Bifidobacterium Is Enriched in Gut Microbiome of Kashmiri Women with Polycystic Ovary Syndrome
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Cohort and Recruitment
2.2. Blood Biochemistry, Hormonal Tests and Anthropometry
2.3. Stool Samples
2.4. Metagenomic DNA Extraction and 16S rRNA Sequencing Data Generation
2.5. Bioinformatic Analysis and Quality Control
2.6. Statistical Analysis
2.6.1. Association Analyses between Individual Species, PCOS and Hormone Levels
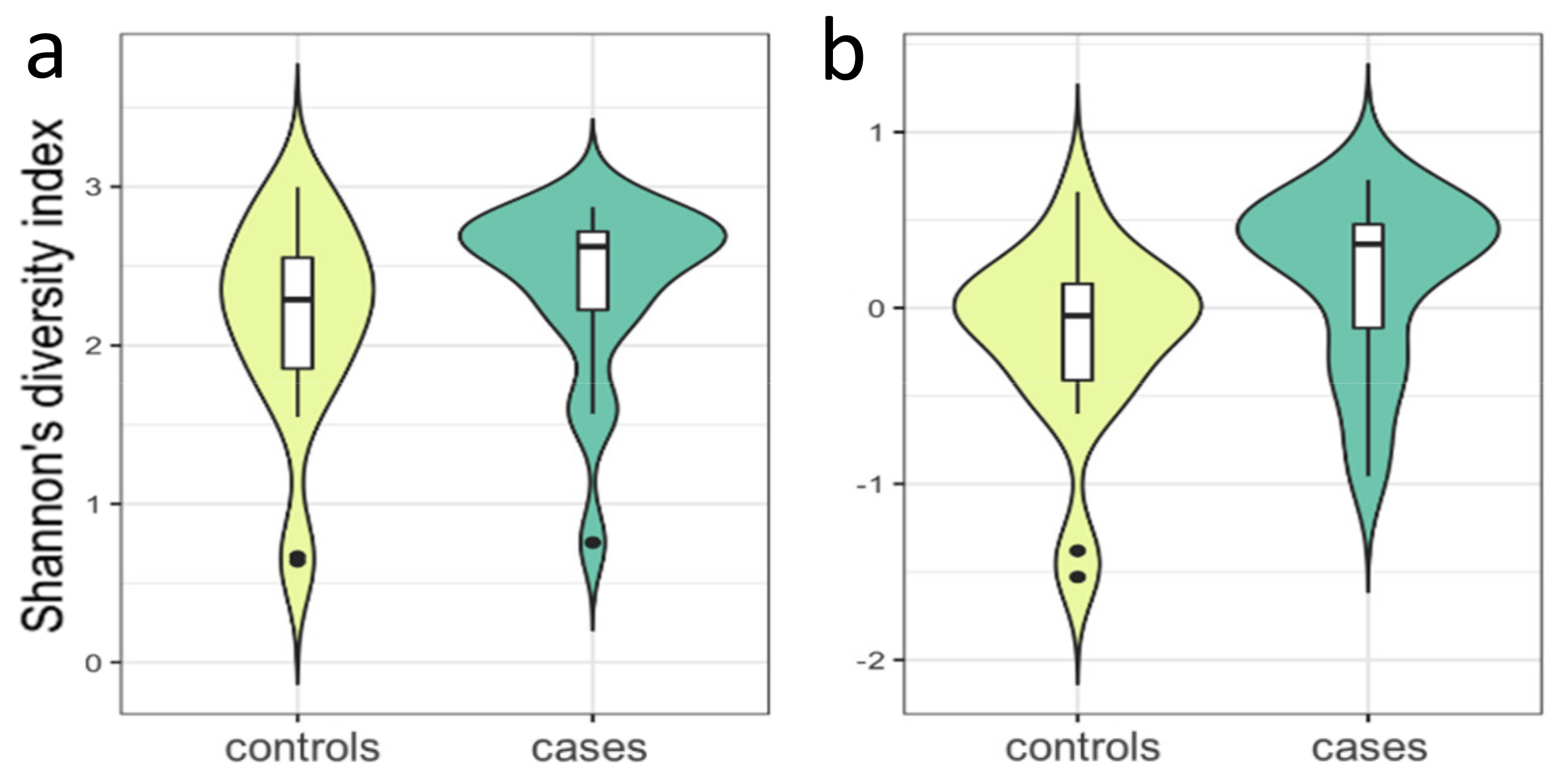
2.6.2. Microbial Distance and α Diversity
3. Results
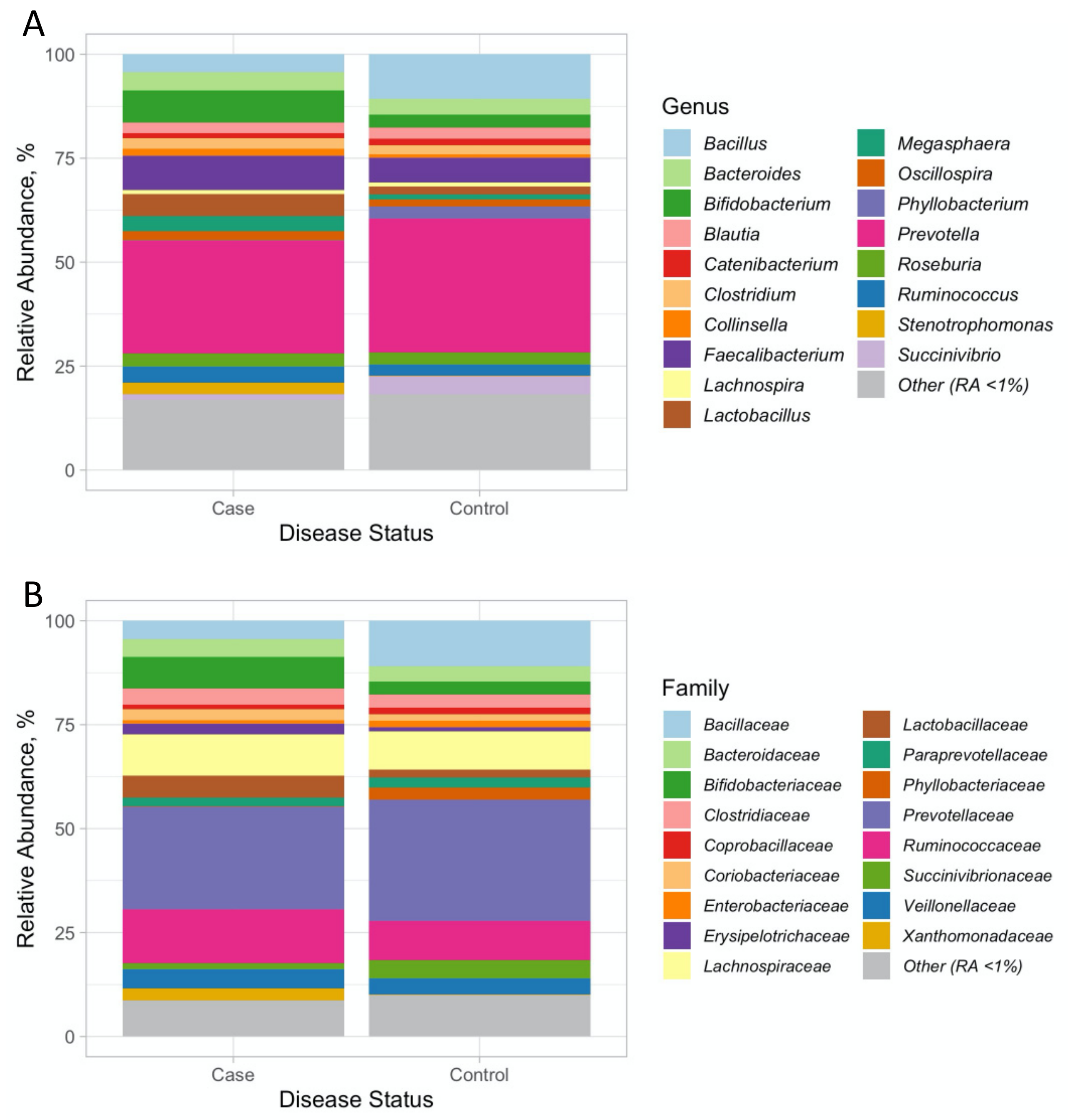
3.1. 16 SrRNA Gut Microbiome in PCOS
3.2. Associations with PCOS at the Genus and Family Level
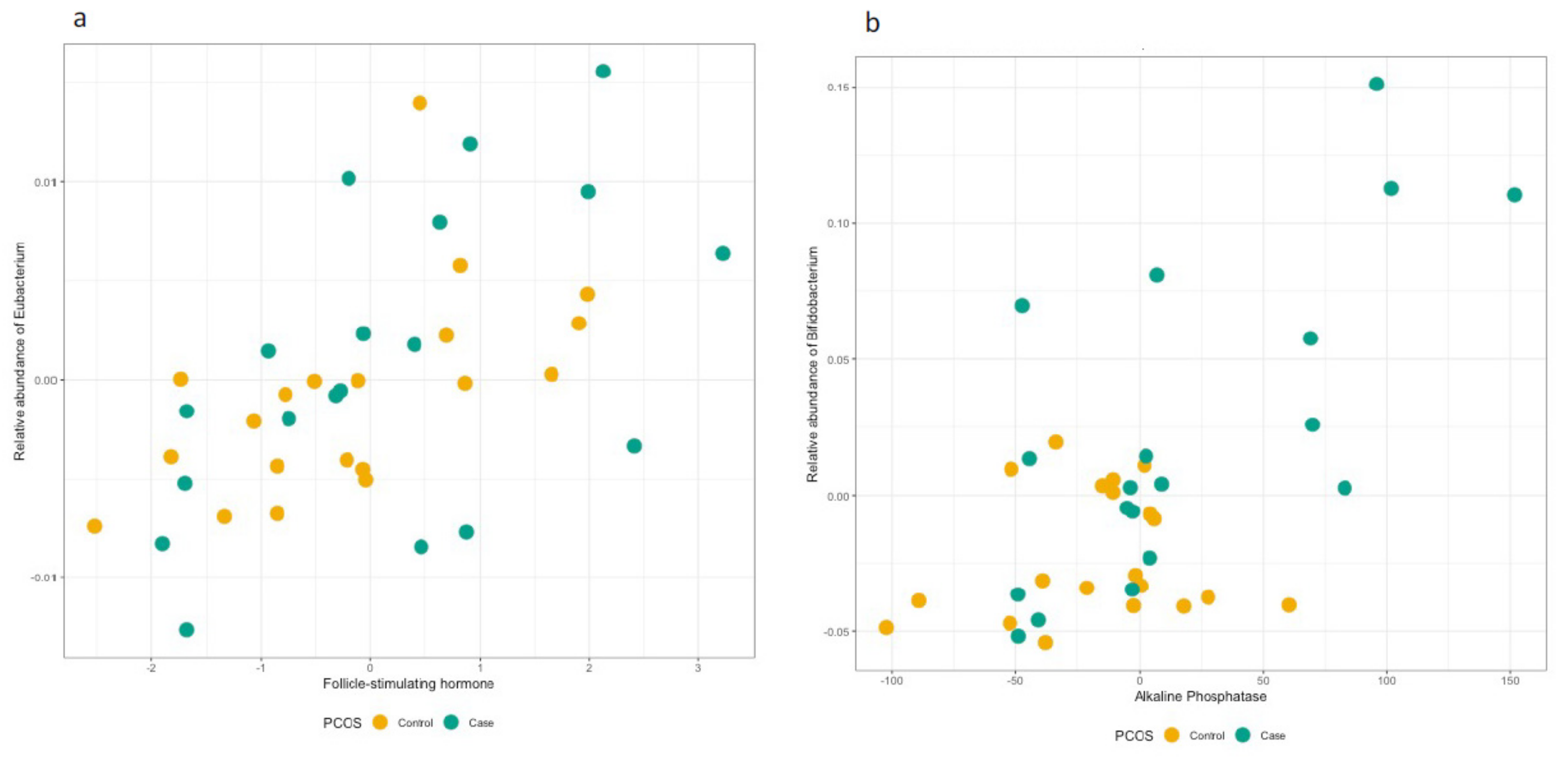
3.3. Hormonal Profiles at the Genus and Family Level
3.4. Follow-Up of the Identified OTUs at the Genus and Family Level
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rotterdam ESHRE/ASRM-Sponsored PCOS Consensus Workshop Group. Revised 2003 consensus on diagnostic criteria and long-term health risks related to polycystic ovary syndrome (PCOS). Hum. Reprod. 2004, 19, 41–47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- March, W.A.; Moore, V.M.; Willson, K.J.; Phillips, D.I.; Norman, R.J.; Davies, M.J. The prevalence of polycystic ovary syndrome in a community sample assessed under contrasting diagnostic criteria. Hum. Reprod. 2010, 25, 544–551. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Azziz, R.; Carmina, E.; Dewailly, D.; Diamanti-Kandarakis, E.; Escobar-Morreale, H.F.; Futterweit, W.; Janssen, O.E.; Legro, R.S.; Norman, R.J.; Taylor, A.E.; et al. Positions statement: Criteria for defining polycystic ovary syndrome as a predominantly hyperandrogenic syndrome: An Androgen Excess Society guideline. J. Clin. Endocrinol. Metab. 2006, 91, 4237–4245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bharathi, R.V.S.; Neerajaa, J.; Madhavica, J.V.; Janani, D.M.; Rekha, S.N.; Ramya, S.; Usha, B. An epidemiological survey: Effect of predisposing factors for PCOS in Indian urban and rural population. Middle East. Fertil. Soc. J. 2017, 22, 313–316. [Google Scholar] [CrossRef]
- Norman, R.J.; Mahabeer, S.; Masters, S. Ethnic differences in insulin and glucose response to glucose between white and Indian women with polycystic ovary syndrome. Fertil. Steril. 1995, 63, 58–62. [Google Scholar] [CrossRef]
- Wijeyaratne, C.N.; Balen, A.H.; Barth, J.H.; Belchetz, P.E. Clinical manifestations and insulin resistance (IR) in polycystic ovary syndrome (PCOS) among South Asians and Caucasians: Is there a difference? Clin. Endocrinol. 2002, 57, 343–350. [Google Scholar] [CrossRef] [PubMed]
- Ganie, M.A.; Rashid, A.; Sahu, D.; Nisar, S.; Wani, I.A.; Khan, J. Prevalence of polycystic ovary syndrome (PCOS) among reproductive age women from Kashmir valley: A cross-sectional study. Int. J. Gynaecol. Obstet. 2020, 149, 231–236. [Google Scholar] [CrossRef]
