Extensive Diversity and Prevalent Fluconazole Resistance among Environmental Yeasts from Tropical China
Abstract
:1. Introduction
2. Materials and Methods
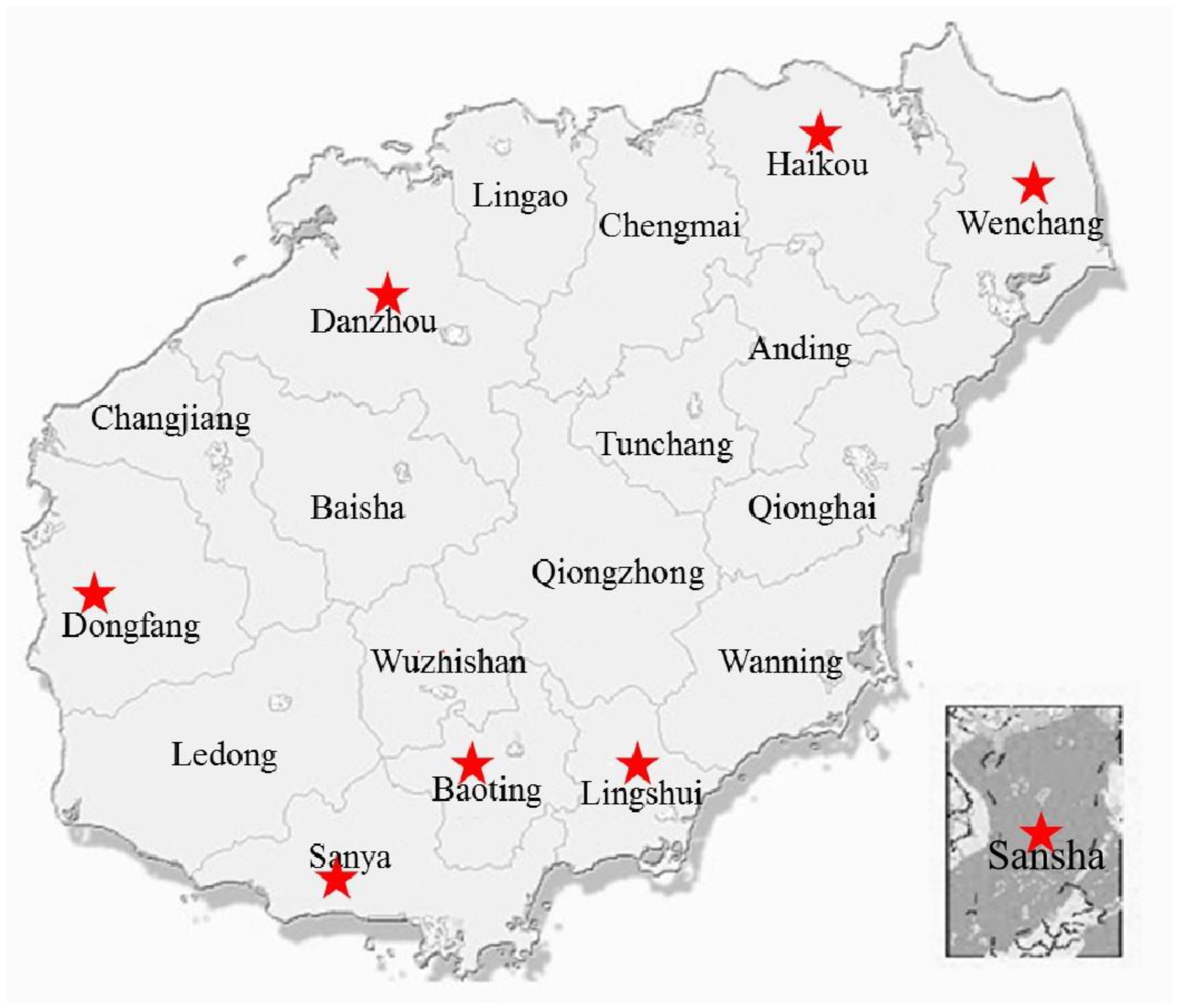
2.1. Sample Collection
2.2. Yeast Isolation
2.3. Identification of Yeast Species
2.4. Fluconazole Susceptibility Testing
2.5. Multi-Locus Sequence Analyses of C. tropicalis
2.6. Statistical Analyses
3. Results
3.1. Yeast Isolation and Species Identification
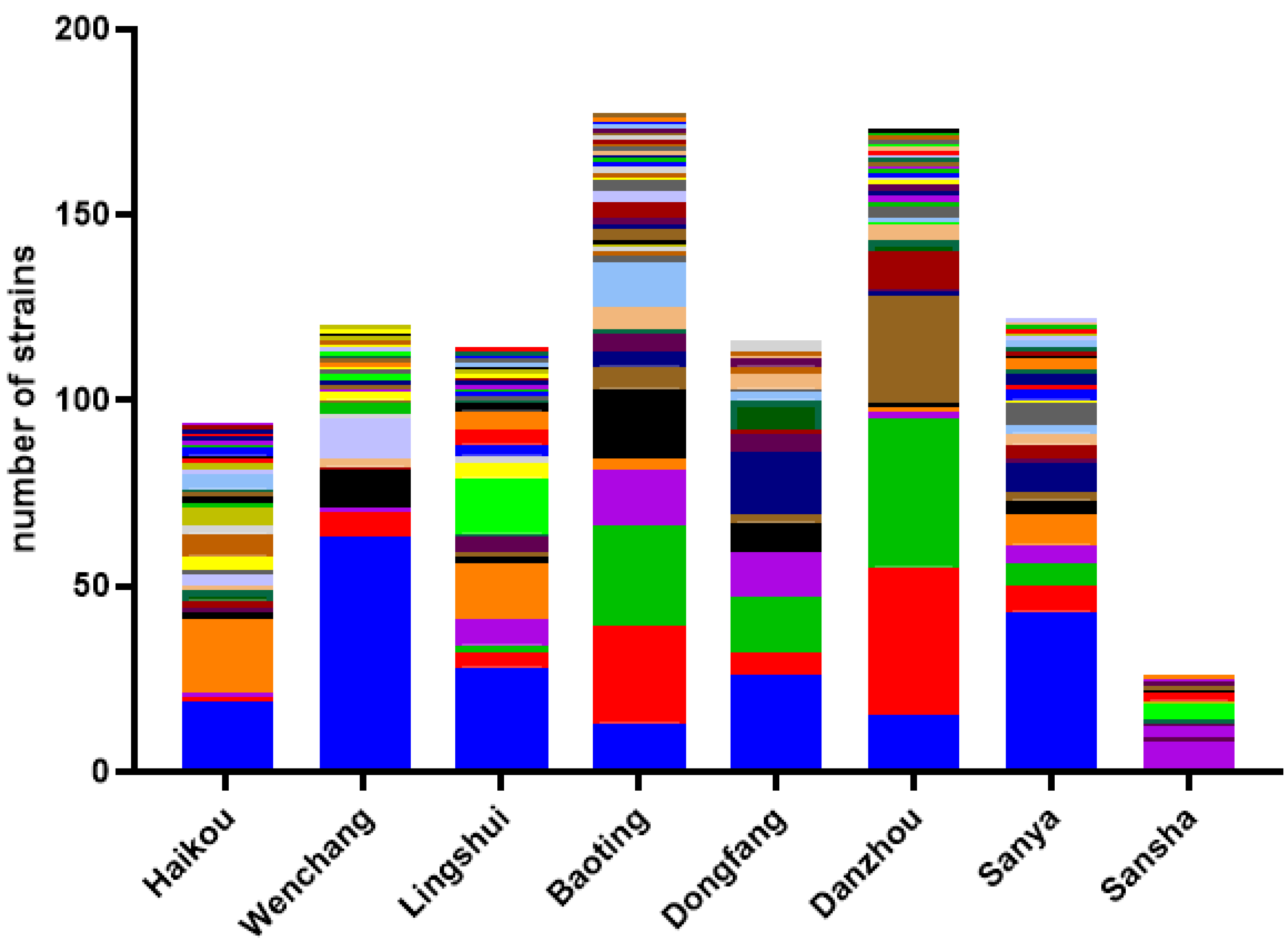
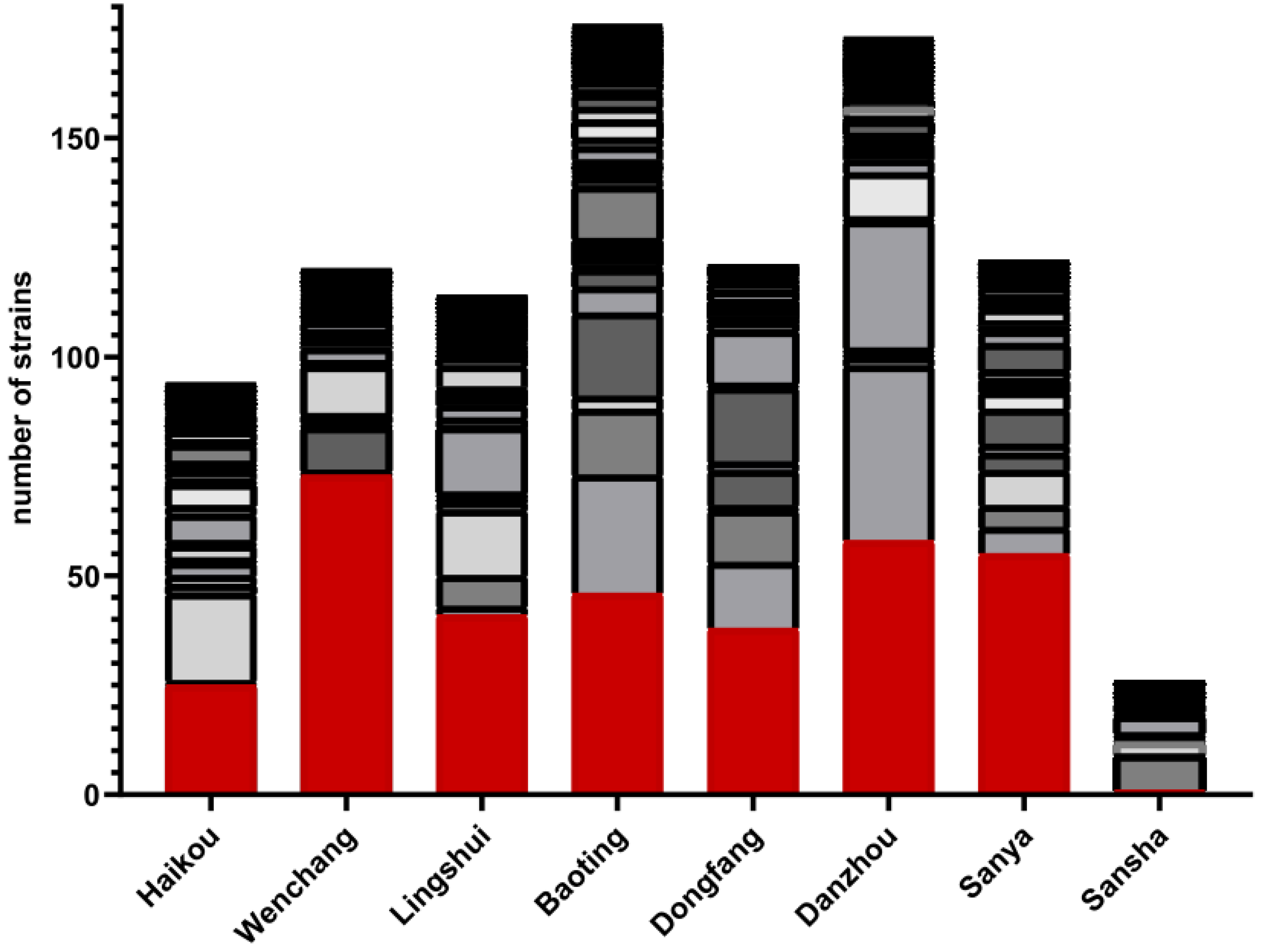
3.2. Diversity and Abundance of Culturable Environment Yeast Populations from Each Geographic Region
3.2.1. Haikou
3.2.2. Wenchang
3.2.3. Lingshui
3.2.4. Baoting
3.2.5. Dongfang
3.2.6. Danzhou
3.2.7. Sanya
3.2.8. Sansha
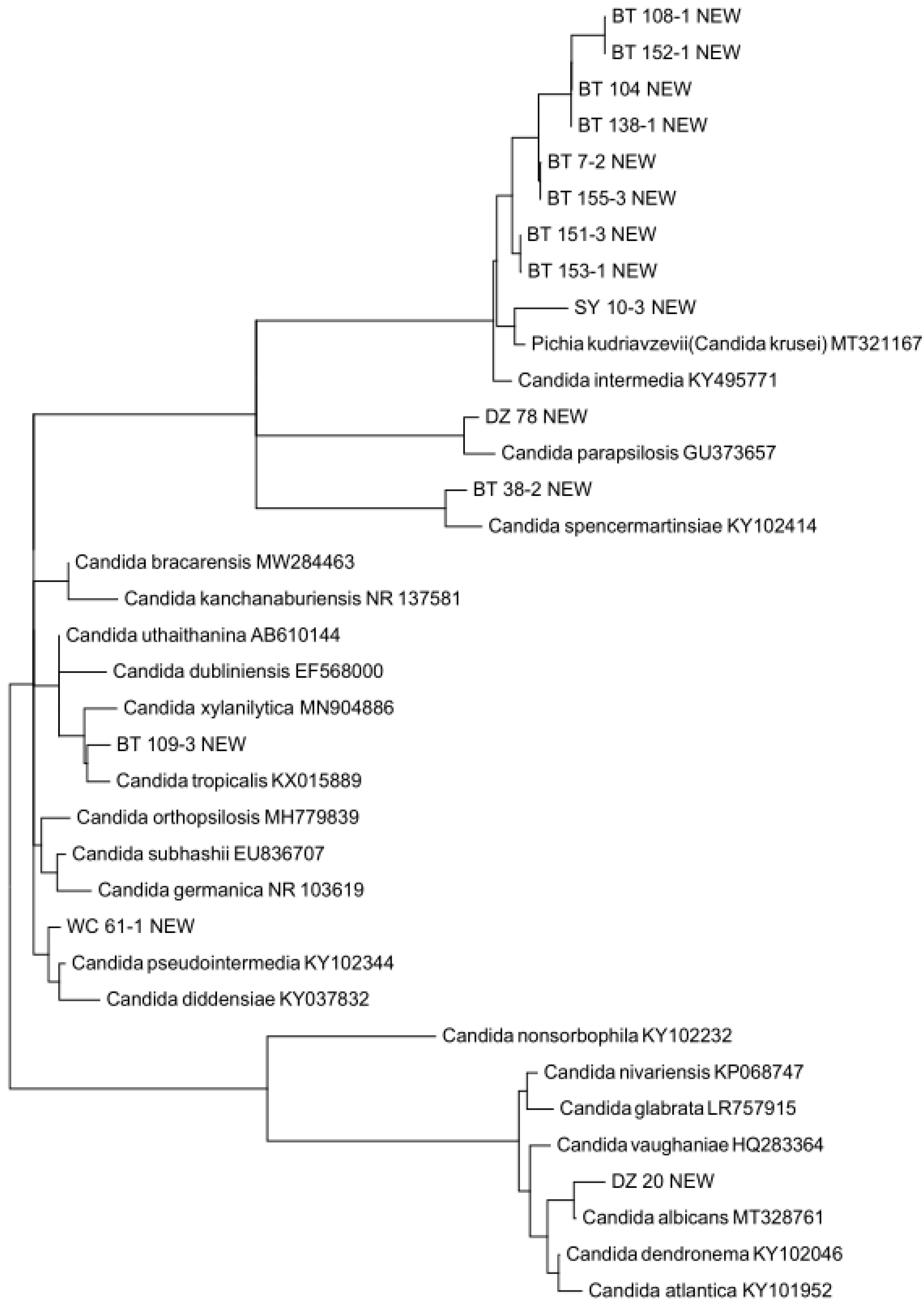
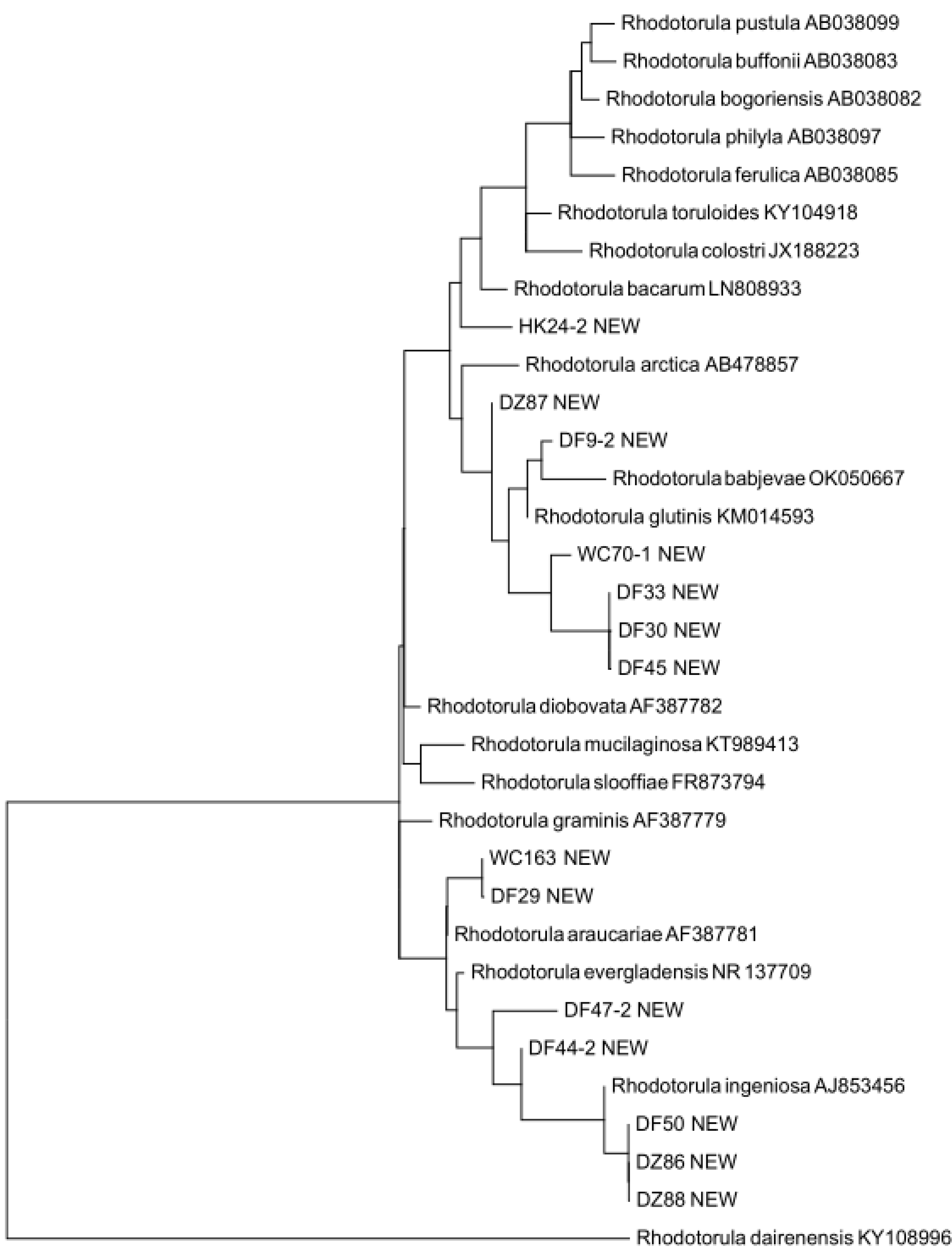
3.3. Putative Novel Yeast Species
3.4. Pathogenic Yeast Species
