics. Stem. Cells 2015, 33, 1642–1656. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- An, J.; Chen, X.; Chen, W.; Liang, R.; Reinach, P.S.; Yan, D.; Tu, L. MicroRNA Expression Profile and the Role of miR-204 in Corneal Wound Healing. Invest Ophthalmol. Vis. Sci. 2015, 56, 3673–3683. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Funari, V.A.; Winkler, M.; Brown, J.; Dimitrijevich, S.D.; Ljubimov, A.V.; Saghizadeh, M. Differentially expressed wound healing-related microRNAs in the human diabetic cornea. PLoS ONE 2013, 8, e84425. [Google Scholar] [CrossRef] [Green Version]
- Amit, C.; Padmanabhan, P.; Narayanan, J. Deciphering the mechanoresponsive role of β-catenin in keratoconus epithelium. Sci. Rep. 2020, 10, 21382. [Google Scholar] [CrossRef]
- Wang, Y.M.; Ng, T.K.; Choy, K.W.; Wong, H.K.; Chu, W.K.; Pang, C.P.; Jhanji, V. Histological and microRNA Signatures of Corneal Epithelium in Keratoconus. J. Refract. Surg. 2018, 34, 201–211. [Google Scholar] [CrossRef] [Green Version]
- Duan, R.; Pak, C.; Jin, P. Single nucleotide polymorphism associated with mature miR-125a alters the processing of pri-miRNA. Hum. Mol. Genet 2007, 16, 1124–1131. [Google Scholar] [CrossRef] [Green Version]
- Mencía, A.; Modamio-Høybjør, S.; Redshaw, N.; Morín, M.; Mayo-Merino, F.; Olavarrieta, L.; Luis, A.A.; Ignacio, C.; Karen, P.S.; Tamas, D.; et al. Mutations in the seed region of human miR-96 are responsible for nonsyndromic progressive hearing loss. Nat. Genet 2009, 41, 609–613. [Google Scholar] [CrossRef]
- Lewis, M.A.; Quint, E.; Glazier, A.M.; Fuchs, H.; De Angelis, M.H.; Langford, C.; van Dongen, S.; Abreu-Goodger, C.; Piipari, M.; Redshaw, N.; et al. An ENU-induced mutation of miR-96 associated with progressive hearing loss in mice. Nat. Genet. 2009, 41, 614–618. [Google Scholar] [CrossRef] [Green Version]
- Lechner, J.; Bae, H.A.; Guduric-Fuchs, J.; Rice, A.; Govindarajan, G.; Siddiqui, S.; Abi Farraj, L.; Yip, S.P.; Yap, M.; Das, M.; et al. Mutational analysis of MIR184 in sporadic keratoconus and myopia. Invest Ophthalmol. Vis. Sci. 2013, 54, 5266–5272. [Google Scholar] [CrossRef] [Green Version]
- Bykhovskaya, Y.; Caiado Canedo, A.L.; Wright, K.W.; Rabinowitz, Y.S. C.57 C > T Mutation in MIR 184 is Responsible for Congenital Cataracts and Corneal Abnormalities in a Five-generation Family from Galicia, Spain. Ophthalmic Genet. 2015, 36, 244–247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iliff, B.W.; Riazuddin, S.A.; Gottsch, J.D. A single-base substitution in the seed region of miR-184 causes EDICT syndrome. Invest Ophthalmol. Vis. Sci. 2012, 53, 348–353. [Google Scholar] [CrossRef] [PubMed]
- Jun, A.S.; Broman, K.W.; Do, D.V.; Akpek, E.K.; Stark, W.J.; Gottsch, J.D. Endothelial dystrophy, iris hypoplasia, congenital cataract, and stromal thinning (edict) syndrome maps to chromosome 15q22.1-q25.3. Am. J. Ophthalmol. 2002, 134, 172–176. [Google Scholar] [CrossRef]
- Tian, R.; Wang, L.; Zou, H.; Song, M.; Liu, L.; Zhang, H. Role of the XIST-miR-181a-COL4A1 axis in the development and progression of keratoconus. Mol. Vis. 2020, 26, 1–13. [Google Scholar]
- Kim, W.J.; Rabinowitz, Y.S.; Meisler, D.M.; Wilson, S.E. Keratocyte apoptosis associated with keratoconus. Exp. Eye Res. 1999, 69, 475–481. [Google Scholar] [CrossRef]
- Crespo Millas, S.; López, J.C.; García-Lagarto, E.; Obregón, E.; Hileeto, D.; Maldonado, M.J.; Pastor, J.C. Histological Patterns of Epithelial Alterations in Keratoconus. J. Ophthalmol. 2020, 7, 1468258. [Google Scholar] [CrossRef]