- Joshi, B.; Mukherjee, S.; Patil, A.; Purandare, A.; Chauhan, S.; Vaidya, R. A cross-sectional study of polycystic ovarian syndrome among adolescent and young girls in Mumbai, India. Indian J. Endocrinol. Metab. 2014, 18, 317–324. [Google Scholar] [CrossRef]
- Gill, H.; Tiwari, P.; Dabadghao, P. Prevalence of polycystic ovary syndrome in young women from North India: A Community-based study. Indian J. Endocrinol. Metab. 2012, 16, S389–S392. [Google Scholar] [CrossRef]
- Nidhi, R.; Padmalatha, V.; Nagarathna, R.; Amritanshu, R. Prevalence of polycystic ovarian syndrome in Indian adolescents. J. Pediatr. Adolesc. Gynecol. 2011, 24, 223–227. [Google Scholar] [CrossRef]
- Stein, I.F. Amenorrhea associated with bilateral polycystic ovaries. Am. J. Obstet. Gynecol. 1935, 29, 181–191. [Google Scholar] [CrossRef]
- Dunaif, A.; Segal, K.R.; Futterweit, W.; Dobrjansky, A. Profound peripheral insulin resistance, independent of obesity, in polycystic ovary syndrome. Diabetes 1989, 38, 1165–1174. [Google Scholar] [CrossRef] [PubMed]
- Teede, H.; Deeks, A.; Moran, L. Polycystic ovary syndrome: A complex condition with psychological, reproductive and metabolic manifestations that impacts on health across the lifespan. BMC Med. 2010, 8, 41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wild, R.A.; Applebaum-Bowden, D.; Demers, L.M.; Bartholomew, M.; Landis, J.R.; Hazzard, W.R.; Santen, R.J. Lipoprotein lipids in women with androgen excess: Independent associations with increased insulin and androgen. Clin. Chem. 1990, 36, 283–289. [Google Scholar] [CrossRef]
- Kelly, C.C.; Lyall, H.; Petrie, J.R.; Gould, G.W.; Connell, J.M.; Sattar, N. Low grade chronic inflammation in women with polycystic ovarian syndrome. J. Clin. Endocrinol. Metab. 2001, 86, 2453–2455. [Google Scholar] [CrossRef]
- Repaci, A.; Gambineri, A.; Pasquali, R. The role of low-grade inflammation in the polycystic ovary syndrome. Mol. Cell. Endocrinol. 2011, 335, 30–41. [Google Scholar] [CrossRef]
- Wild, R.A.; Carmina, E.; Diamanti-Kandarakis, E.; Dokras, A.; Escobar-Morreale, H.F.; Futterweit, W.; Lobo, R.; Norman, R.J.; Talbott, E.; Dumesic, D.A. Assessment of cardiovascular risk and prevention of cardiovascular disease in women with the polycystic ovary syndrome: A consensus statement by the Androgen Excess and Polycystic Ovary Syndrome (AE-PCOS) Society. J. Clin. Endocrinol. Metab. 2010, 95, 2038–2049. [Google Scholar] [CrossRef] [Green Version]
- Dumesic, D.A.; Oberfield, S.E.; Stener-Victorin, E.; Marshall, J.C.; Laven, J.S.; Legro, R.S. Scientific Statement on the Diagnostic Criteria, Epidemiology, Pathophysiology, and Molecular Genetics of Polycystic Ovary Syndrome. Endocr. Rev. 2015, 36, 487–525. [Google Scholar] [CrossRef] [Green Version]