3.5. Profile of Fluconazole Susceptibilities
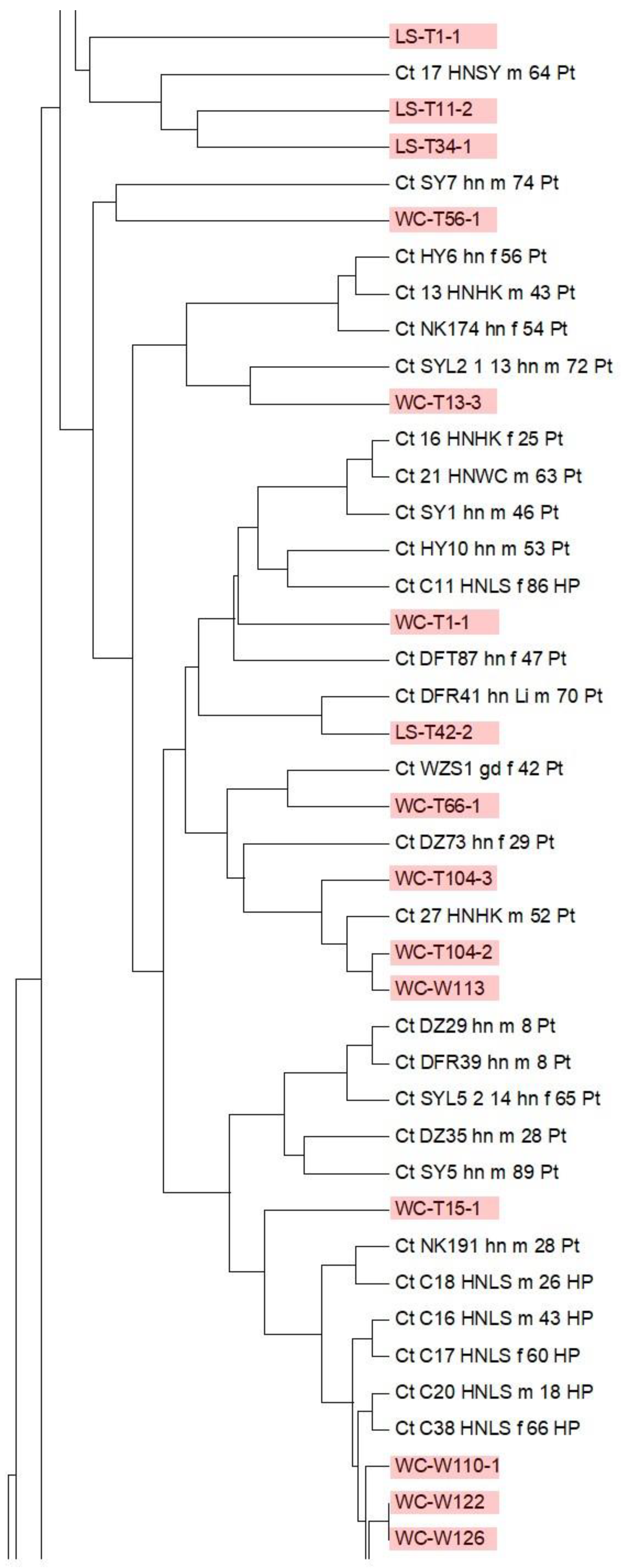
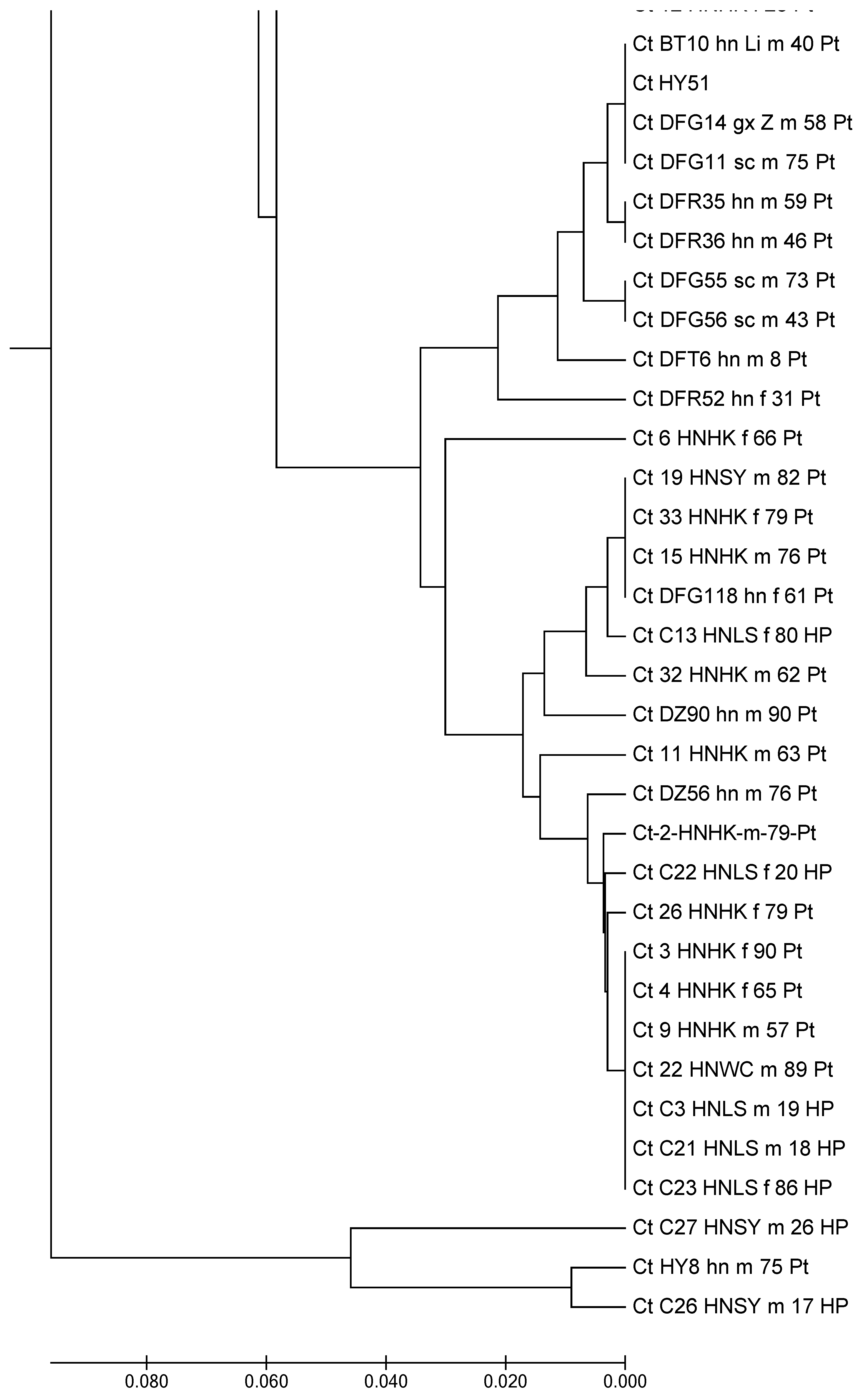
3.6. Multi-Locus Sequence Typing of C. tropicalis in the Environment
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fell, J.W. Collection and identification of marine yeasts. Methods Microbiol. 2001, 30, 347–356. [Google Scholar] [CrossRef]
- Samarasinghe, H.; Lu, Y.; Aljohani, R.; Al-Amad, A.; Yoell, H.; Xu, J. Global patterns in culturable soil yeast diversity. IScience 2021, 24, 103098. [Google Scholar] [CrossRef] [PubMed]
- Monapathi, M.E.; Bezuidenhout, C.C.; James Rhode, O.H. Aquatic yeasts: Diversity, characteristics and potential health implications. J. Water Health 2020, 18, 91–105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kutty, S.N.; Philip, R. Marine yeasts—A review. Yeast 2008, 25, 465–483. [Google Scholar] [CrossRef] [PubMed]
