Variable Clinical Appearance of the Kir2.1 Rare Variants in Russian Patients with Long QT Syndrome
Abstract
1. Introduction
2. Materials and Methods
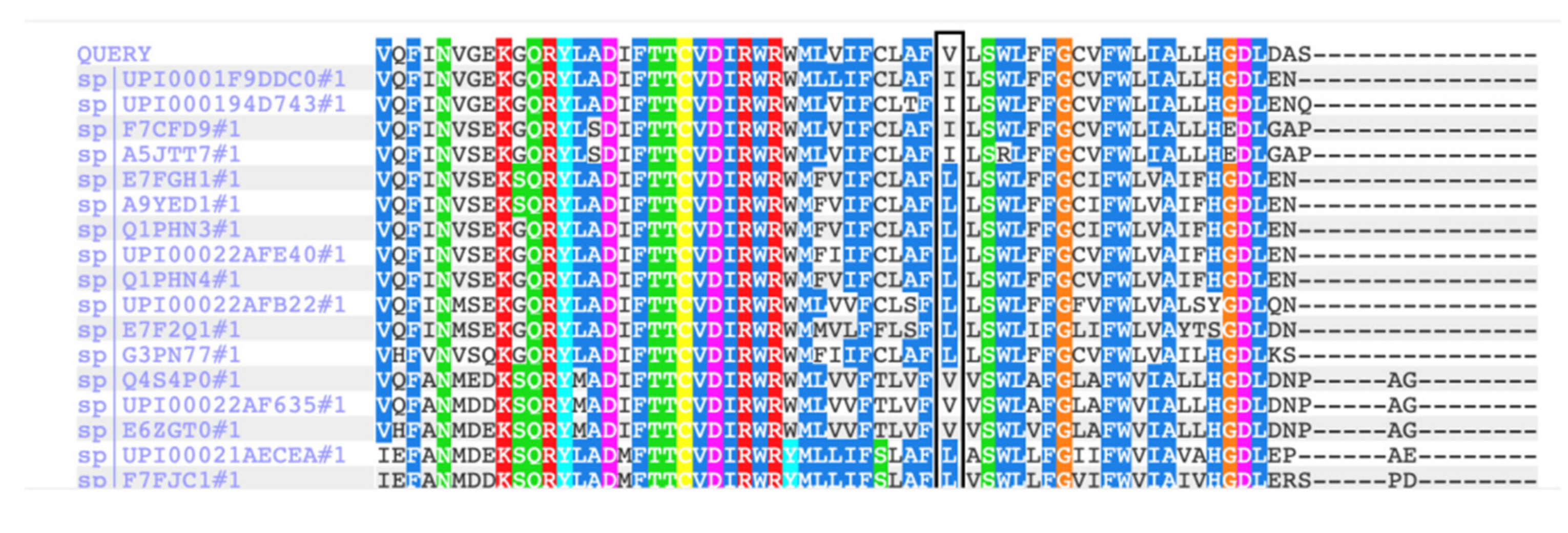
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Xia, M.; Jin, Q.; Bendahhou, S.; He, Y.; Larroque, M.; Chen, Y.; Zhou, Q.; Yang, Y.; Liu, Y.; Liu, B.; et al. A Kir2.1 gain-of-function mutation underlies familial atrial fibrillation. Biochem. Biophys. Res. Commun. 2005, 332, 1012–1019. [Google Scholar] [CrossRef]
- Wilde, A.A.M.; Amin, A.S.; Postema, P.G. Diagnosis, management and therapeutic strategies for congenital long QT syndrome. Heart 2022, 108, 332–338. [Google Scholar] [CrossRef] [PubMed]
- OMIM: On-Line Mendelian Inheritance in Man. Available online: https://omim.org/entry/600681?search=KCNJ2&highlight=kcnj2 (accessed on 31 January 2022).