- Najmi, H.; Mobarki, Y.; Mania, K.; Altowairqi, B.; Basehi, M.; Mahfouz, M.S.; Elmahdy, M. The correlation between keratoconus and eye rubbing: A review. Int. J. Ophthalmol. 2019, 12, 1775–1781. [Google Scholar] [CrossRef]
- Sahebjada, S.; Al-Mahrouqi, H.H.; Moshegov, S.; Panchatcharam, S.M.; Chan, E.; Daniell, M.; Baird, P.N. Eye rubbing in the aetiology of keratoconus: A systematic review and meta-analysis. Graefes Arch. Clin. Exp. Ophthalmol. 2021, 259, 2057–2067. [Google Scholar] [CrossRef]
- Fast, I.; Hewel, C.; Wester, L.; Schumacher, J.; Gebert, D.; Zischler, H.; Berger, C.; Rosenkranz, D. Temperature-responsive miRNAs in Drosophila orchestrate adaptation to different ambient temperatures. RNA 2017, 23, 1352–1364. [Google Scholar] [CrossRef] [Green Version]
- May, P.; Liao, W.; Wu, Y.; Shuai, B.; McCombie, W.R.; Zhang, M.Q.; Liu, Q.A. The effects of carbon dioxide and temperature on microRNA expression in Arabidopsis development. Nat. Commun. 2013, 4, 2145. [Google Scholar] [CrossRef] [Green Version]
- Roberts, J.C.; Warren, R.B.; Griffiths, C.E.; Ross, K. Expression of microRNA-184 in keratinocytes represses argonaute 2. J. Cell Physiol. 2013, 228, 2314–2323. [Google Scholar] [CrossRef] [PubMed]
- Ljubimov, A.V.; Saghizadeh, M. Progress in corneal wound healing. Prog. Retin. Eye Res. 2015, 49, 17–45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, J.; Peng, H.; Ruan, Q.; Fatima, A.; Getsios, S.; Lavker, R.M. MicroRNA-205 promotes keratinocyte migration via the lipid phosphatase SHIP2. FASEB J. 2010, 24, 3950–3959. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alsharafi, W.; Xiao, B. Dynamic Expression of MicroRNAs (183, 135a, 125b, 128, 30c and 27a) in the Rat Pilocarpine Model and Temporal Lobe Epilepsy Patients. CNS Neurol. Disord. Drug Targets 2015, 14, 1096–1102. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Xuan, F.; Ge, X.; Zhu, J.; Zhang, W. Dynamic mRNA and miRNA expression analysis in response to hypoxia and reoxygenation in the blunt snout bream (Megalobrama amblycephala). Sci. Rep. 2017, 71, 12846. [Google Scholar] [CrossRef]
- Beauchemin, M.; Smith, A.; Yin, V.P. Dynamic microRNA-101a and Fosab expression controls zebrafish heart regeneration. Development 2015, 142, 4026–4037. [Google Scholar] [CrossRef] [Green Version]
- Kirigin, F.F.; Lindstedt, K.; Sellars, M.; Ciofani, M.; Low, S.L.; Jones, L.; Bell, F.; Pauli, F.; Bonneau, R.; Myers, R.M.; et al. Dynamic microRNA gene transcription and processing during T cell development. J. Immunol. 2012, 188, 3257–3267. [Google Scholar] [CrossRef]
- El Wardani, M.; Hashemi, K.; Aliferis, K.; Kymionis, G. Topographic changes simulating keratoconus in patients with irregular inferior epithelial thickening documented by anterior segment optical coherence tomography. Clin. Ophthalmol. 2019, 13, 2103–2110. [Google Scholar] [CrossRef] [Green Version]
- Tuori, A.J.; Virtanen, I.; Aine, E.; Kalluri, R.; Miner, J.H.; Uusitalo, H.M. The immunohistochemical composition of corneal basement membrane in keratoconus. Curr. Eye Res. 1997, 16, 792–801. [Google Scholar] [CrossRef]
- Ljubimov, A.V.; Burgeson, R.E.; Butkowski, R.J.; Michael, A.F.; Sun, T.T.; Kenney, M.C. Human corneal basement membrane heterogeneity: Topographical differences in the expression of type IV collagen and laminin isoforms. Lab. Invest 1995, 72, 461–473. [Google Scholar]