- Kollmann, M.; Klaritsch, P.; Martins, W.P.; Guenther, F.; Schneider, V.; Herzog, S.A.; Craciunas, L.; Lang, U.; Obermayer-Pietsch, B.; Lerchbaum, E.; et al. Maternal and neonatal outcomes in pregnant women with PCOS: Comparison of different diagnostic definitions. Hum. Reprod. 2015, 30, 2396–2403. [Google Scholar] [CrossRef] [Green Version]
- Lerchbaum, E.; Schwetz, V.; Giuliani, A.; Obermayer-Pietsch, B. Assessment of glucose metabolism in polycystic ovary syndrome: HbA1c or fasting glucose compared with the oral glucose tolerance test as a screening method. Hum. Reprod. 2013, 28, 2537–2544. [Google Scholar] [CrossRef]
- Wehr, E.; Gruber, H.J.; Giuliani, A.; Moller, R.; Pieber, T.R.; Obermayer-Pietsch, B. The lipid accumulation product is associated with impaired glucose tolerance in PCOS women. J. Clin. Endocrinol. Metab. 2011, 96, E986–E990. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shabir, I.; Ganie, M.A.; Zargar, M.A.; Bhat, D.; Mir, M.M.; Jan, A.; Shah, Z.A.; Jan, V.; Rasool, R.; Naqati, A. Prevalence of metabolic syndrome in the family members of women with polycystic ovary syndrome from North India. Indian J. Endocrinol. Metab. 2014, 18, 364–369. [Google Scholar] [CrossRef] [PubMed]
- Vink, J.M.; Sadrzadeh, S.; Lambalk, C.B.; Boomsma, D.I. Heritability of polycystic ovary syndrome in a Dutch twin-family study. J. Clin. Endocrinol. Metab. 2006, 91, 2100–2104. [Google Scholar] [CrossRef] [Green Version]
- Abbott, D.H.; Dumesic, D.A.; Eisner, J.R.; Colman, R.J.; Kemnitz, J.W. Insights into the development of polycystic ovary syndrome (PCOS) from studies of prenatally androgenized female rhesus monkeys. Trends Endocrinol. Metab. 1998, 9, 62–67. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, Y.; Liu, X.; Xu, L.; Zhou, L.; Tang, L.; Zhuang, J.; Guo, W.; Hu, R. High Intake of Energy and Fat in Southwest Chinese Women with PCOS: A Population-Based Case-Control Study. PLoS ONE 2015, 10, e0127094. [Google Scholar] [CrossRef] [PubMed]
- Lindheim, L.; Bashir, M.; Munzker, J.; Trummer, C.; Zachhuber, V.; Leber, B.; Horvath, A.; Pieber, T.R.; Gorkiewicz, G.; Stadlbauer, V.; et al. Alterations in Gut Microbiome Composition and Barrier Function Are Associated with Reproductive and Metabolic Defects in Women with Polycystic Ovary Syndrome (PCOS): A Pilot Study. PLoS ONE 2017, 12, e0168390. [Google Scholar] [CrossRef]
- Heintz-Buschart, A.; Wilmes, P. Human Gut Microbiome: Function Matters. Trends Microbiol. 2018, 26, 563–574. [Google Scholar] [CrossRef]
- Insenser, M.; Murri, M.; Del Campo, R.; Martinez-Garcia, M.A.; Fernandez-Duran, E.; Escobar-Morreale, H.F. Gut Microbiota and the Polycystic Ovary Syndrome: Influence of Sex, Sex Hormones, and Obesity. J. Clin. Endocrinol. Metab. 2018, 103, 2552–2562. [Google Scholar] [CrossRef]
- Liu, R.; Zhang, C.; Shi, Y.; Zhang, F.; Li, L.; Wang, X.; Ling, Y.; Fu, H.; Dong, W.; Shen, J.; et al. Dysbiosis of Gut Microbiota Associated with Clinical Parameters in Polycystic Ovary Syndrome. Front. Microbiol. 2017, 8, 324. [Google Scholar] [CrossRef]