- Tristani-Firouzi, M.; Jensen, J.L.; Donaldson, M.R.; Sansone, V.; Meola, G.; Hahn, A.; Bendahhou, S.; Kwiecinski, H.; Fidzianska, A.; Plaster, N.; et al. Functional and clinical characterization of KCNJ2 mutations associated with LQT7 (Andersen syndrome). J. Clin. Investig. 2002, 110, 381–388. [Google Scholar] [CrossRef] [PubMed]
- Zangerl-Plessl, E.-M.; Qile, M.; Bloothooft, M.; Stary-Weinzinger, A.; van der Heyden, M.A.G. Disease Associated Mutations in KIR Proteins Linked to Aberrant Inward Rectifier Channel Trafficking. Biomolecules 2019, 9, 650. [Google Scholar] [CrossRef]
- Adams, D.S.; Uzel, S.G.; Akagi, J.; Wlodkowic, D.; Andreeva, V.; Yelick, P.C.; Devitt-Lee, A.; Pare, J.F.; Levin, M. Bioelectric signalling via potassium channels: A mechanism for craniofacial dysmorphogenesis in KCNJ2-associated Andersen-Tawil Syndrome. J. Physiol. 2016, 594, 3245–3270. [Google Scholar] [CrossRef] [PubMed]
- UniProtKB: UniProt Knowledge Database. Available online: https://www.uniprot.org/uniprot/Q14524 (accessed on 31 January 2022).
- Plaster, N.M.; Tawil, R.; Tristani-Firouzi, M.; Canún, S.; Bendahhou, S.; Tsunoda, A.; Donaldson, M.R.; Iannaccone, S.T.; Brunt, E.; Barohn, R.; et al. Mutations in Kir2.1 cause the developmental and episodic electrical phenotypes of Andersen’s syndrome. Cell 2001, 105, 511–519. [Google Scholar] [CrossRef]
- Priori, S.G.; Pandit, S.V.; Rivolta, I.; Berenfeld, O.; Ronchetti, E.; Dhamoon, A.; Napolitano, C.; Anumonwo, J.; di Barletta, M.R.; Gudapakkam, S.; et al. A novel form of short QT syndrome (SQT3) is caused by a mutation in the KCNJ2 gene. Circ. Res. 2005, 96, 800–807. [Google Scholar] [CrossRef]
- Barrón-Díaz, D.R.; Totomoch-Serra, A.; Escobar-Cedillo, R.E.; García-Gutierrez, A.; Reyes-Quintero, Á.E.; Villegas Davirán, S.E.; Ibarra-Miramón, C.B.; Márquez, M.F. Andersen-Tawil Syndrome with High Risk of Sudden Cardiac Death in Four Mexican Patients. Cardiac and Extra-Cardiac Phenotypes. Rev. Investig. Clin. 2020, 73, 145–153. [Google Scholar] [CrossRef]
- Vivekanandam, V.; Männikkö, R.; Skorupinska, I.; Germain, L.; Gray, B.; Wedderburn, S.; Kozyra, D.; Sud, R.; James, N.; Holmes, S.; et al. Andersen-Tawil syndrome: Deep phenotyping reveals significant cardiac and neuromuscular morbidity. Brain 2021. Epub ahead of print. [Google Scholar] [CrossRef]
- Priori, S.G.; Blomström-Lundqvist, C.; Mazzanti, A.; Blom, N.; Borggrefe, M.; Camm, J.; Elliott, P.M.; Fitzsimons, D.; Hatala, R.; Hindricks, G.; et al. 2015 ESC Guidelines for the management of patients with ventricular arrhythmias and the prevention of sudden cardiac death: The Task Force for the Management of Patients with Ventricular Arrhythmias and the Prevention of Sudden Cardiac Death of the European Society of Cardiology (ESC). Endorsed by: Association for European Paediatric and Congenital Cardiology (AEPC). Eur. Heart J. 2015, 36, 2793–2867. [Google Scholar] [CrossRef]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–423. [Google Scholar] [CrossRef] [PubMed]
- Polyak, M.; Shestak, A.; Podolyak, D.; Zaklyazminskaya, E. Compound heterozygous mutations in KCNJ2 and KCNH2 in a patient with severe Andersen-Tawil syndrome. BMJ Case Rep. 2020, 13, e235703. [Google Scholar] [CrossRef]
- Walsh, R.; Lahrouchi, N.; Tadros, R.; Kyndt, F.; Glinge, C.; Postema, P.; Amin, A.; Nannenberg, E.; Ware, J.; Whiffin, N.; et al. Enhancing rare variant interpretation in inherited arrhythmias through quantitative analysis of consortium disease cohorts and population controls. Genet. Med. 2021, 23, 47–58. [Google Scholar] [CrossRef] [PubMed]
