Genetic Architecture of Maize Stalk Diameter and Rind Penetrometer Resistance in a Recombinant Inbred Line Population
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Phenotyping
2.3. Data Analysis and QTL Mapping
2.4. Validation and Fine Mapping of qSD9-2
3. Results
3.1. Phenotypic Variation in SD and RPR
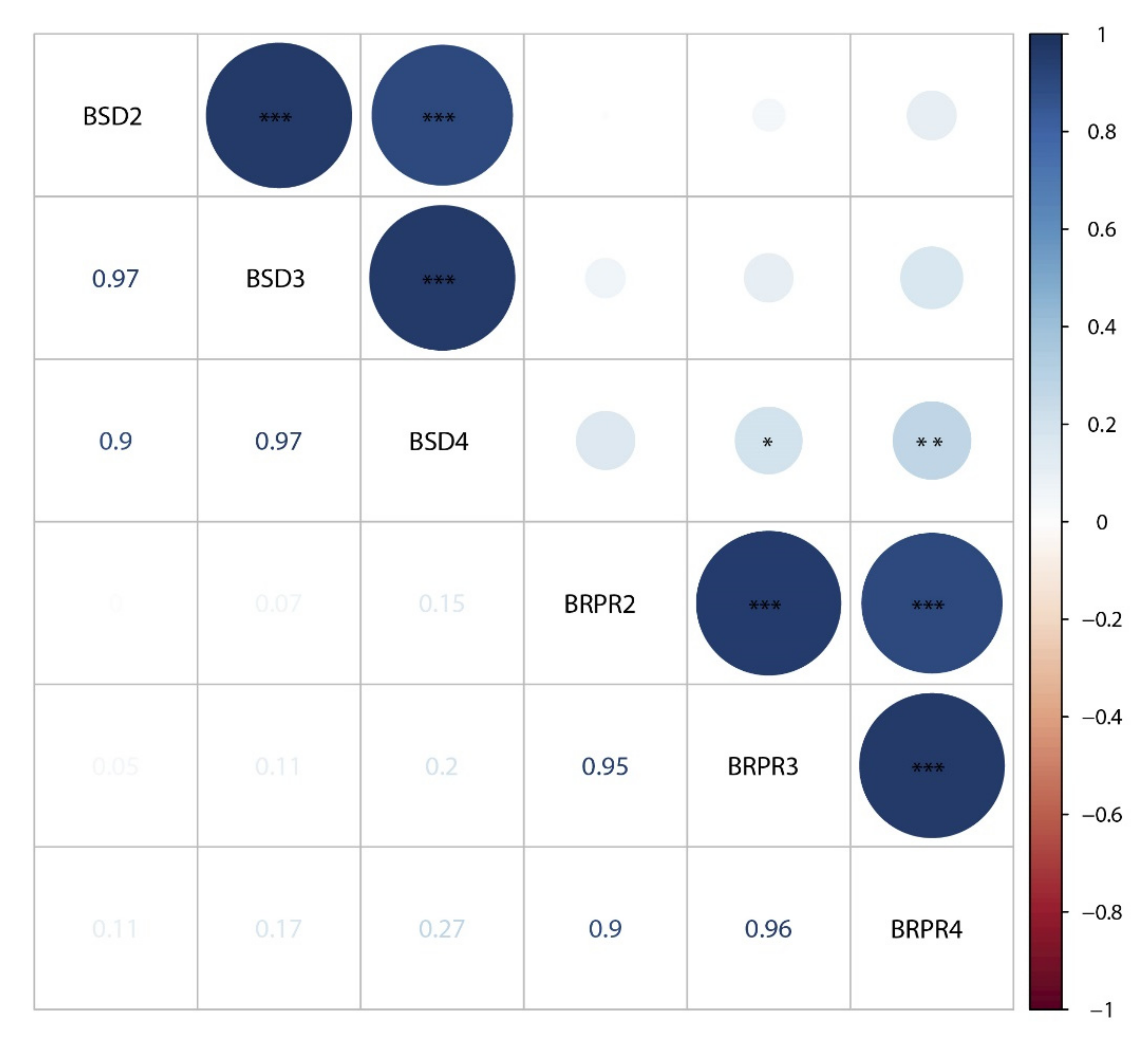
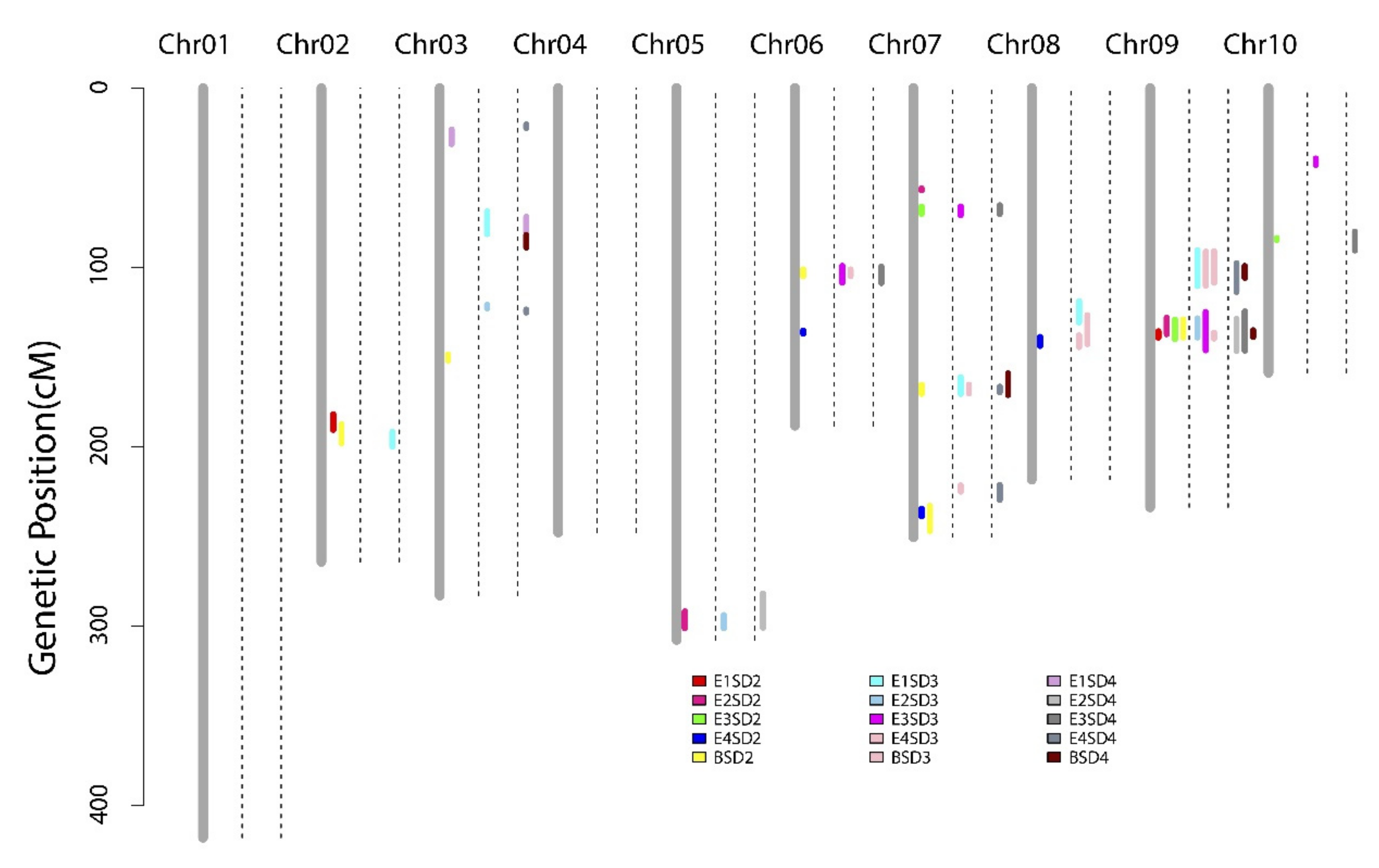
3.2. QTL Mappings of Stalk Diameters across Four Environments and the BLUP Model
3.3. QTL Mapping of RPR across Four Environments and the BLUP Model
3.4. Validation and Fine Mapping of qSD9-2
3.5. Candidate Gene Prediction of RPR-Related QTLs
4. Discussion
4.1. Genetic Basis of Lodging-Related Traits at Different Internodes
4.2. Pleiotropic QTL Influencing Stalk Lodging Resistance-Related Traits in Maize
4.3. Comparison with Previous QTL Mapping Results
4.4. Cloned Genes Related to Stalk Phenotype within the QTL Intervals
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Feng, G.; Huang, C.; Xing, J. The research progress in lodging resistance of maize. Crops 2008, 24, 12–14. (In Chinese) [Google Scholar]
- Gou, L.; Zhao, M.; Huang, J.; Zhang, B.; Tao, L.; Sun, R. Bending Mechanical Properties of Stalk and Lodging-Resistance of Maize (Zea mays L.). Acta Agron. Sin. 2008, 34, 653–661. (In Chinese) [Google Scholar]
- Wang, H.; Wu, R.; Zhu, K.; Zhang, Y.; Zhang, Y.; Sun, J. Reviews of Causes and Control of Maize Lodging. J. Henan Agric. Sci. 2011, 40, 1–5. (In Chinese) [Google Scholar]
- Flint-Garcia, S.A.; Jampatong, C.; Darrah, L.L.; McMullen, M.D. Quantitative Trait Locus Analysis of Stalk Strength in Four Maize Populations. Crop. Sci. 2003, 43, 13–22. [Google Scholar]
- Xue, J.; Li, L.; Xie, R.; Wang, K.; Li, S. Effect of Lodging on Maize Grain Losing and Harvest Efficiency in Mechanical Grain Harvest. Acta Agron. Sin. 2018, 44, 1774. [Google Scholar]
- Li, K.; Yan, J.; Li, J.; Yang, X. Genetic architecture of rind penetrometer resistance in two maize recombinant inbred line populations. BMC Plant. Biol. 2014, 14, 152. [Google Scholar]
- Xia Qing, W.; Wei, S.; Ru Yang, Z.; Yi Ning, C.; Xuan, S.; Jiu Ran, Z. Genetic Research Advances on Maize Stalk Lodging Resistance. Sci. Agric. Sin. 2021, 54, 2261–2272. (In Chinese) [Google Scholar]
- Tesso, T.; Ejeta, G. Stalk strength and reaction to infection by Macrophomina phaseolina of brown midrib maize (Zea mays) and sorghum (Sorghum bicolor). Field Crops Res. 2011, 120, 271–275. [Google Scholar]
- Xu, C.; Gao, Y.; Tian, B.; Ren, J.; Meng, Q.; Wang, P. Effects of EDAH, a novel plant growth regulator, on mechanical strength, stalk vascular bundles and grain yield of summer maize at high densities. Field Crops Res. 2017, 200, 71–79. [Google Scholar]
- Zhang, Y.; Liang, T.; Chen, M.; Zhang, Y.; Wang, T.; Lin, H.; Rong, T.; Zou, C.; Liu, P.; Lee, M.; et al. Genetic dissection of stalk lodging-related traits using an IBM Syn10 DH population in maize across three environments (Zea mays L.). Mol. Genet. Genom. 2019, 294, 1277–1288. [Google Scholar]
- Xue, J.; Gao, S.; Fan, Y.; Li, L.; Ming, B.; Wang, K.; Xie, R.; Hou, P.; Li, S. Traits of plant morphology, stalk mechanical strength, and biomass accumulation in the selection of lodging-resistant maize cultivars. Eur. J. Agron. 2020, 117, 126073. [Google Scholar]
- Zhang, P.; Gu, S.; Wang, Y.; Yang, R.; Yan, Y.; Zhang, S.; Sheng, D.; Cui, T.; Huang, S.; Wang, P. Morphological and mechanical variables associated with lodging in maize (Zea mays L.). Field Crops Res. 2021, 269, 108178. [Google Scholar]