- Naranjo-Ortiz, M.A.; Gabaldón, T. Fungal evolution: Diversity, taxonomy and phylogeny of the Fungi. Biol. Rev. Camb. Philos. Soc. 2019, 94, 2101–2137. [Google Scholar] [CrossRef]
- Vuyst, L.D.; Harth, H.; Kerrebroeck, S.V.; Leroy, F. Yeast diversity of sourdoughs and associated metabolic properties and functionalities. Int. J. Food Microbiol. 2016, 239, 26–34. [Google Scholar] [CrossRef]
- Ahmad, K.M.; Kokošar, J.; Guo, X.; Gu, Z.; Ishchuk, O.P.; Piškur, J. Genome structure and dynamics of the yeast pathogen Candida glabrata. FEMS Yeast Res. 2014, 14, 529–535. [Google Scholar] [CrossRef] [Green Version]
- Dujon, B.; Sherman, D.; Fischer, G.; Durrens, P.; Casaregola, S.; Lafontaine, I.; de Montigny, J.; Marck, C.; Neuvéglise, C.; Talla, E.; et al. Genome evolution in yeasts. Nature 2004, 430, 35–44. [Google Scholar] [CrossRef]
- Timmermans, B.; De Las Peñas, A.; Castaño, I.; Van Dijck, P. Adhesins in Candida glabrata. J. Fungi 2018, 4, 60. [Google Scholar] [CrossRef] [Green Version]