- Obeyesekere, M.; Klein, G.; Modi, S.; Leong-Sit, P.; Gula, L.; Yee, R.; Skanes, A.; Krahn, A. How to Perform and Interpret Provocative Testing for the Diagnosis of Brugada Syndrome, Long-QT Syndrome, and Catecholaminergic Polymorphic Ventricular Tachycardia. Circ. Arrhythm. Electrophysiol. 2011, 4, 958–964. [Google Scholar] [CrossRef] [PubMed]
- Olesen, M.; Nielsen, M.; Haunsø, S.; Svendsen, J. Atrial fibrillation: The role of common and rare genetic variants. Eur. J. Hum. Genet. 2014, 22, 297–306. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Glukhov, G.; Pustovit, K.; Kacher, Y.; Rusinova, V.; Kiseleva, I.; Komolyatova, V.N.; Makarov, L.M.; Zaklyazminskaya, E.V.; Sokolova, O.S. Phenotypic manifestations of Val93Ile missense mutation and its influence on Kir2.1 channel functioning. Moscow Univ. Biol. Sci. Bull. 2021, 76, 142–146. [Google Scholar] [CrossRef]
- Veerapandiyan, A.; Statland, J.; Tawil, R. Andersen-Tawil Syndrome. 2004 Nov 22 [Updated 2018 Jun 7]; Adam, M., Ardinger, H., Pagon, R., Eds.; GeneReviews® [Internet]; University of Washington: Seattle, WA, USA, 1993–2022. Available online: https://www.ncbi.nlm.nih.gov/books/NBK1264/ (accessed on 31 January 2022).
- Eckhardt, L.; Farley, A.; Rodriguez, E.; Ruwaldt, K.; Hammill, D.; Tester, D.; Ackerman, M.; Makielski, J. KCNJ2 mutations in arrhythmia patients referred for LQT testing: A mutation T305A with novel effect on rectification properties. Heart Rhythm. 2007, 4, 323–329. [Google Scholar] [CrossRef]
- Burns, C.; Ingles, J.; Davis, A.; Connell, V.; Gray, B.; Hunt, L.; McGaughran, J.; Semsarian, C. Clinical and genetic features of Australian families with long QT syndrome: A registry-based study. J. Arrhythm. 2016, 32, 456–461. [Google Scholar] [CrossRef]
- Schwartz, P.J.; Stramba-Badiale, M.; Crotti, L.; Pedrazzini, M.; Besana, A.; Bosi, G.; Gabbarini, F.; Goulene, K.; Insolia, R.; Mannarino, S.; et al. Prevalence of the congenital long-QT syndrome. Circulation 2009, 120, 1761–1767. [Google Scholar] [CrossRef]
- Maltese, P.E.; Orlova, N.; Krasikova, E.; Emelyanchik, E.; Cheremisina, A.; Kuscaeva, A.; Salmina, A.; Miotto, R.; Bonizzato, A.; Guerri, G.; et al. Gene-Targeted Analysis of Clinically Diagnosed Long QT Russian Families. Int. Heart J. 2017, 58, 81–87. [Google Scholar] [CrossRef][Green Version]
- Reilly, L.; Eckhardt, L.L. Cardiac potassium inward rectifier Kir2: Review of structure, regulation, pharmacology, and arrhythmogenesis. Heart Rhythm. 2021, 18, 1423–1434. [Google Scholar] [CrossRef] [PubMed]
- Kimura, H.; Zhou, J.; Kawamura, M.; Itoh, H.; Mizusawa, Y.; Ding, W.G.; Wu, J.; Ohno, S.; Makiyama, T.; Miyamoto, A.; et al. Phenotype variability in patients carrying KCNJ2 mutations. Circ. Cardiovasc. Genet. 2012, 5, 344–353. [Google Scholar] [CrossRef] [PubMed]
- Krych, M.; Biernacka, E.K.; Ponińska, J.; Kukla, P.; Filipecki, A.; Gajda, R.; Hasdemir, C.; Antzelevitch, C.; Kosiec, A.; Szperl, M.; et al. Andersen-Tawil syndrome: Clinical presentation and predictors of symptomatic arrhythmias-Possible role of polymorphisms K897T in KCNH2 and H558R in SCN5A gene. J. Cardiol. 2017, 70, 504–510. [Google Scholar] [CrossRef] [PubMed]
- Bates, E.A. A potential molecular target for morphological defects of fetal alcohol syndrome: Kir2.1. Curr. Opin. Genet. Dev. 2013, 23, 324–329. [Google Scholar] [CrossRef]
- Locus Specific Mutation Databases. Available online: https://grenada.lumc.nl/LSDB_list/lsdbs/KCNJ2 (accessed on 31 January 2022).