- Robertson, D.J.; Julias, M.; Lee, S.Y.; Cook, D.D. Maize stalk lodging: Morphological determinants of stalk strength. Crop Sci. 2017, 57, 926–934. [Google Scholar]
- Albrecht, K.; Zuber, M.; Grogan, C. Selection reversal in strains of corn previously long-term selected for chemical composition. Crop Sci. 1986, 26, 1051–1055. [Google Scholar]
- Martin, M.J.; Russell, W.A. Correlated Responses of Yield and Other Agronomic Traits to Recurrent Selection for Stalk Quality in a Maize Synthetic. Crop Sci. 1984, 24, 746–750. [Google Scholar]
- Yang, J.; Geng, W.; Zhang, J.; Ren, B.; Wang, L. Responses of the Lodging Resistance of Summer Maize with Different Gene Types to Plant Density. Agronomy 2021, 12, 10. [Google Scholar]
- Robertson, D.; Smith, S.; Gardunia, B.; Cook, D. An Improved Method for Accurate Phenotyping of Corn Stalk Strength. Crop Sci. 2014, 54, 2038–2044. [Google Scholar]
- Sekhon, R.S.; Joyner, C.N.; Ackerman, A.J.; McMahan, C.S.; Cook, D.D.; Robertson, D.J. Stalk Bending Strength is Strongly Associated with Maize Stalk Lodging Incidence Across Multiple Environments. Field Crops Res. 2020, 249, 107737. [Google Scholar]
- Hu, H.; Liu, W.; Fu, Z.; Homann, L.; Technow, F.; Wang, H.; Song, C.; Li, S.; Melchinger, A.E.; Chen, S. QTL mapping of stalk bending strength in a recombinant inbred line maize population. Theor. Appl. Genet. 2013, 126, 2257–2266. [Google Scholar]
- Zhang, Y.; Liu, P.; Zhang, X.; Zheng, Q.; Chen, M.; Ge, F.; Li, Z.; Sun, W.; Guan, Z.; Liang, T.; et al. Multi-Locus Genome-Wide Association Study Reveals the Genetic Architecture of Stalk Lodging Resistance-Related Traits in Maize. Front. Plant Sci. 2018, 9, 611. [Google Scholar]
- Sibale, E.M.; Darrah, L.L.; Zuber, M.S. Comparison of two rind penetrometers for measurement of stalk strength in maize. Maydica 1992, 37, 111–114. [Google Scholar]
- Thompson, D.L. Recurrent Selection for Lodging Susceptibility and Resistance in Corn. Crop Sci. 1972, 12, 631–634. [Google Scholar]
- Kang, M.S.; Din, A.K.; Zhang, Y.; Magari, R. Combining ability for rind-puncture resistance in maize. Crop Sci. 1999, 39, 368–371. [Google Scholar]
- Hu, H.; Meng, Y.; Wang, H.; Liu, H.; Chen, S. Identifying quantitative trait loci and determining closely related stalk traits for rind penetrometer resistance in a high-oil maize population. Theor. Appl. Genet. 2012, 124, 1439–1447. [Google Scholar] [PubMed]
- Peiffer, J.A.; Flint-Garcia, S.A.; Leon, N.D.; McMullen, M.D.; Kaeppler, S.M.; Buckler, E.S. The genetic architecture of maize stalk strength. PLoS ONE 2013, 8, e67066. [Google Scholar]
- Liu, F.; Qu, W.; Fang, H.; Li, L.; Jin, F.; Wu, W. Analysis of Meta-QTL and Candidate Genes Related to Stem Diameter in Maize. J. Northeast Agric. Sci. 2019, 44, 30–33. (In Chinese) [Google Scholar]
- Liu, X.; Hu, X.; Li, K.; Liu, Z.; Wu, Y.; Wang, H.; Huang, C. Genetic mapping and genomic selection for maize stalk strength. BMC Plant Biol. 2020, 20, 196. [Google Scholar]
- Zhang, Z.; Zhang, X.; Lin, Z.; Wang, J.; Liu, H.; Zhou, L.; Zhong, S.; Li, Y.; Zhu, C.; Lai, J.; et al. A large transposon insertion in the stiff1 promoter increases stalk strength in maize. Plant Cell 2020, 32, 152–165. [Google Scholar] [PubMed]
- Sindhu, A.; Langewisch, T.; Olek, A.; Multani, D.S.; McCann, M.C.; Vermerris, W.; Carpita, N.C.; Johal, G. Maize Brittle stalk2 Encodes a COBRA-Like Protein Expressed in Early Organ Development but Required for Tissue Flexibility at Maturity. Plant Physiol. 2007, 145, 1444–1459. [Google Scholar]
- Jiao, S.; Hazebroek, J.P.; Chamberlin, M.A.; Perkins, M.; Sandhu, A.S.; Gupta, R.; Simcox, K.D.; Li, Y.; Prall, A.; Heetland, L.; et al. Chitinase-like1 Plays a Role in Stalk Tensile Strength in Maize. Plant Physiol. 2019, 181, 00615.02019. [Google Scholar]