- Turnbaugh, P.J.; Hamady, M.; Yatsunenko, T.; Cantarel, B.L.; Duncan, A.; Ley, R.E.; Sogin, M.L.; Jones, W.J.; Roe, B.A.; Affourtit, J.P.; et al. A core gut microbiome in obese and lean twins. Nature 2009, 457, 480–484. [Google Scholar] [CrossRef] [Green Version]
- Turnbaugh, P.J.; Ley, R.E.; Mahowald, M.A.; Magrini, V.; Mardis, E.R.; Gordon, J.I. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 2006, 444, 1027–1031. [Google Scholar] [CrossRef]
- Vrieze, A.; Van Nood, E.; Holleman, F.; Salojarvi, J.; Kootte, R.S.; Bartelsman, J.F.; Dallinga-Thie, G.M.; Ackermans, M.T.; Serlie, M.J.; Oozeer, R.; et al. Transfer of intestinal microbiota from lean donors increases insulin sensitivity in individuals with metabolic syndrome. Gastroenterology 2012, 143, 913–916. [Google Scholar] [CrossRef] [PubMed]
- Barber, T.M.; Dimitriadis, G.K.; Andreou, A.; Franks, S. Polycystic ovary syndrome: Insight into pathogenesis and a common association with insulin resistance. Clin. Med. 2016, 16, 262–266. [Google Scholar] [CrossRef] [Green Version]
- Markle, J.G.; Frank, D.N.; Mortin-Toth, S.; Robertson, C.E.; Feazel, L.M.; Rolle-Kampczyk, U.; von Bergen, M.; McCoy, K.D.; Macpherson, A.J.; Danska, J.S. Sex differences in the gut microbiome drive hormone-dependent regulation of autoimmunity. Science 2013, 339, 1084–1088. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, Y.; Qi, Y.; Yang, X.; Zhao, L.; Wen, S.; Liu, Y.; Tang, L. Association between Polycystic Ovary Syndrome and Gut Microbiota. PLoS ONE 2016, 11, e0153196. [Google Scholar] [CrossRef] [Green Version]
- Sherman, S.B.; Sarsour, N.; Salehi, M.; Schroering, A.; Mell, B.; Joe, B.; Hill, J.W. Prenatal androgen exposure causes hypertension and gut microbiota dysbiosis. Gut Microbes 2018, 9, 400–421. [Google Scholar] [CrossRef] [Green Version]
- Le Roy, C.I.; Beaumont, M.; Jackson, M.A.; Steves, C.J.; Spector, T.D.; Bell, J.T. Heritable components of the human fecal microbiome are associated with visceral fat. Gut Microbes 2018, 9, 61–67. [Google Scholar] [CrossRef] [PubMed]
- Lim, M.Y.; You, H.J.; Yoon, H.S.; Kwon, B.; Lee, J.Y.; Lee, S.; Song, Y.M.; Lee, K.; Sung, J.; Ko, G. The effect of heritability and host genetics on the gut microbiota and metabolic syndrome. Gut 2017, 66, 1031–1038. [Google Scholar] [CrossRef]
- Kurilshikov, A.; Medina-Gomez, C.; Bacigalupe, R.; Radjabzadeh, D.; Wang, J.; Demirkan, A.; Le Roy, C.I.; Raygoza Garay, J.A.; Finnicum, C.T.; Liu, X.; et al. Large-scale association analyses identify host factors influencing human gut microbiome composition. Nat. Genet. 2021, 53, 156–165. [Google Scholar] [CrossRef] [PubMed]
- Tremellen, K.; Pearce, K. Dysbiosis of Gut Microbiota (DOGMA)--a novel theory for the development of Polycystic Ovarian Syndrome. Med. Hypotheses 2012, 79, 104–112. [Google Scholar] [CrossRef]
- Frohlich, E.; Wahl, R. Microbiota and Thyroid Interaction in Health and Disease. Trends Endocrinol. Metab. 2019, 30, 479–490. [Google Scholar] [CrossRef] [Green Version]