- Douglass, A.P.; Offei, B.; Braun-Galleani, S.; Coughlan, A.Y.; Martos, A.A.; Ortiz-Merino, R.A.; Byrne, K.P.; Wolfe, K.H. Population genomics shows no distinction between pathogenic Candida krusei and environmental Pichia kudriavzevii: One species, four names. PLoS Pathog. 2018, 14, e1007138. [Google Scholar] [CrossRef]
- Arora, P.; Singh, P.; Wang, Y.; Yadav, A.; Pawar, K.; Singh, A.; Padmavati, G.; Xu, J.; Chowdhary, A. Environmental isolation of Candida auris from the coastal wetlands of Andaman Islands, India. MBio 2021, 12, e1007138. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Gan, C.; Li, J.; Liu, Y.; Chen, Z.; Zhang, Y.; Yi, G.; Sui, J.; Xu, J. Species Diversity and Antifungal Susceptibilities of Oral Yeasts from Patients with Head and Neck Cancer. Infect. Drug Resist. 2021, 14, 2279–2288. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Guo, H.; Yi, G.; Zhou, L.; He, X.; Huang, X.; Wang, H.; Xue, W.; Xu, J. Prevalent drug resistance among oral yeasts from asymptomatic patients in Hainan, China. Mycopathologia 2014, 177, 299–307. [Google Scholar] [CrossRef]
- Chowdhary, A.; Kathuria, S.; Xu, J.; Sharma, C.; Sundar, G.; Singh, P.K.; Gaur, S.N.; Hagen, F.; Klaassen, C.H.; Meis, J.F. Clonal expansion and emergence of environmental multiple-triazole-resistant Aspergillus fumigatus strains carrying the TR34/L98H mutations in the cyp51A gene in India. PLoS ONE 2012, 7, e52871. [Google Scholar] [CrossRef] [PubMed]
- Chowdhary, A.; Kathuria, S.; Xu, J.; Meis, J.F. Emergence of azole resistant Aspergillus fumigatus strains due to agricultural azole use creates an increasing threat to human health. PLoS Pathog. 2013, 9, e1003633. [Google Scholar] [CrossRef]
- National Bureau of Statistics, PRC. China Statistical Yearbook; China Statistics Press: Beijing, China, 2020.