- Halpin, C.; Holt, K.; Chojecki, J.; Oliver, D.; Chabbert, B.; Monties, B.; Edwards, K.; Barakate, A.; Foxon, G.A. Brown-midrib maize (bm1)—A mutation affecting the cinnamyl alcohol dehydrogenase gene. Plant J. 1998, 14, 545–553. [Google Scholar] [PubMed]
- Tang, H.M.; Liu, S.; Hill-Skinner, S.; Wu, W.; Reed, D.; Yeh, C.-T.; Nettleton, D.; Schnable, P.S. The maize brown midrib2 (bm2) gene encodes a methylenetetrahydrofolate reductase that contributes to lignin accumulation. Plant J. 2014, 77, 380–392. [Google Scholar] [PubMed] [Green Version]
- Vignols, F.; Rlgau, J.; Torres, M.A.; Capellades, M.; Puigdomènech, P. The Brown Midrib3 (Bm3) Mutation in Maize Occurs in the Gene Encoding Caffeic Acid O-Methyltransferase. Plant Cell 1995, 7, 407–416. [Google Scholar] [PubMed] [Green Version]
- Li, L.; Hill-Skinner, S.; Liu, S.; Beuchle, D.; Tang, H.; Yeh, C.-T.; Nettleton, D.; Schnable, P.S. The maize brown midrib4 (bm4) gene encodes a functional folylpolyglutamate synthase. Plant J. 2015, 81, 493–504. [Google Scholar] [PubMed] [Green Version]
- Xiong, W.; Wu, Z.; Liu, Y.; Li, Y.; Su, K.; Bai, Z.; Guo, S.; Hu, Z.; Zhang, Z.; Bao, Y.; et al. Mutation of 4-coumarate: Coenzyme A ligase 1 gene affects lignin biosynthesis and increases the cell wall digestibility in maize brown midrib5 mutants. Biotechnol. Biofuels 2019, 12, 82. [Google Scholar] [PubMed] [Green Version]
- Bensen, R.J.; Johal, G.S.; Crane, V.C.; Tossberg, J.T.; Schnable, P.S.; Meeley, R.; Briggs, S.P. Cloning and characterization of the maize An1 gene. Plant Cell 1995, 7, 75–84. [Google Scholar]
- Winkler, R.G.; Helentjaris, T. The maize Dwarf3 gene encodes a cytochrome P450-mediated early step in Gibberellin biosynthesis. Plant Cell 1995, 7, 1307–1317. [Google Scholar]
- Cassani, E.; Bertolini, E.; Badone, F.C.; Landoni, M.; Gavina, D.; Sirizzotti, A.; Pilu, R. Characterization of the first dominant dwarf maize mutant carrying a single amino acid insertion in the VHYNP domain of the dwarf8 gene. Mol. Breed. 2009, 24, 375–385. [Google Scholar]
- Avila, L.M.; Cerrudo, D.; Swanton, C.; Lukens, L. Brevis plant1, a putative inositol polyphosphate 5-phosphatase, is required for internode elongation in maize. J. Exp. Bot. 2016, 67, 1577–1588. [Google Scholar]
- Xing, A.; Gao, Y.; Ye, L.; Zhang, W.; Cai, L.; Ching, A.; Llaca, V.; Johnson, B.; Liu, L.; Yang, X.; et al. A rare SNP mutation in Brachytic2 moderately reduces plant height and increases yield potential in maize. J. Exp. Bot. 2015, 66, 3791–3802. [Google Scholar]
- Multani, D.S.; Briggs, S.P.; Chamberlin, M.A.; Blakeslee, J.J.; Murphy, A.S.; Johal, G.S. Loss of an MDR Transporter in Compact Stalks of Maize br2 and Sorghum dw3 Mutants. Science 2003, 302, 81–84. [Google Scholar] [PubMed]
- Makarevitch, I.; Thompson, A.; Muehlbauer, G.J.; Springer, N.M. Brd1 Gene in Maize Encodes a Brassinosteroid C-6 Oxidase. PLoS ONE 2012, 7, e30798. [Google Scholar]
- Xiao, W.; Yang, Y.; Yu, J. ZmNST3 and ZmNST4 are master switches for secondary wall deposition in maize (Zea mays L.). Plant Sci. 2018, 266, 83–94. [Google Scholar] [PubMed]
- Sun, Q.; Liu, X.; Yang, J.; Liu, W.; Du, Q.; Wang, H.; Fu, C.; Li, W. microRNA528 Affects Lodging Resistance of Maize by Regulating Lignin Biosynthesis under Nitrogen-Luxury Conditions. Mol. Plant 2018, 11, 806–814. [Google Scholar] [PubMed] [Green Version]
- Bates, D. lme4: Linear Mixed-Effects Models Using S4 Classes; R Package Version 0.999375-37; R Foundation: Vienna, Austria, 2010; Available online: https://www.R-project.org/ (accessed on 21 February 2022).