- Torricelli, A.A.; Singh, V.; Santhiago, M.R.; Wilson, S.E. The corneal epithelial basement membrane: Structure, function, and disease. Invest Ophthalmol. Vis. Sci. 2013, 54, 6390–6400. [Google Scholar] [CrossRef]
- Chaurasia, S.S.; Kaur, H.; de Medeiros, F.W.; Smith, S.D.; Wilson, S.E. Reprint of Dynamics of the expression of intermediate filaments vimentin and desmin during myofibroblast differentiation after corneal injury. Exp. Eye Res. 2009, 89, 590–596. [Google Scholar] [CrossRef]
- Singh, V.; Santhiago, M.R.; Barbosa, F.L.; Agrawal, V.; Singh, N.; Ambati, B.K.; Wilson, S.E. Effect of TGFβ and PDGF-B blockade on corneal myofibroblast development in mice. Exp. Eye Res. 2011, 93, 810–817. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Netto, M.V.; Mohan, R.R.; Sinha, S.; Sharma, A.; Dupps, W.; Wilson, S.E. Stromal haze, myofibroblasts, and surface irregularity after PRK. Exp. Eye Res. 2006, 82, 788–797. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaur, H.; Chaurasia, S.S.; Agrawal, V.; Suto, C.; Wilson, S.E. Corneal myofibroblast viability: Opposing effects of IL-1 and TGF beta1. Exp. Eye Res. 2009, 89, 152–158. [Google Scholar] [CrossRef] [Green Version]
- Saghizadeh, M.; Brown, J.; Ljubimov, A.V.; Funari, V.A. miRNA expression profiling in central and limbal diabetic and normal human corneas using deep sequencing. Investig. Ophthalmol. Vis. Sci. 2013, 54, 3229. [Google Scholar]
- Teng, Y.; Wong, H.K.; Jhanji, V.; Chen, J.H.; Young, A.L.; Zhang, M.; Choy, K.W.; Mehta, J.S.; Pang, C.P.; Yam, G.H. Signature microRNAs in human cornea limbal epithelium. Funct. Integr. Genomics 2015, 15, 277–294. [Google Scholar] [CrossRef]
- Winkler, M.A.; Dib, C.; Ljubimov, A.V.; Saghizadeh, M. Targeting miR-146a to treat delayed wound healing in human diabetic organ-cultured corneas. PLoS ONE 2014, 9, e114692. [Google Scholar] [CrossRef] [Green Version]
- Lin, D.; Halilovic, A.; Yue, P.; Bellner, L.; Wang, K.; Wang, L.; Zhang, C. Inhibition of miR-205 impairs the wound-healing process in human corneal epithelial cells by targeting KIR4.1 (KCNJ10). Invest Ophthalmol. Vis. Sci. 2013, 54, 6167–6178. [Google Scholar] [CrossRef] [Green Version]
- Bertero, T.; Gastaldi, C.; Bourget-Ponzio, I.; Imbert, V.; Loubat, A.; Selva, E.; Busca, R.; Mari, B.; Hofman, P.; Barbry, P.; et al. miR-483-3p controls proliferation in wounded epithelial cells. FASEB J. 2011, 25, 3092–3105. [Google Scholar] [CrossRef]
- Gao, J.N.; Wang, Y.; Zhao, X.; Chen, P.; Xie, L. MicroRNA-204-5p-mediated regulation of SIRT1 contributes to the delay of epithelial cell-cycle traversal in diabetic corneas. Invest Ophthalmol. Vis. Sci. 2015, 56, 1493–14504. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roy, S.; Sen, C.K. MiRNA in innate immune responses: Novel players in wound inflammation. Physiol. Genomics. 2011, 43, 557–565. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stunf Pukl, S. Are miRNAs Dynamic Biomarkers in Keratoconus? A Review of the Literature. Genes 2022, 13, 588. https://doi.org/10.3390/genes13040588
Stunf Pukl S. Are miRNAs Dynamic Biomarkers in Keratoconus? A Review of the Literature. Genes. 2022; 13(4):588. https://doi.org/10.3390/genes13040588
Chicago/Turabian StyleStunf Pukl, Spela. 2022. "Are miRNAs Dynamic Biomarkers in Keratoconus? A Review of the Literature" Genes 13, no. 4: 588. https://doi.org/10.3390/genes13040588
APA StyleStunf Pukl, S. (2022). Are miRNAs Dynamic Biomarkers in Keratoconus? A Review of the Literature. Genes, 13(4), 588. https://doi.org/10.3390/genes13040588