- Singla, R.; Gupta, Y.; Khemani, M.; Aggarwal, S. Thyroid disorders and polycystic ovary syndrome: An emerging relationship. Indian J. Endocrinol. Metab. 2015, 19, 25–29. [Google Scholar] [CrossRef]
- Ferriman, D.; Gallwey, J.D. Clinical assessment of body hair growth in women. J. Clin. Endocrinol. Metab. 1961, 21, 1440–1447. [Google Scholar] [CrossRef] [PubMed]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glockner, F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013, 41, e1. [Google Scholar] [CrossRef] [PubMed]
- Fadrosh, D.W.; Ma, B.; Gajer, P.; Sengamalay, N.; Ott, S.; Brotman, R.M.; Ravel, J. An improved dual-indexing approach for multiplexed 16S rRNA gene sequencing on the Illumina MiSeq platform. Microbiome 2014, 2, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [Green Version]
- DeSantis, T.Z.; Hugenholtz, P.; Larsen, N.; Rojas, M.; Brodie, E.L.; Keller, K.; Huber, T.; Dalevi, D.; Hu, P.; Andersen, G.L. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006, 72, 5069–5072. [Google Scholar] [CrossRef] [Green Version]
- Macklin, M.T.; Mann, H.B. Fallacies inherent in the proband method of analysis of human pedigrees for inheritance of recessive traits; two methods of correction of the formula. Am. J. Dis. Child. 1947, 74, 456–467. [Google Scholar] [CrossRef]
- McIver, L.J.; Abu-Ali, G.; Franzosa, E.A.; Schwager, R.; Morgan, X.C.; Waldron, L.; Segata, N.; Huttenhower, C. bioBakery: A meta’omic analysis environment. Bioinformatics 2018, 34, 1235–1237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Oksanen, J.K.R.; Legendre, P.; O’Hara, B.; Simpson, G.; Henry, M.; Stevens, H.; Wagner, H. Community Ecology Package (Vegan); R Foundation for Statistical Computing: Vienna, Austria, 2008. [Google Scholar]
- Das, B.; Ghosh, T.S.; Kedia, S.; Rampal, R.; Saxena, S.; Bag, S.; Mitra, R.; Dayal, M.; Mehta, O.; Surendranath, A.; et al. Analysis of the Gut Microbiome of Rural and Urban Healthy Indians Living in Sea Level and High Altitude Areas. Sci. Rep. 2018, 8, 10104. [Google Scholar] [CrossRef] [PubMed]
- Jobira, B.; Frank, D.N.; Pyle, L.; Silveira, L.J.; Kelsey, M.M.; Garcia-Reyes, Y.; Robertson, C.E.; Ir, D.; Nadeau, K.J.; Cree-Green, M. Obese Adolescents With PCOS Have Altered Biodiversity and Relative Abundance in Gastrointestinal Microbiota. J. Clin. Endocrinol. Metab. 2020, 105, e2134–e2144. [Google Scholar] [CrossRef] [PubMed]
- Torres, P.J.; Siakowska, M.; Banaszewska, B.; Pawelczyk, L.; Duleba, A.J.; Kelley, S.T.; Thackray, V.G. Gut Microbial Diversity in Women With Polycystic Ovary Syndrome Correlates With Hyperandrogenism. J. Clin. Endocrinol. Metab. 2018, 103, 1502–1511. [Google Scholar] [CrossRef] [PubMed]
- Abdelazim, I.A.; Amer, O.O.; Farghali, M. Common endocrine disorders associated with the polycystic ovary syndrome. Prz. Menopauzalny 2020, 19, 179–183. [Google Scholar] [CrossRef] [PubMed]