- Yang, Q.; Zhang, D.; Xu, M. A Sequential Quantitative Trait Locus Fine-Mapping Strategy Using Recombinant-Derived Progeny. J. Integr. Plant Biol. 2012, 54, 228–237. [Google Scholar]
- Kumar, R.; Gyawali, A.; Morrison, G.D.; Saski, C.A.; Robertson, D.J.; Cook, D.D.; Tharayil, N.; Schaefer, R.J.; Beissinger, T.M.; Sekhon, R.S. Genetic architecture of maize rind strength revealed by the analysis of divergently selected populations. Plant Cell Physiol. 2021, 62, 1199–1214. [Google Scholar]
- Tan, J.; Wang, M.; Shi, Z.; Miao, X. OsEXPA10 mediates the balance between growth and resistance to biotic stress in rice. Plant Cell Rep. 2018, 37, 993–1002. [Google Scholar]
- Berthet, S.; Demont-Caulet, N.; Pollet, B.; Bidzinski, P.; Cézard, L.; Bris, P.L.; Borrega, N.; Hervé, J.; Blondet, E.; Balzergue, S.; et al. Disruption of LACCASE4 and 17 results in tissue-specific alterations to lignification of Arabidopsis thaliana stems. Plant Cell 2011, 23, 1124–1137. [Google Scholar]
- Zhao, Q.; Nakashima, J.; Chen, F.; Yin, Y.; Fu, C.; Yun, J.; Shao, H.; Wang, X.; Wang, Z.-Y.; Dixon, R.A. Laccase is necessary and nonredundant with peroxidase for lignin polymerization during vascular development in Arabidopsis. Plant Cell 2013, 25, 3976–3987. [Google Scholar]
- Shang, Q.; Zhang, D.; Li, R.; Wang, K.; Cheng, Z.; Zhou, Z.; Hao, Z.; Pan, J.; Li, X.; Shi, L. Mapping quantitative trait loci associated with stem-related traits in maize (Zea mays L.). Plant Mol. Biol. 2020, 104, 583–595. [Google Scholar]
- Yu, G.; Yumei, H.; Huan, C.; Pengshuai, Y.; Qingguo, D.; Yafei, W.; Hongqiu, W.; Zhonghua, W.; Dingming, K.; WenXue, L. Identification of traits and genes associated with lodging resistance in maize. Crop J. 2021, 9, 1408–1417. [Google Scholar]
- Zuber, M.S.; Colbert, T.R.; Darrah, L.L. Effect of Recurrent Selection for Crushing Strength on Several Stalk Components in Maize. Crop Sci. 1980, 20, 711–717. [Google Scholar]
- Mazaheri, M.; Heckwolf, M.; Vaillancourt, B.; Gage, J.L.; Burdo, B.; Heckwolf, S.; Barry, K.; Lipzen, A.; Ribeiro, C.B.; Kono, T.J.Y.; et al. Genome-wide association analysis of stalk biomass and anatomical traits in maize. BMC Plant Biol. 2019, 19, 45. [Google Scholar]
- Hartwig, T.; Chuck, G.S.; Fujioka, S.; Klempien, A.; Weizbauer, R.; Potluri, D.P.V.; Choe, S.; Johal, G.S.; Schulz, B. Brassinosteroid control of sex determination in maize. Proc. Natl. Acad. Sci. USA 2011, 108, 19814–19819. [Google Scholar] [PubMed] [Green Version]



| Trait | Env. | Parent | RIL Population | ||||||
|---|---|---|---|---|---|---|---|---|---|
| YS501 | LDC-1 | Mean ± SDa | Skew | Kurtosis | Range | CV (%) | H2 (%) | ||
| SD2 | E1 | 18.74 ± 1.35 | 21.08 ± 1.63 ** | 18.94 ± 2.40 | 0.06 | −0.32 | 13.10–25.50 | 12.67 | 63.77 |
| E2 | 17.99 ± 2.30 | 20.17 ± 1.16 * | 18.91 ± 3.09 | 0.33 | 0.50 | 10.20–28.60 | 16.35 | ||
| E3 | 14.73 ± 0.49 | 19.68 ± 1.42 ** | 14.68 ± 1.73 | 0.13 | −0.03 | 9.60–19.40 | 11.79 | ||
| E4 | 18.26 ± 1.48 | 25.10 ± 0.48 ** | 18.98 ± 2.67 | 0.10 | −0.02 | 10.80–25.90 | 14.06 | ||
| BLUP | 17.25 ± 2.25 | 20.93 ± 2.15 ** | 17.89 ± 2.88 | 0.21 | −0.30 | 11.50–27.80 | 16.07 | ||
| SD3 | E1 | 18.14 ± 1.96 | 19.82 ± 1.31 * | 17.97 ± 2.45 | −0.18 | −0.13 | 10.70–24.20 | 13.62 | 63.82 |
| E2 | 17.91 ± 1.82 | 19.68 ± 0.88 ** | 18.72 ± 3.01 | 0.43 | 0.72 | 11.00–29.40 | 16.07 | ||
| E3 | 14.17 ± 0.57 | 19.30 ± 0.78 ** | 14.91 ± 1.71 | 0.17 | 0.17 | 9.70–21.00 | 11.46 | ||
| E4 | 18.16 ± 1.28 | 24.58 ± 0.47 ** | 18.98 ± 2.69 | 0.13 | −0.02 | 11.80–26.80 | 14.17 | ||
| BLUP | 16.86 ± 2.34 | 20.23 ± 1.94 ** | 17.66 ± 2.73 | 0.30 | −0.14 | 11.10–26.80 | 15.49 | ||