- Scherer, S.; Stevens, D.A. A Candida albicans dispersed, repeated gene family and its epidemiologic applications. Proc. Natl. Acad. Sci. USA 1988, 85, 1452–1456. [Google Scholar] [CrossRef] [Green Version]
- Samarasinghe, H.; Aljohani, R.; Jimenez, C.; Xu, J. Fantastic yeasts and where to find them: The discovery of a predominantly clonal Cryptococcus deneoformans population in Saudi Arabian soils. FEMS Microbiol. Ecol. 2019, 95, fiz122. [Google Scholar] [CrossRef]
- Vu, D.; Groenewald, M.; Szöke, S.; Cardinali, G.; Eberhardt, U.; Stielow, B.; de Vries, M.; Crous, P.W.; Boekhout, T.; Robert, V. DNA barcoding analysis of more than 9000 yeast isolates contributes to quantitative thresholds for yeast species and genera delimitation. Stud. Mycol. 2016, 85, 91–105. [Google Scholar] [CrossRef]
- Institute Cals. M44-A2 Reference Method for Antifungaldisk Diffusion Susceptibility Testing of Yeasts, 2nd ed.; Approved Guideline; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2009. [Google Scholar]
- Anonymous. NEO-SENSITABS User’s Guide in Antifungal Susceptibility Testing of Yeast; Rosco Diagno’stica A/S: Albertslund, Denmark, 2011. [Google Scholar]
- Wu, J.; Guo, H.; Wang, H.; Yi, G.; Zhou, L.; He, X.; Zhang, Y.; Xu, J. Multilocus sequence analyses reveal extensive diversity and multiple origins of fluconazole resistance in Candida tropicalis from tropical China. Sci. Rep. 2017, 7, 42537. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [Green Version]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research—An update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; Mcglinn, D.; Minchin, P.R.; O’hara, R.B.; Simpson, G.L.; Solymos, P.; et al. Package “Vegan”. Community Ecol. Package Version 2020, 2, 5–7. [Google Scholar]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed]
- Opulente, D.A.; Langdon, Q.K.; Buh, V.K.; Haase, M.A.; Sylvester, K.; Moriarty, V.R.; Jarzyna, M.; Considine, S.L.; Schneider, R.M.; Hittinger, C.T. Pathogenic budding yeasts isolated outside of clinical settings. FEMS Yeast Res. 2019, 19, foz032. [Google Scholar] [CrossRef]
- Frąc, M.; Hannula, S.E.; Bełka, M.; Jędryczka, M. Fungal biodiversity and their role in soil health. Front. Microbiol. 2018, 9, 707. [Google Scholar] [CrossRef] [Green Version]
- Bajpai, V.K.; Khan, I.; Shukla, S.; Kumar, P.; Rather, I.A.; Park, Y.; Huh, Y.S.; Han, Y. Invasive fungal infections and their epidemiology: Measures in the clinical scenario. Biotechnol. Bioproc. E 2019, 24, 436–444. [Google Scholar] [CrossRef]
- Castano, G.; Yarrarapu, S.N.; Mada, P.K. Trichosporonosis. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2021. [Google Scholar]
- Maciel, N.O.; Johann, S.; Brandão, L.R.; Kucharíková, S.; Morais, C.G.; Oliveira, A.P.; Freitas, G.J.; Borelli, B.M.; Pellizzari, M.F.; Santos, A.D.; et al. Occurrence, antifungal susceptibility, and virulence factors of opportunistic yeasts isolated from Brazilian beaches. Mem. Inst. Oswaldo. Cruz. 2019, 114, e180566. [Google Scholar] [CrossRef]
- Ruosta, F.N.; Charsizadeh, A.; Ghahri, M.; Jafari, Z.; Mirhendi, H. Frequency of uncommon clinical yeast species confirmed by ITS-Sequencing. Arch. Clin. Infect. Dis. 2019, 14, e62816. [Google Scholar] [CrossRef] [Green Version]
- Ayaz, C.; Gülmez, D.; Arikan Akdağli, S.E.; Uzun, Ö. Nadir Etken Bir Maya: Üçüncü Basamak Üniversite Hastanesinde İzlenen Rhodotorula mucilaginosa Enfeksiyonu Olguları. Mikrobiyol. Bul. 2021, 55, 91–98. [Google Scholar] [CrossRef]
- Chakrabarti, A.; Rudramurthy, S.M.; Kale, P.; Hariprasath, P.; Dhaliwal, M.; Singhi, S.; Rao, K.L. Epidemiological study of a large cluster of fungaemia cases due to Kodamaea ohmeri in an Indian tertiary care centre. Clin. Microbiol. Infect. 2014, 20, O83–O89. [Google Scholar] [CrossRef] [Green Version]
- Das, K.; Bhattacharyya, A.; MammenChandy, M.; Roy, M.K.; Goel, G.; Hmar, L.; Bhattacharya, S.; Roy, P.; Chakrabarti, A. Infection control challenges of infrequent and rare fungal pathogens: Lessons from disseminated Fusarium and Kodamaea ohmeri infections. Infect. Control. Hosp. Epidemiol. 2015, 36, 866–868. [Google Scholar] [CrossRef] [Green Version]