- Barlow, G.M.; Yu, A.; Mathur, R. Role of the Gut Microbiome in Obesity and Diabetes Mellitus. Nutr. Clin. Pract. 2015, 30, 787–797. [Google Scholar] [CrossRef]
- Guirro, M.; Costa, A.; Gual-Grau, A.; Herrero, P.; Torrell, H.; Canela, N.; Arola, L. Effects from diet-induced gut microbiota dysbiosis and obesity can be ameliorated by fecal microbiota transplantation: A multiomics approach. PLoS ONE 2019, 14, e0218143. [Google Scholar] [CrossRef] [Green Version]
- Saad, M.J.; Santos, A.; Prada, P.O. Linking Gut Microbiota and Inflammation to Obesity and Insulin Resistance. Physiology 2016, 31, 283–293. [Google Scholar] [CrossRef]
- Zhang, J.; Sun, Z.; Jiang, S.; Bai, X.; Ma, C.; Peng, Q.; Chen, K.; Chang, H.; Fang, T.; Zhang, H. Probiotic Bifidobacterium lactis V9 Regulates the Secretion of Sex Hormones in Polycystic Ovary Syndrome Patients through the Gut-Brain Axis. mSystems 2019, 4, 17–19. [Google Scholar] [CrossRef] [Green Version]
- Hall, A.B.; Tolonen, A.C.; Xavier, R.J. Human genetic variation and the gut microbiome in disease. Nat. Rev. Genet. 2017, 18, 690–699. [Google Scholar] [CrossRef]
- Lerchbaum, E.; Giuliani, A.; Gruber, H.J.; Pieber, T.R.; Obermayer-Pietsch, B. Adult-type hypolactasia and calcium intake in polycystic ovary syndrome. Clin. Endocrinol. 2012, 77, 834–843. [Google Scholar] [CrossRef]
- Aoki, R.; Kamikado, K.; Suda, W.; Takii, H.; Mikami, Y.; Suganuma, N.; Hattori, M.; Koga, Y. A proliferative probiotic Bifidobacterium strain in the gut ameliorates progression of metabolic disorders via microbiota modulation and acetate elevation. Sci. Rep. 2017, 7, 43522. [Google Scholar] [CrossRef]
- Fawley, J.; Gourlay, D.M. Intestinal alkaline phosphatase: A summary of its role in clinical disease. J. Surg. Res. 2016, 202, 225–234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vassilatou, E. Nonalcoholic fatty liver disease and polycystic ovary syndrome. World J. Gastroenterol. 2014, 20, 8351–8363. [Google Scholar] [CrossRef]
- Gambarin-Gelwan, M.; Kinkhabwala, S.V.; Schiano, T.D.; Bodian, C.; Yeh, H.C.; Futterweit, W. Prevalence of nonalcoholic fatty liver disease in women with polycystic ovary syndrome. Clin. Gastroenterol. Hepatol. 2007, 5, 496–501. [Google Scholar] [CrossRef] [PubMed]
- Ma, R.C.; Liu, K.H.; Lam, P.M.; Cheung, L.P.; Tam, W.H.; Ko, G.T.; Chan, M.H.; Ho, C.S.; Lam, C.W.; Chu, W.C.; et al. Sonographic measurement of mesenteric fat predicts presence of fatty liver among subjects with polycystic ovary syndrome. J. Clin. Endocrinol. Metab. 2011, 96, 799–807. [Google Scholar] [CrossRef] [Green Version]
- Zhu, L.; Baker, S.S.; Gill, C.; Liu, W.; Alkhouri, R.; Baker, R.D.; Gill, S.R. Characterization of gut microbiomes in nonalcoholic steatohepatitis (NASH) patients: A connection between endogenous alcohol and NASH. Hepatology 2013, 57, 601–609. [Google Scholar] [CrossRef]