| SD4 | E1 | 15.44 ± 2.00 | 16.66 ± 1.21 * | 16.11 ± 2.70 | −0.15 | −0.03 | 7.50–23.70 | 16.74 | 64.38 |
| E2 | 17.59 ± 2.21 | 19.02 ± 0.80 | 18.19 ± 3.04 | 0.51 | 1.17 | 9.80–30.60 | 16.69 | ||
| E3 | 14.53 ± 0.27 | 19.82 ± 1.31 ** | 14.87 ± 1.64 | 0.05 | −0.28 | 10.00–19.10 | 11.05 | ||
| E4 | 17.74 ± 1.31 | 23.46+ ± 0.54 ** | 18.18 ± 2.53 | 0.22 | −0.04 | 11.50–25.40 | 13.94 | ||
| BLUP | 16 ± 2.09 | 19.11 ± 2.38 ** | 16.85 ± 2.63 | 0.35 | 0.19 | 7.50–25.50 | 15.59 | ||
| RPR2 | E1 | 318.65 ± 40.5 | 382.86 ± 48.44 ** | 328.67 ± 53.30 | 0.18 | −0.21 | 189.10–456.80 | 16.22 | 83.58 |
| E2 | 318.92 ± 31.84 | 446.87 ± 48.25 ** | 392.21 ± 67.87 | 0.29 | 0.45 | 163.00–608.50 | 17.30 | ||
| E3 | 304.9 ± 19.96 | 556.19 ± 46.62 ** | 448.23 ± 73.33 | 0.80 | 0.53 | 286.20–684.20 | 16.36 | ||
| E4 | 271.8 ± 14.75 | 413.36 ± 26.92 ** | 376.89 ± 54.43 | 0.71 | 0.87 | 253.60–586.00 | 14.44 | ||
| BLUP | 308.48 ± 33.10 | 456.52 ± 84.00 ** | 386.32 ± 69.19 | 0.47 | 0.48 | 190.60–659.70 | 17.91 | ||
| RPR3 | E1 | 294.03 ± 30.18 | 322.2 ± 47.81 * | 295.71 ± 50.15 | 0.28 | 0.34 | 166.40–457.00 | 16.96 | 79.58 |
| E2 | 285.45 ± 45.04 | 395.10 ± 33.05 ** | 342.35 ± 65.89 | 0.38 | 0.36 | 152.80–546.50 | 19.25 | ||
| E3 | 278.88 ± 20.15 | 512.06 ± 34.15 ** | 397.78 ± 60.30 | 0.58 | 0.23 | 254.30–605.80 | 15.16 | ||
| E4 | 249.48 ± 15.37 | 372.32 ± 26.40 ** | 319.12 ± 45.51 | 0.49 | 0.33 | 216.10–481.10 | 14.26 | ||
| BLUP | 281.45 ± 32.82 | 405.74 ± 85.33 ** | 306.87 ± 53.85 | 0.61 | 0.77 | 179.10–488.70 | 17.55 | ||
| RPR4 | E1 | 259.65 ± 42.31 | 245.94 ± 41.18 | 271.05 ± 46.40 | 0.29 | 0.31 | 129.70–420.50 | 17.12 | 78.10 |
| E2 | 225.74 ± 31.21 | 356.59 ± 22.67 ** | 302.01 ± 60.10 | 0.71 | 0.65 | 167.60–519.70 | 19.90 | ||
| E3 | 252.16 ± 14.50 | 464.58 ± 33.61 ** | 351.32 ± 53.40 | 0.51 | 0.60 | 225.30–548.40 | 15.20 | ||
| E4 | 232.22 ± 12.87 | 330.78 ± 18.68 ** | 277.64 ± 38.22 | 0.40 | 0.13 | 180.10–401.30 | 13.77 | ||
| BLUP | 245.13 ± 32.10 | 357.59 ± 86.80 ** | 300.71 ± 54.43 | 0.65 | 0.56 | 157.50–519.10 | 18.10 | ||
| Trait | QTL | Chr | Env. | LOD | PVE (%) | A | Peak (cM) | Interval (CM) | Peak (Mb) | Physical Location (Mb) |
|---|---|---|---|---|---|---|---|---|---|---|
| SD2 | q2SD2-1 | 2 | E1 | 3.42 | 6.86 | −0.60 | 182.61 | 181.90–190.60 | 215.37 | 215.16–220.88 |
| BLUP | 2.82 | 4.89 | −0.22 | 194.51 | 187.40–198.10 | 224.43 | 217.21–225.33 | |||
| q2SD3-1 | 3 | BLUP | 3.07 | 4.82 | 0.21 | 150.31 | 148.50–152.00 | 182.51 | 181.14–183.88 | |
| q2SD5-1 | 5 | E2 | 3.49 | 7.16 | −0.65 | 294.31 | 291.80–301.20 | 220.94 | 220.49–221.52 | |
| q2SD6-1 | 6 | BLUP | 3.24 | 5.14 | 0.22 | 101.91 | 101.10–104.90 | 144.58 | 143.82–151.51 | |
| q2SD6-2 | 6 | E4 | 2.68 | 5.06 | 0.53 | 135.81 | 135.20–136.90 | 162.43 | 161.55–162.81 | |
| q2SD7-1 | 7 | E2 | 2.58 | 5.17 | −0.55 | 56.51 | 56.00–57.00 | 8.76 | 8.52–9.14 | |
| q2SD7-2 | 7 | E3 | 3.13 | 5.38 | 0.35 | 68.81 | 66.20–70.10 | 15.41 | 13.64–18.33 | |
| q2SD7-3 | 7 | BLUP | 4.86 | 7.93 | 0.27 | 167.21 | 165.50–170.50 | 161.42 | 159.78–162.19 | |
| q2SD7-4 | 7 | E4 | 3.28 | 6.25 | 0.61 | 236.51 | 234.50–238.80 | 177.96 | 177.67–178.65 | |
| BLUP | 3.64 | 5.93 | 0.24 | 236.51 | 233.00–246.90 | 177.96 | 178.65–180.75 | |||
| q2SD8-1 | 8 | E4 | 2.84 | 4.19 | 0.20 | 141.81 | 138.70–143.80 | 162.92 | 161.26–164.25 | |
| q2SD9-1 | 9 | E1 | 6.51 | 13.65 | −1.16 | 136.31 | 135.60–139.10 | 125.77 | 121.73–129.87 | |
| E2 | 3.16 | 6.44 | −0.61 | 129.41 | 128.00–137.20 | 117.68 | 117.07–127.65 | |||
| E3 | 9.69 | 18.09 | −0.63 | 136.31 | 129.20–139.90 | 125.77 | 117.68–130.34 | |||
| BLUP | 6.03 | 9.99 | −0.31 | 136.31 | 129.00–139.00 | 125.77 | 117.07–129.87 | |||