- Hu, D.; Barajas-Martinez, H.; Burashnikov, E.; Springer, M.; Wu, Y.; Varro, A.; Pfeiffer, R.; Koopmann, T.T.; Cordeiro, J.M.; Guerchicoff, A.; et al. A mutation in the beta 3 subunit of the cardiac sodium channel associated with Brugada ECG phenotype. Circ. Cardiovasc. Genet. 2009, 2, 270–278. [Google Scholar] [CrossRef]
- Lau, K.; Martin, P.; Chia, K.; Mcgaughran, J.; Atherton, J. Long QT Syndrome Associated With a Mutation in the β3 Subunit of the Cardiac Sodium Channel (SCN3B) Gene in Two Siblings. Heart Lung Circ. 2016, 25, S284–S285. [Google Scholar] [CrossRef]
- Costa, S.; Medeiros-Domingo, A.; Gasperetti, A.; Akdis, D.; Berger, W.; James, C.A.; Ruschitzka, F.; Brunckhorst, C.B.; Duru, F.; Saguner, A.M. Impact of Genetic Variant Reassessment on the Diagnosis of Arrhythmogenic Right Ventricular Cardiomyopathy Based on the 2010 Task Force Criteria. Circ. Genom. Precis. Med. 2021, 14, e003047. [Google Scholar] [CrossRef]
- Norambuena, P.A.; Tomasov, P.; Bonaventura, J.; Geryk, J.; Veselka, J.; Macek, M. 4291 The importance of variant re-evaluation in massive parallel sequencing: Results from hypertrophic cardiomyopathy genetic testing as an example. Eur. Heart J. 2018, 39, 866–867. [Google Scholar] [CrossRef][Green Version]
- Giudicessi, J.R.; Roden, D.M.; Wilde, A.A.M.; Ackerman, M.J. Classification and Reporting of Potentially Proarrhythmic Common Genetic Variation in Long QT Syndrome Genetic Testing. Circulation 2018, 137, 619–630. [Google Scholar] [CrossRef]