- Jobira, B.; Frank, D.N.; Silveira, L.J.; Pyle, L.; Kelsey, M.M.; Garcia-Reyes, Y.; Robertson, C.E.; Ir, D.; Nadeau, K.J.; Cree-Green, M. Hepatic steatosis relates to gastrointestinal microbiota changes in obese girls with polycystic ovary syndrome. PLoS ONE 2021, 16, e0245219. [Google Scholar] [CrossRef]
- Zhang, B.; Shen, S.; Gu, T.; Hong, T.; Liu, J.; Sun, J.; Wang, H.; Bi, Y.; Zhu, D. Increased circulating conjugated primary bile acids are associated with hyperandrogenism in women with polycystic ovary syndrome. J. Steroid Biochem. Mol. Biol. 2019, 189, 171–175. [Google Scholar] [CrossRef]
- Ridlon, J.M.; Kang, D.J.; Hylemon, P.B. Bile salt biotransformations by human intestinal bacteria. J. Lipid Res. 2006, 47, 241–259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tanaka, H.; Doesburg, K.; Iwasaki, T.; Mierau, I. Screening of lactic acid bacteria for bile salt hydrolase activity. J. Dairy Sci. 1999, 82, 2530–2535. [Google Scholar] [CrossRef]
- Moreno-Indias, I.; Sanchez-Alcoholado, L.; Sanchez-Garrido, M.A.; Martin-Nunez, G.M.; Perez-Jimenez, F.; Tena-Sempere, M.; Tinahones, F.J.; Queipo-Ortuno, M.I. Neonatal Androgen Exposure Causes Persistent Gut Microbiota Dysbiosis Related to Metabolic Disease in Adult Female Rats. Endocrinology 2016, 157, 4888–4898. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lagou, V.; Jiang, L.; Ulrich, A.; Zudina, L.; González, K.S.G.; Balkhiyarova, Z.; Faggian, A.; Chen, S.; Todorov, P.; Sharapov, S.; et al. Random glucose GWAS in 493,036 individuals provides insights into diabetes pathophysiology, complications and treatment stratification. MedRxiv 2021. [Google Scholar] [CrossRef]
- Hiam, D.; Moreno-Asso, A.; Teede, H.J.; Laven, J.S.E.; Stepto, N.K.; Moran, L.J.; Gibson-Helm, M. The Genetics of Polycystic Ovary Syndrome: An Overview of Candidate Gene Systematic Reviews and Genome-Wide Association Studies. J. Clin. Med. 2019, 8, 1606. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Movva, V.C.; Williams, M.S.; Lee, M.T.M. Polycystic Ovary Syndrome Susceptibility Loci Inform Disease Etiological Heterogeneity. J. Clin. Med. 2021, 10, 2688. [Google Scholar] [CrossRef] [PubMed]
- Al Rasheed, M.R.; Senseng, C.G. Sarcina ventriculi: Review of the Literature. Arch. Pathol. Lab. Med. 2016, 140, 1441–1445. [Google Scholar] [CrossRef] [PubMed]
- Lull, K.; Arffman, R.K.; Sola-Leyva, A.; Molina, N.M.; Aasmets, O.; Herzig, K.H.; Plaza-Diaz, J.; Franks, S.; Morin-Papunen, L.; Tapanainen, J.S.; et al. The Gut Microbiome in Polycystic Ovary Syndrome and Its Association with Metabolic Traits. J. Clin. Endocrinol. Metab. 2021, 106, 858–871. [Google Scholar] [CrossRef] [PubMed]
- Lindheim, L.; Bashir, M.; Munzker, J.; Trummer, C.; Zachhuber, V.; Pieber, T.R.; Gorkiewicz, G.; Obermayer-Pietsch, B. The Salivary Microbiome in Polycystic Ovary Syndrome (PCOS) and Its Association with Disease-Related Parameters: A Pilot Study. Front. Microbiol. 2016, 7, 1270. [Google Scholar] [CrossRef]