| q2SD10-1 | 10 | E3 | 2.83 | 4.85 | 0.34 | 83.61 | 83.60–84.70 | 133.44 | 133.44–138.54 | |
| SD3 | q3SD2-1 | 2 | E1 | 3.13 | 6.16 | −0.59 | 195.51 | 191.60–199.90 | 224.43 | 221.4–226.88 |
| q3SD3-1 | 3 | E1 | 2.92 | 5.46 | 0.56 | 72.01 | 68.80–81.40 | 30.98 | 25.35–45.72 | |
| q3SD3-2 | 3 | E2 | 4.48 | 9.26 | 0.77 | 122.01 | 120.80–122.80 | 167.66 | 164.41–168.35 | |
| q3SD5-1 | 5 | E2 | 3.15 | 6.32 | −0.60 | 299.41 | 293.80–301.20 | 221.36 | 220.49–221.52 | |
| q3SD6-1 | 6 | E3 | 3.70 | 5.98 | 0.36 | 103.81 | 99.10–108.40 | 149.26 | 141.26–152.61 | |
| BLUP | 3.34 | 5.29 | 0.22 | 103.81 | 101.10–104.90 | 149.26 | 143.82–151.51 | |||
| q3SD7-1 | 7 | E3 | 3.35 | 5.39 | 0.34 | 68.81 | 66.00–70.80 | 15.41 | 13.64–21.49 | |
| q3SD7-2 | 7 | E1 | 3.37 | 6.31 | 0.59 | 167.21 | 161.30–170.50 | 161.42 | 158.52–162.19 | |
| BLUP | 5.30 | 8.66 | 0.29 | 167.21 | 165.40–170.30 | 161.42 | 159.78–162.19 | |||
| q3SD7-3 | 7 | E4 | 3.39 | 6.34 | 0.61 | 222.21 | 221.50–225.00 | 175.02 | 175.63–176.82 | |
| q3SD8-1 | 8 | E1 | 3.36 | 6.34 | 0.61 | 119.31 | 118.80–130.60 | 145.98 | 145.83–154.57 | |
| E4 | 2.92 | 5.46 | 0.55 | 141.71 | 137.90–144.40 | 162.92 | 161.02–164.25 | |||
| BLUP | 3.29 | 6.17 | 0.59 | 128.71 | 126.60–142.70 | 150.93 | 150.21–163.87 | |||
| q3SD9-1 | 9 | E1 | 4.53 | 8.42 | −0.68 | 100.81 | 90.40–110.40 | 85.54 | 36.17–103.21 | |
| E4 | 3.78 | 7.14 | −0.65 | 101.41 | 91.20–110.20 | 92.35 | 38.64–103.21 | |||
| BLUP | 5.09 | 8.14 | −0.35 | 100.81 | 91.00–108.60 | 85.54 | 37.79–100.82 | |||
| q3SD9-2 | 9 | E2 | 4.06 | 8.18 | −0.68 | 136.31 | 128.40–139.00 | 125.77 | 117.07–129.87 | |
| E3 | 9.70 | 16.84 | −0.60 | 135.81 | 124.80–146.20 | 124.08 | 114.3–131.5 | |||
| BLUP | 3.97 | 6.67 | −0.42 | 137.11 | 136.40–139.70 | 127 | 125.77–129.87 | |||
| q3SD10-1 | 10 | E3 | 3.12 | 5.01 | 0.33 | 40.31 | 39.30–42.90 | 8.79 | 5.9–10.42 | |
| SD4 | q4SD2-1 | 2 | E1 | 5.10 | 9.59 | −0.77 | 25.71 | 23.10–31.20 | 4.28 | 3.88–6.16 |
| q4SD3-1 | 3 | E4 | 2.82 | 4.76 | −0.51 | 21.31 | 20.30–22.30 | 3.14 | 3.09–4.76 | |
| q4SD3-2 | 3 | E1 | 2.76 | 5.47 | 0.56 | 72.01 | 71.80–80.50 | 30.98 | 29.43–44.15 | |
| q4SD3-3 | 3 | BLUP | 3.29 | 5.68 | 0.24 | 85.91 | 81.90–88.90 | 55.4 | 45.73–102.71 | |
| q4SD3-4 | 3 | E4 | 2.70 | 4.94 | 0.50 | 124.21 | 123.50–125.10 | 168.35 | 167.66–168.77 | |
| q4SD5-1 | 5 | E2 | 3.05 | 7.13 | −0.60 | 294.31 | 281.80–300.90 | 220.94 | 218.52–221.52 | |
| q4SD6-1 | 6 | E3 | 3.63 | 5.41 | 0.33 | 103.81 | 99.60–108.70 | 149.26 | 141.26–152.61 | |
| q4SD7-1 | 7 | E3 | 3.59 | 5.33 | 0.33 | 68.81 | 65.30–70.20 | 15.41 | 13.64–21.49 | |
| q4SD7-2 | 7 | E4 | 3.24 | 5.92 | 0.55 | 167.21 | 166.50–169.60 | 161.42 | 160.9–162.19 | |
| BLUP | 5.86 | 9.70 | 0.31 | 167.21 | 159.10–171.10 | 161.42 | 157.09–162.19 | |||
| q4SD7-3 | 7 | E4 | 3.25 | 5.88 | 0.54 | 222.21 | 221.30–229.60 | 175.02 | 175.63–179.03 | |
| q4SD9-1 | 9 | E4 | 5.62 | 10.44 | −0.73 | 101.41 | 97.60–113.60 | 91.64 | 70.77–105.86 | |
| BLUP | 3.36 | 5.32 | −0.27 | 100.81 | 99.20–105.80 | 85 | 82.57–94.9 | |||
| q4SD9-2 | 9 | E2 | 5.60 | 11.55 | −0.82 | 136.31 | 128.60–146.60 | 125.77 | 117.07–131.5 | |
| E3 | 12.98 | 21.72 | −0.66 | 136.31 | 124.50–146.40 | 125.77 | 114.3–131.5 | |||
| BLUP | 2.91 | 4.62 | −0.26 | 135.81 | 135.00–138.80 | 122.42 | 121.73–129 | |||
| q4SD10-1 | 10 | E3 | 4.45 | 6.69 | 0.37 | 83.61 | 80.00–90.50 | 133.44 | 126.32–138.54 | |
| RPR2 | q2RPR1-1 | 1 | BLUP | 2.78 | 4.03 | 7.52 | 240.71 | 238.90–241.30 | 197.5 | 194.7–197.84 |
| q2RPR1-2 | 1 | E2 | 5.40 | 9.80 | 17.25 | 259.41 | 251.20–264.00 | 211.12 | 202.08–214.25 | |
| q2RPR1-3 | 1 | E1 | 5.05 | 9.04 | 14.85 | 275.21 | 274.60–280.60 | 220.78 | 219.09–225.03 | |