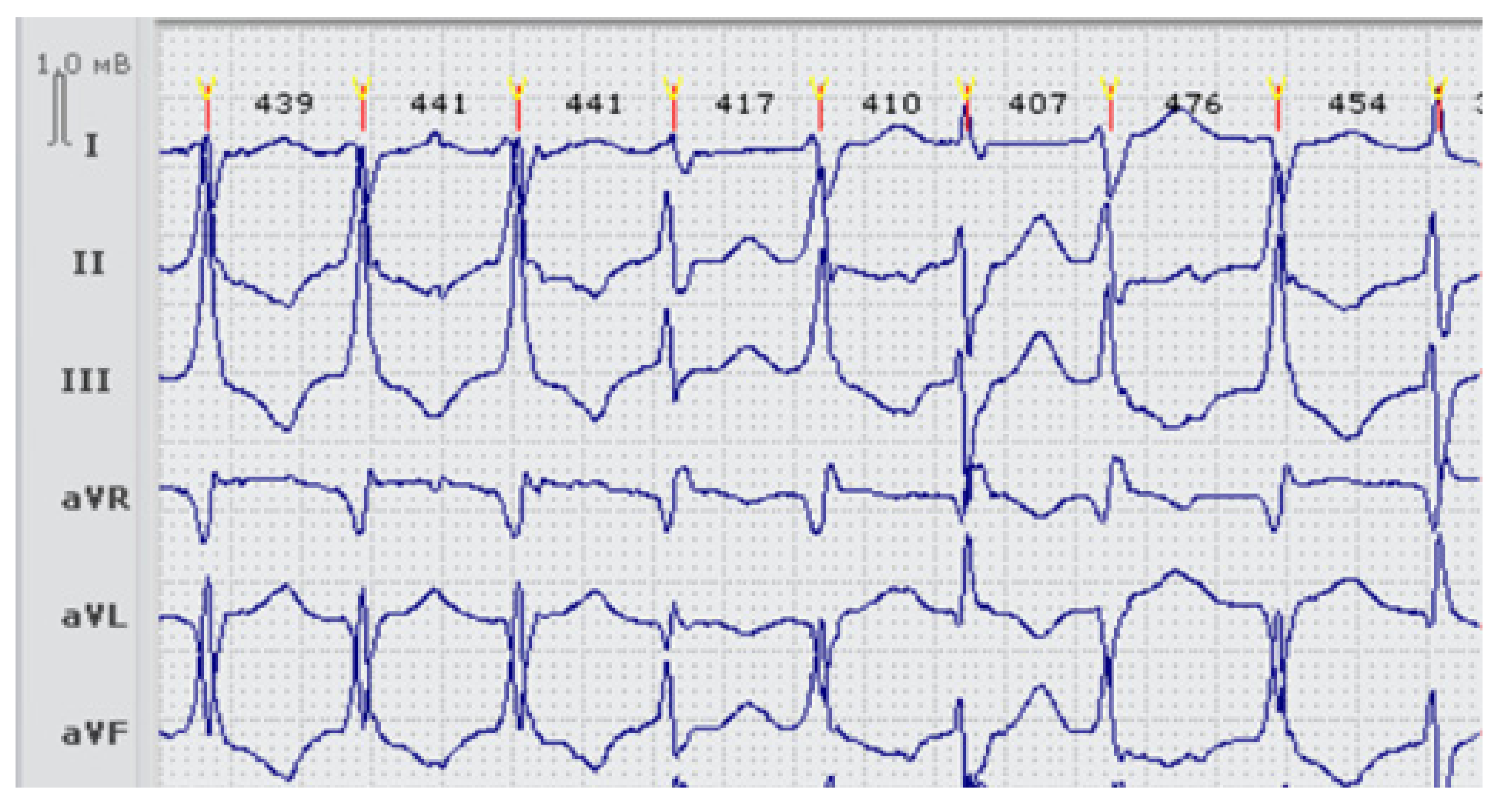
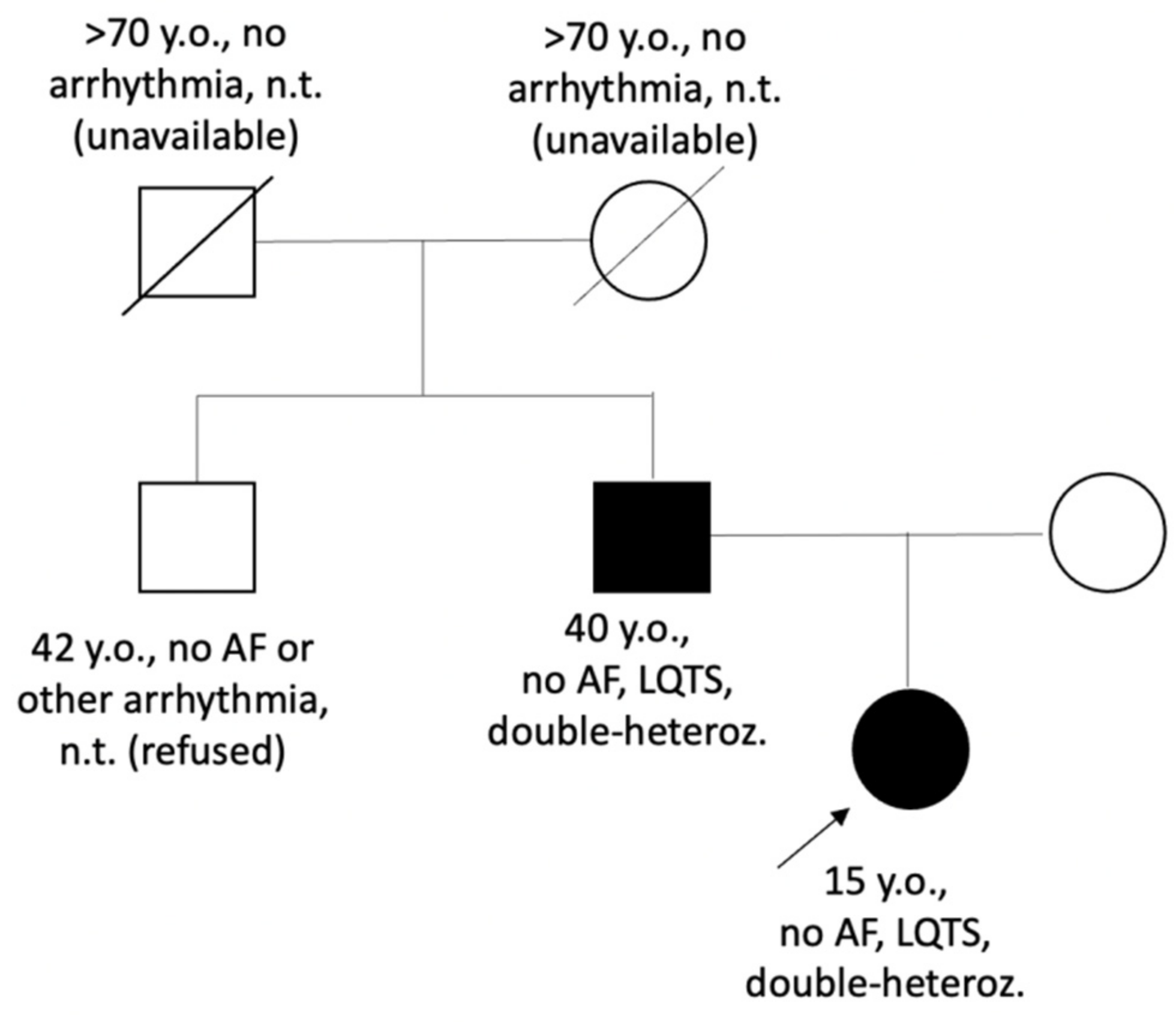
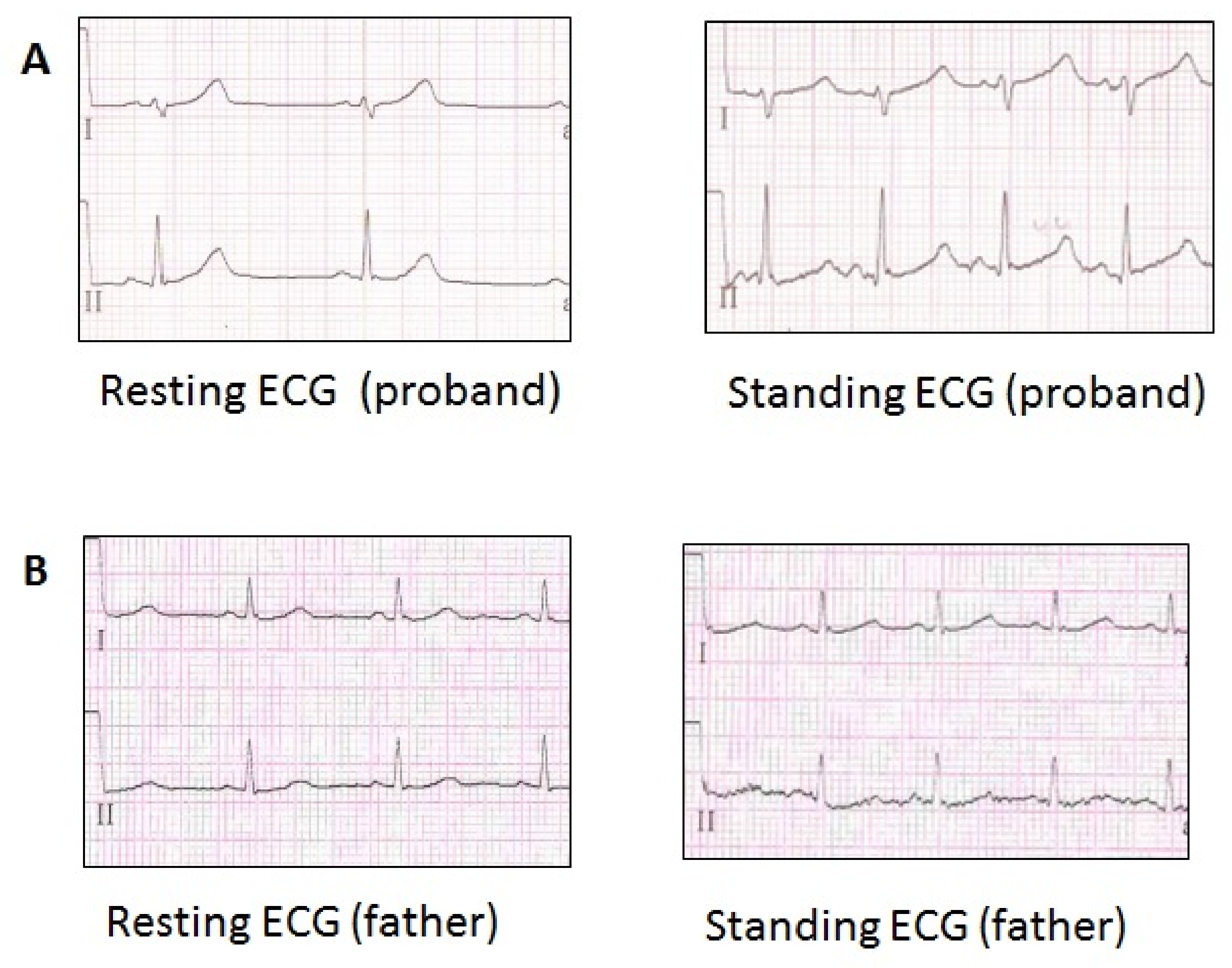
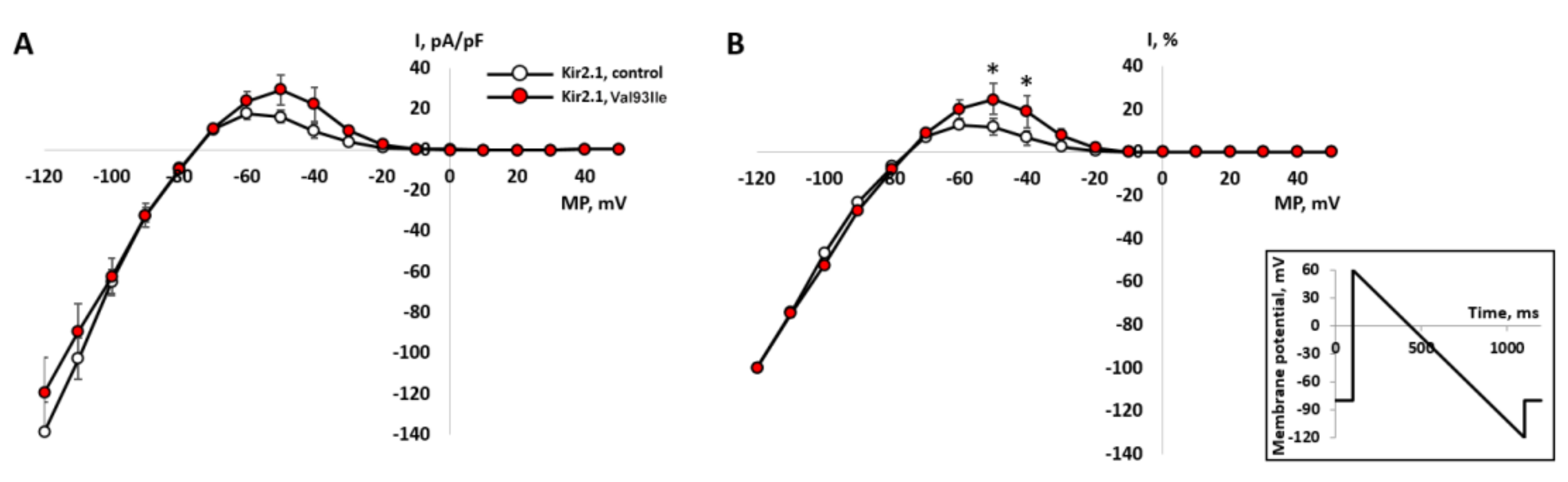
| Family (Ptnt) | Age *, Years | Variant/ (Class of Pathogenicity) | Origin | Kir2.1 Current Effect | QTc Resting | QTc Standing | Syncope | Andersen-Tawil Phenotype | Arrhythmia | Therapy |
|---|---|---|---|---|---|---|---|---|---|---|
| LQTS_96 | 29 | p.Arg67Trp (V) | De novo | Decreased [9] | 494–500 | 540 | Yes | + | VT, PVB, bigeminy | BB, ICD |
| LQTS_139 | 15 | p.Val93Ile (III), p.Arg132Trp in SCN3B (III) | Inherited | Increased [11,13] | 449–480 | 568 | No | no | no | BB recommended (refused) |