- Paul, S.; Kannan, I. Molecular identification and antifungal susceptibility pattern of Candida species isolated from HIV infected Patients with candidiasis. Curr. Med. Mycol. 2019, 5, 21–26. [Google Scholar] [CrossRef] [PubMed]
- Ioannou, P.; Papakitsou, I. Kodamaea ohmeri infections in humans: A systematic review. Mycoses 2020, 63, 636–643. [Google Scholar] [CrossRef] [PubMed]
- Potenza, L.; Chitasombat, M.N.; Klimko, N.; Bettelli, F.; Dragonetti, G.; Del Principe, M.I.; Nucci, M.; Busca, A.; Fracchiolla, N.; Sciumè, M. Rhodotorula infection in haematological patient: Risk factors and outcome. Mycoses 2019, 62, 223–229. [Google Scholar] [CrossRef] [PubMed]
- Pristov, K.E.; Ghannoum, M.A. Resistance of Candida to azoles and echinocandins worldwide. Clin. Microbiol. Infect. 2019, 25, 792–798. [Google Scholar] [CrossRef]
- Arrua, J.M.; Rodrigues, L.A.; Pereira, F.O.; Lima, E.O. Prevalence of Candida tropicalis and Candida krusei in onychomycosis in João Pessoa, Paraiba, Brazil from 1999 to 2010. An. Acad. Bras. Cienc. 2015, 87, 1819–1822. [Google Scholar] [CrossRef] [Green Version]
- Kumar, A. A fungus among us: The emerging opportunistic pathogen Candida tropicalis and PKA signaling. Virulence 2018, 9, 659–661. [Google Scholar] [CrossRef] [Green Version]
- Wu, J.; Zhou, D.; Zhang, Y.; Mi, F.; Xu, J. Analyses of the global multilocus genotypes of the human pathogenic yeast Candida tropicalis. Front. Microbiol. 2019, 10, 900. [Google Scholar] [CrossRef] [Green Version]
- Xu, J. Fungal species concepts in the genomics era. Genome 2020, 63, 459–468. [Google Scholar] [CrossRef]


| Sample Types | Sample Characteristics | No. of Samples | No. of Samples with Yeasts (% Oositive/Total) | Yeast Isolates | Known Species | Putative novel Species | Private Species | Shannon Diversity Index |
|---|---|---|---|---|---|---|---|---|
| Ecological | Soil | 516 | 369(71.5) | 512 | 88 | 27 | 46 | 3.50 |
| niches | Freshwater | 273 | 234(85.7) | 361 | 52 | 14 | 11 | 2.90 |
| Seawater along beaches | 179 | 78(43.6) | 111 | 21 | 2 | - | 2.41 | |
| Geographic | Haikou | 120 | 70(58.3) | 97 | 32 | 3 | 9 | 2.88 |
| regions | Wenchang | 131 | 93(71) | 125 | 27 | 4 | 12 | 2.96 |
| Lingshui | 119 | 88(74) | 114 | 31 | - | 7 | 3.04 | |
| Baoting | 135 | 116(85.9) | 191 | 39 | 15 | 9 | 2.55 | |
| Dongfang | 127 | 95(74.8) | 128 | 20 | 8 | 4 | 2.49 | |
| Danzhou | 153 | 112(73.2) | 179 | 34 | 6 | 7 | 2.66 | |
| Sanya | 133 | 88(66.2) | 124 | 29 | 2 | 5 | 2.22 | |
| Sansha | 50 | 19(38) | 26 | 13 | - | 4 | 2.10 | |
| Total | 968 | 681(70.4) | 984 | 106 | 38 | 57 |
| Species | Total | S | I | R | I+R | (% Total) | Not Tested |
|---|---|---|---|---|---|---|---|
| C. tropicalis | 207 | 150 | 8 | 43 | 51 | 25.4% | 6 |
| C. krusei | 90 | 29 | 32 | 22 | 56 | 65.9% | 7 |
| Torulaspora delbrueckii | 90 | 21 | 4 | 59 | 63 | 75.0% | 6 |
| Meyerozyma caribbica | 51 | 45 | 0 | 5 | 5 | 10.0% | 1 |
| C. pseudolambica | 48 | 11 | 11 | 24 | 35 | 76.1% | 2 |
| Trichosporon asahii | 46 | 40 | 0 | 6 | 6 | 13.0% | 0 |
| Diutina rugosa | 40 | 35 | 0 | 3 | 3 | 7.9% | 2 |
| Wickerhamomyces sydowiorum | 30 | 24 | 0 | 6 | 6 | 20.0% | 0 |
| Rhodotorula toruloides | 20 | 2 | 0 | 17 | 17 | 89.5% | 1 |
| Dirkmeia churashimaensis | 18 | 4 | 0 | 9 | 9 | 69.2% | 5 |
| C. guilliermondii | 18 | 12 | 2 | 4 | 6 | 33.3% | 0 |
| C. quercitrusa | 17 | 8 | 5 | 3 | 8 | 50% | 1 |
| C. intermedia | 16 | 13 | 0 | 2 | 2 | 13.3% | 1 |
| Kodamaea ohmeri | 16 | 14 | 1 | 1 | 2 | 12.5% | 0 |
| C. palmioleophila | 14 | 1 | 0 | 13 | 13 | 92.9% | 0 |
| Rhodotorula paludigena | 13 | 1 | 0 | 8 | 8 | 88.9% | 4 |
| Diutina mesorugosa | 7 | 5 | 0 | 1 | 1 | 16.7% | 1 |
| C. gosingica | 6 | 4 | 1 | 0 | 1 | 20% | 1 |
| Cyberlindnera saturnus | 6 | 1 | 3 | 0 | 3 | 75.0% | 2 |
| Rhodotorula mucilaginosa | 6 | 0 | 0 | 6 | 6 | 100% | 0 |
| Aureobasidium melanogenum | 5 | 2 | 1 | 1 | 2 | 50% | 1 |