| BLUP | 2.99 | 4.31 | 7.70 | 275.81 | 274.40–278.50 | 222.27 | 219.09–223.6 | |||
| q2RPR2-1 | 2 | E2 | 3.83 | 7.66 | 15.58 | 69.41 | 63.10–73.70 | 16.39 | 12.57–19.34 | |
| E3 | 2.76 | 4.92 | 17.55 | 72.51 | 64.90–73.70 | 18.76 | 12.57–19.34 | |||
| E4 | 4.88 | 8.94 | 14.17 | 65.01 | 61.70–70.90 | 15.01 | 12.57–18.31 | |||
| BLUP | 7.55 | 12.38 | 13.24 | 65.01 | 61.40–72.90 | 15.01 | 12.57–19.34 | |||
| q2RPR2-2 | 2 | E1 | 3.08 | 5.42 | 11.55 | 99.11 | 96.80–102.50 | 43.98 | 41.24–55.85 | |
| q2RPR3-1 | 3 | E1 | 2.92 | 5.17 | 11.61 | 71.11 | 70.10–72.00 | 29.43 | 27.55–33.2 | |
| BLUP | 7.15 | 11.71 | 13.19 | 71.11 | 60.60–80.90 | 29.43 | 16.96–45.43 | |||
| q2RPR3-2 | 3 | E2 | 6.51 | 11.72 | 20.04 | 105.91 | 94.60–115.10 | 153.57 | 128.63–163.19 | |
| E4 | 3.78 | 6.91 | 12.70 | 103.11 | 93.10–109.40 | 149.05 | 119.21–158.12 | |||
| q2RPR4-1 | 4 | E3 | 2.76 | 5.07 | 17.37 | 192.61 | 192.00–193.60 | 231.9 | 231.9–235.51 | |
| q2RPR5-1 | 5 | E1 | 4.29 | 7.65 | −13.71 | 89.81 | 85.90–101.70 | 26.06 | 19.66–36.42 | |
| q2RPR5-2 | 5 | E2 | 6.37 | 11.37 | −18.58 | 128.91 | 122.70–139.20 | 82.97 | 76.22–145.91 | |
| BLUP | 5.27 | 8.40 | −10.97 | 128.91 | 119.00–138.90 | 82.97 | 68.75–145.91 | |||
| q2RPR6-1 | 6 | E4 | 2.76 | 4.93 | −10.49 | 96.31 | 90.20–97.50 | 136.98 | 133.53–141.26 | |
| q2RPR6-2 | 6 | BLUP | 3.47 | 5.41 | −8.76 | 134.71 | 132.70–137.00 | 161.55 | 160.9–162.81 | |
| q2RPR7-1 | 7 | E4 | 3.16 | 5.66 | 11.16 | 159.21 | 156.00–160.40 | 157.7 | 155–158.52 | |
| BLUP | 4.76 | 7.61 | 10.26 | 158.51 | 147.50–166.30 | 157.7 | 151.21–160.9 | |||
| q2RPR8-1 | 8 | E3 | 3.24 | 6.39 | 19.68 | 174.31 | 171.10–177.20 | 173.03 | 171.99–174.33 | |
| q2RPR9-1 | 9 | BLUP | 2.89 | 4.17 | 7.64 | 5.31 | 1.80–7.20 | 4.44 | 1.84–5.08 | |
| q2RPR10-1 | 10 | E3 | 3.68 | 6.71 | 20.69 | 60.61 | 56.20–67.30 | 39.07 | 19.92–90.13 | |
| q2RPR10-2 | 10 | E4 | 2.68 | 4.83 | 10.47 | 129.81 | 128.00–130.50 | 145.97 | 145.56–146.56 | |
| RPR3 | q3RPR1-1 | 1 | E1 | 4.90 | 8.93 | 13.25 | 239.11 | 228.90–250.90 | 195.09 | 186.38–203.7 |
| E2 | 4.18 | 8.05 | 15.39 | 253.51 | 237.00–256.00 | 204.96 | 193.34–208.08 | |||
| BLUP | 2.74 | 4.24 | 6.41 | 240.41 | 240.10–247.00 | 197.05 | 195.68–201.74 | |||
| q3RPR2-1 | 2 | E2 | 6.27 | 12.95 | 22.55 | 65.01 | 61.00–68.90 | 15.01 | 15.01–18.31 | |
| E3 | 7.25 | 12.47 | 17.81 | 65.01 | 61.50–73.70 | 15.01 | 12.57–19.34 | |||
| E4 | 6.34 | 11.63 | 12.85 | 65.01 | 61.90–68.60 | 15.01 | 12.57–18.31 | |||
| BLUP | 10.19 | 17.25 | 12.91 | 65.01 | 54.00–75.70 | 15.01 | 11.17–19.71 | |||
| q3RPR2-2 | 2 | E1 | 3.07 | 5.58 | 10.41 | 99.71 | 98.70–104.40 | 49.26 | 43.54–59.84 | |
| q3RPR3-1 | 3 | E2 | 4.04 | 7.75 | 16.32 | 65.51 | 64.60–73.50 | 22.67 | 21.00–32.39 | |
| BLUP | 4.60 | 7.27 | 8.58 | 71.11 | 65.60–76.50 | 29.43 | 27.55–37.81 | |||
| q3RPR3-2 | 3 | E3 | 4.36 | 7.29 | 13.85 | 88.41 | 78.00–98.30 | 63.86 | 38.78–140.85 | |
| q3RPR4-1 | 4 | E3 | 2.69 | 4.41 | 10.66 | 188.61 | 187.90–189.40 | 230.7 | 226.19–231.58 | |
| q3RPR5-1 | 5 | E1 | 5.30 | 9.80 | −13.83 | 104.51 | 93.40–107.40 | 41.98 | 31.81–51.91 | |
| q3RPR5-2 | 5 | E2 | 3.73 | 7.03 | −14.33 | 128.41 | 118.40–133.50 | 82.99 | 68.35–93.1 | |
| BLUP | 2.84 | 4.11 | −6.29 | 128.91 | 126.50–130.60 | 82.97 | 70.34–88.29 | |||
| q3RPR6-1 | 6 | E1 | 2.94 | 5.31 | −10.28 | 58.81 | 58.30–59.60 | 95.99 | 95.86–98.07 | |
| q3RPR6-2 | 6 | E3 | 2.95 | 4.85 | −11.15 | 134.71 | 133.70–136.00 | 161.55 | 160.9–162.81 | |
| BLUP | 5.64 | 8.94 | −9.27 | 135.81 | 130.60–144.80 | 162.43 | 159.79–165.36 | |||
| q3RPR7-1 | 7 | E3 | 4.38 | 7.33 | 13.50 | 159.21 | 156.00–171.40 | 157.7 | 155.00–162.19 | |