| LQTS_33 | 23 | p.Arg218Gln (V), p.Thr983Ile in KCNH2 (III) | DNA samples from parents unavailable | Decreased [14] | 600–620 | 650 | Yes | + | PVB, paroxysmal VT, VF, appropriate shocks | BB, ICD |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zaklyazminskaya, E.; Polyak, M.; Shestak, A.; Sadekova, M.; Komoliatova, V.; Kiseleva, I.; Makarov, L.; Podolyak, D.; Glukhov, G.; Zhang, H.; et al. Variable Clinical Appearance of the Kir2.1 Rare Variants in Russian Patients with Long QT Syndrome. Genes 2022, 13, 559. https://doi.org/10.3390/genes13040559
Zaklyazminskaya E, Polyak M, Shestak A, Sadekova M, Komoliatova V, Kiseleva I, Makarov L, Podolyak D, Glukhov G, Zhang H, et al. Variable Clinical Appearance of the Kir2.1 Rare Variants in Russian Patients with Long QT Syndrome. Genes. 2022; 13(4):559. https://doi.org/10.3390/genes13040559
Chicago/Turabian StyleZaklyazminskaya, Elena, Margarita Polyak, Anna Shestak, Mariam Sadekova, Vera Komoliatova, Irina Kiseleva, Leonid Makarov, Dmitriy Podolyak, Grigory Glukhov, Han Zhang, and et al. 2022. "Variable Clinical Appearance of the Kir2.1 Rare Variants in Russian Patients with Long QT Syndrome" Genes 13, no. 4: 559. https://doi.org/10.3390/genes13040559
APA StyleZaklyazminskaya, E., Polyak, M., Shestak, A., Sadekova, M., Komoliatova, V., Kiseleva, I., Makarov, L., Podolyak, D., Glukhov, G., Zhang, H., Abramochkin, D., & Sokolova, O. S. (2022). Variable Clinical Appearance of the Kir2.1 Rare Variants in Russian Patients with Long QT Syndrome. Genes, 13(4), 559. https://doi.org/10.3390/genes13040559