| C. albicans | 5 | 4 | 0 | 1 | 1 | 20% | 0 |
| Diutina catenulata | 5 | 3 | 1 | 1 | 2 | 40% | 0 |
| Moesziomyces antarcticus | 5 | 0 | 0 | 2 | 2 | 100% | 3 |
| C. dubliniensis | 5 | 3 | 0 | 2 | 2 | 40% | 0 |
| C. stellimalicola | 4 | 2 | 0 | 1 | 1 | 33.3% | 1 |
| Rhodosporidiobolus ruineniae | 4 | 1 | 0 | 3 | 3 | 75% | 0 |
| Rhodotorula glutinis | 4 | 0 | 0 | 1 | 1 | 100% | 3 |
| Wickerhamomyces rabaulensis | 4 | 1 | 0 | 3 | 3 | 75% | 0 |
| C. nivariensis | 3 | 2 | 0 | 1 | 1 | 33.3% | 0 |
| Hanseniaspora opuntiae | 3 | 2 | 0 | 1 | 1 | 33.3% | 0 |
| Papiliotrema laurentii | 3 | 0 | 1 | 2 | 3 | 100% | 0 |
| Pichia manshurica | 4 | 0 | 0 | 1 | 3 | 100% | 0 |
| C. inconspicua | 2 | 0 | 1 | 0 | 1 | 100% | 1 |
| Debaryomyces castellii | 2 | 0 | 1 | 1 | 2 | 100% | 0 |
| Kwoniella heveanensis | 1 | 1 | 0 | 0 | 1 | 50% | 0 |
| Lachancea dasiensis | 2 | 0 | 1 | 0 | 1 | 100% | 1 |
| Moesziomyces aphidis | 2 | 0 | 0 | 1 | 1 | 100% | 1 |
| Pichia kluyveri | 2 | 0 | 0 | 2 | 2 | 100% | 0 |
| Rhodosporidium toruloides | 2 | 0 | 0 | 2 | 2 | 100% | 0 |
| Saccharomyces cerevisiae | 2 | 1 | 0 | 1 | 1 | 50% | 0 |
| Ustilago sparsa | 2 | 1 | 0 | 1 | 1 | 50% | 0 |
| Wickerhamomyces edaphicus | 2 | 1 | 0 | 1 | 1 | 50% | 0 |
| Apiotrichum laibachii | 1 | 0 | 1 | 0 | 1 | 100% | 0 |
| Apiotrichum mycotoxinovorans | 1 | 0 | 0 | 1 | 1 | 100% | 0 |
| C. fennica | 1 | 0 | 0 | 1 | 1 | 100% | 0 |
| C. sonorensis | 1 | 0 | 0 | 1 | 1 | 100% | 0 |
| Clavispora opuntiae | 1 | 0 | 0 | 1 | 1 | 100% | 0 |
| Cryptococcus flavescens | 1 | 0 | 1 | 0 | 1 | 100% | 0 |
| Moesziomyces rugulosus | 1 | 0 | 0 | 1 | 1 | 100% | 0 |
| Pichia bruneiensis | 1 | 0 | 0 | 1 | 1 | 100% | 0 |
| Pichia fermentans | 1 | 0 | 0 | 1 | 1 | 100% | 0 |
| Pichia membranifaciens | 1 | 0 | 0 | 1 | 1 | 100% | 0 |
| Schwanniomyces vanrijiae | 1 | 0 | 1 | 0 | 1 | 100% | 0 |
| Wickerhamiella infanticola | 1 | 0 | 0 | 1 | 1 | 100% | 0 |
| Yamadazyma cocois | 1 | 0 | 0 | 1 | 1 | 100% | 0 |
| Candida sp. NEW | 14 | 8 | 0 | 6 | 8 | 100% | 0 |
| Saturnispora sp. NEW | 1 | 0 | 1 | 0 | 1 | 100% | 0 |
| Kalmanozyma sp. NEW | 1 | 0 | 0 | 1 | 1 | 100% | 0 |
| Rhodotorula sp. NEW | 15 | 1 | 0 | 11 | 11 | 91.7% | 2 |
| Apiotrichum sp. NEW | 1 | 0 | 0 | 1 | 1 | 100% | 0 |
| Meyerozyma sp. NEW | 1 | 0 | 0 | 1 | 1 | 100% | 0 |
| Gene Loci | Clinical Sample (116 Isolates) G/P (Ratio) | Environmental Sample (44 Isolates) G/P (Ratio) | Total (160) |
|---|---|---|---|
| ICL1 | 8/6 (1.3) | 6/3 (2.0) | 10/6 (1.7) |
| MDR1 | 37/21 (1.8) | 21/12 (1.8) | 47/22 (2.1) |
| SAPT2 | 11/7 (1.6) | 8/6 (1.3) | 14/10 (1.4) |
| SAPT4 | 21/18 (1.2) | 12/14 (0.9) | 28/18 (1.6) |
| XYR1 | 33/16 (2.4) | 22/17 (1.3) | 45/18 (2.5) |
| ZWFa1 | 14/11 (1.3) | 9/5 (1.8) | 17/11 (1.6) |
| Total | 94/79 (1.2) | 33/57 (0.6) | 124/85 (1.5) |
| Source | df | SS | MS | Est. Var. | % |
|---|---|---|---|---|---|
| Among Pops | 1 | 41.609 | 41.609 | 0.516 | 6% *** |
| Within Pops | 158 | 1369.484 | 8.668 | 8.668 | 94% *** |
| Total | 159 | 1411.094 | 9.184 | 100% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Y.; Chen, Z.; Li, J.; Zhu, Z.; Pang, S.; Xu, J.; Wu, J. Extensive Diversity and Prevalent Fluconazole Resistance among Environmental Yeasts from Tropical China. Genes 2022, 13, 444. https://doi.org/10.3390/genes13030444
Liu Y, Chen Z, Li J, Zhu Z, Pang S, Xu J, Wu J. Extensive Diversity and Prevalent Fluconazole Resistance among Environmental Yeasts from Tropical China. Genes. 2022; 13(3):444. https://doi.org/10.3390/genes13030444
Chicago/Turabian StyleLiu, Yiwei, Zhongyao Chen, Jingyuan Li, Zhiqing Zhu, Sibei Pang, Jianping Xu, and Jinyan Wu. 2022. "Extensive Diversity and Prevalent Fluconazole Resistance among Environmental Yeasts from Tropical China" Genes 13, no. 3: 444. https://doi.org/10.3390/genes13030444
APA StyleLiu, Y., Chen, Z., Li, J., Zhu, Z., Pang, S., Xu, J., & Wu, J. (2022). Extensive Diversity and Prevalent Fluconazole Resistance among Environmental Yeasts from Tropical China. Genes, 13(3), 444. https://doi.org/10.3390/genes13030444