| q3RPR9-1 | 9 | BLUP | 2.80 | 4.34 | 6.45 | 5.31 | 2.70–7.20 | 4.44 | 3.99–5.08 | |
| q3RPR10-1 | 10 | E3 | 2.94 | 4.50 | 10.89 | 60.61 | 56.00–65.70 | 39.07 | 17.92–86.46 | |
| q3RPR10-2 | 10 | E4 | 2.73 | 5.07 | 8.56 | 127.31 | 126.20–130.10 | 145.56 | 145.47–146.56 | |
| RPR4 | q4RPR1-1 | 1 | E2 | 3.51 | 6.38 | 12.49 | 260.21 | 250.90–265.10 | 210.46 | 203.7–214.51 |
| q4RPR1-2 | 1 | BLUP | 3.60 | 5.07 | 6.14 | 275.81 | 274.10–282.10 | 219.76 | 219.09–227.88 | |
| q4RPR1-3 | 1 | E3 | 3.37 | 5.38 | 10.36 | 327.21 | 317.40–330.20 | 262.09 | 255.17–263.52 | |
| q4RPR2-1 | 2 | E2 | 4.57 | 8.57 | 14.76 | 65.11 | 61.50–70.00 | 15.01 | 12.57–18.31 | |
| E3 | 5.38 | 8.92 | 13.50 | 65.01 | 60.10–71.20 | 15.01 | 12.57–18.31 | |||
| E4 | 6.29 | 11.88 | 11.59 | 65.01 | 61.20–73.50 | 15.01 | 12.57–19.34 | |||
| BLUP | 8.57 | 14.11 | 10.34 | 65.01 | 57.00–75.70 | 15.01 | 12.24–19.71 | |||
| q4RPR2-2 | 2 | E1 | 4.86 | 9.37 | 12.84 | 99.71 | 98.70–104.30 | 49.26 | 43.54–57.89 | |
| q4RPR3-1 | 3 | E1 | 3.95 | 7.54 | 11.86 | 88.41 | 77.70–90.30 | 63.86 | 38.78–114.25 | |
| E2 | 4.74 | 8.73 | 15.55 | 71.11 | 65.10–81.40 | 29.43 | 21.00–54.00 | |||
| BLUP | 5.50 | 8.66 | 8.24 | 89.51 | 80.90–99.80 | 108.5 | 44.15–142.88 | |||
| q4RPR3-2 | 3 | E3 | 4.28 | 6.94 | 11.70 | 130.41 | 124.50–135.60 | 172.48 | 168.35–175.09 | |
| q4RPR4-1 | 4 | E3 | 2.58 | 3.83 | 8.77 | 42.21 | 42.10–42.50 | 10.45 | 6.09–10.76 | |
| q4RPR5-1 | 5 | E2 | 5.26 | 9.51 | −15.32 | 128.91 | 121.90–132.70 | 82.97 | 72.79–95.1 | |
| q4RPR6-1 | 6 | E1 | 3.29 | 6.11 | −10.30 | 123.41 | 117.60–125.30 | 158.32 | 155.77–159.37 | |
| E3 | 3.27 | 5.13 | −10.30 | 116.41 | 112.80–122.90 | 155.77 | 153.76–158.32 | |||
| q4RPR6-2 | 6 | BLUP | 5.08 | 7.94 | −7.69 | 135.81 | 127.80–137.80 | 162.43 | 159.53–162.81 | |
| q4RPR7-1 | 7 | E4 | 3.38 | 6.09 | 8.35 | 146.81 | 142.60–148.20 | 151.21 | 148.97–152.57 | |
| q4RPR7-2 | 7 | E3 | 3.20 | 4.71 | 9.63 | 159.21 | 158.00–169.20 | 157.7 | 157.09–162.19 | |
| q4RPR9-1 | 9 | E3 | 3.09 | 4.94 | 9.82 | 5.31 | 2.50–12.50 | 4.44 | 1.84–6.79 | |
| BLUP | 3.79 | 5.91 | 6.55 | 2.41 | 1.20–8.80 | 3.99 | 2.79–6.69 | |||
| q4RPR9-2 | 9 | E1 | 3.77 | 7.05 | −11.06 | 96.91 | 87.10–106.80 | 70.77 | 30.77–96.81 | |
| q4RPR10-1 | 10 | E3 | 5.57 | 9.18 | 13.77 | 60.61 | 56.40–67.10 | 39.07 | 19.92–90.13 | |
| BLUP | 2.84 | 4.34 | 5.72 | 62.81 | 59.00–63.10 | 72.23 | 27.25–76.55 | |||
| q4RPR10-2 | 10 | E1 | 2.62 | 4.58 | 8.96 | 129.81 | 129.70–130.60 | 145.97 | 145.97–146.56 | |
| E4 | 3.70 | 7.10 | 9.15 | 128.31 | 125.30–132.30 | 145.97 | 145.22–146.56 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, H.; Wang, H.; Shao, C.; Han, Y.; He, Y.; Yin, Z. Genetic Architecture of Maize Stalk Diameter and Rind Penetrometer Resistance in a Recombinant Inbred Line Population. Genes 2022, 13, 579. https://doi.org/10.3390/genes13040579
Liu H, Wang H, Shao C, Han Y, He Y, Yin Z. Genetic Architecture of Maize Stalk Diameter and Rind Penetrometer Resistance in a Recombinant Inbred Line Population. Genes. 2022; 13(4):579. https://doi.org/10.3390/genes13040579
Chicago/Turabian StyleLiu, Huanhuan, Huan Wang, Cong Shao, Youle Han, Yonghui He, and Zhitong Yin. 2022. "Genetic Architecture of Maize Stalk Diameter and Rind Penetrometer Resistance in a Recombinant Inbred Line Population" Genes 13, no. 4: 579. https://doi.org/10.3390/genes13040579
APA StyleLiu, H., Wang, H., Shao, C., Han, Y., He, Y., & Yin, Z. (2022). Genetic Architecture of Maize Stalk Diameter and Rind Penetrometer Resistance in a Recombinant Inbred Line Population. Genes, 13(4), 579. https://doi.org/10.3390/genes13040579