| Cases (n = 19) | Controls (n = 20) | p-Value * | |
|---|---|---|---|
| Mean (SD) or n [%] | Mean (SD) or n [%] | ||
| Anthropometrics and blood measurements | |||
| Age, years | 23.9 (6.9) | 21.1 (2.5) | 0.093 |
| BMI, kg/m2 | 25.4 (3.3) | 23.2 (3.2) | 0.041 |
| Waist–hip ratio | 0.9 (0.1) | 0.9 (0) | 0.73 |
| Systolic blood pressure, mmHg | 124.5 (9.7) | 120.8 (6.1) | 0.16 |
| Diastolic blood pressure, mmHg | 81.6 (7.6) | 80.5 (3.9) | 0.58 |
| Hormonal and Biochemical Parameters | |||
| Luteinizing hormone (LH), IU/L | 5 (2.8) | 4.3 (0.6) | 0.28 |
| Follicle stimulating hormone (FSH), IU/L | 5.5 (1.6) | 5.1 (1.2) | 0.34 |
| LH/FSH ratio | 0.97 (0.70) | 0.90 (0.28) | 0.66 |
| Testosterone, ng/dL | 56.8 (26.6) | 49.6 (12.7) | 0.28 |
| Prolactin, ng/dL | 15.7 (7.2) | 10 (4.7) | 5.64 × 10−3 |
| Fasting blood glucose, mg/dL | 114.2 (8.6) | 106.1 (9.1) | 6.71 × 10−3 |
| Blood glucose 1 h, mg/dL | 126 (11.8) | 120 (24.3) | 0.33 |
| Blood glucose 2 h, mg/dL | 121.2 (10.7) | 121.3 (22.3) | 0.99 |
| Total cholesterol, mg/dL | 164.7 (31.8) | 149.1 (26.9) | 0.11 |
| Triglycerides, mg/dL | 205.9 (105.8) | 144.3 (96.6) | 0.065 |
| High-density lipoprotein (HDL), mg/dL | 39.6 (4.6) | 41.8 (4.4) | 0.14 |
| Low-density lipoprotein (LDL), mg/dL | 110.2 (8.4) | 108.4 (9.5) | 0.54 |
| PCOS-related clinical criteria | |||
| Age at menarche, years | 13.3 (2.2) | 12.2 (1.2) | 0.063 |
| Number of cycles per year | 6.8 (1.9) | 10.7 (1) | 1.69 × 10−9 |
| Hirsutism score | 7.3 (5.5) | 0.4 (1.8) | 6.03 × 10−6 |
| Menstrual cycle irregularity, yes | 19 [100] | 0 [0] | 1.45 × 10−11 |
| Acanthosis nigricans, yes | 5 [26] | 0 [0] | 0.02 |
| Acne, yes | 7 [37] | 2 [10] | 0.06 |
| Alopecia, yes | 9 [47] | 4 [20] | 0.10 |
| Duration of hirsutism, years | 1.8 (0.8) | 0.1 (0.2) | 1.87 × 10−11 |
| Family history of hirsutism, yes | 3 [16] | 2 [10] | 0.66 |
| Family history of menstrual disturbances, yes | 5 [26] | 2 [10] | 0.24 |
| Family history of T2D, yes | 11 [58] | 7 [35] | 0.2 |
| Stool sample collection and sequencing | |||
| Day of menstrual cycle at stool collection | |||
| Day 3 | 7 [37] | 6 [30] | 1.00 |
| Day 4 | 5 [26] | 6 [30] | |
| Day 5 | 6 [32] | 7 [35] | |
| Day 6 | 1 [5] | 1 [5] | |
| Stool consistency | |||
| Sausage shaped with cracks on the surface | 3 [16] | 2 [10] | 0.66 |
| Sausage shaped and smooth soft stool | 12 [63] | 15 [75] | |
| Solid clumpy stool | 3 [16] | 1 [5] | |
| Watery stool | 1 [5] | 2 [10] | |
| Read depth, genus level | 80,434.6 (44,927.2) | 161,279.9 (158,861.1) | 0.039 |
| Read depth, family level | 81,547.8 (45,539.7) | 164,695 (162,499.2) | 0.038 |
| Mann–Whitney U-Test | MaAsLin | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Unadjusted | Model 1 | Model 2 | |||||||||||
| Feature | Case | Control | p-Value | n | Coefficient | p-Value | Q-Value | Coefficient | p-Value | Q-Value | Coefficient | p-Value | Q-Value |
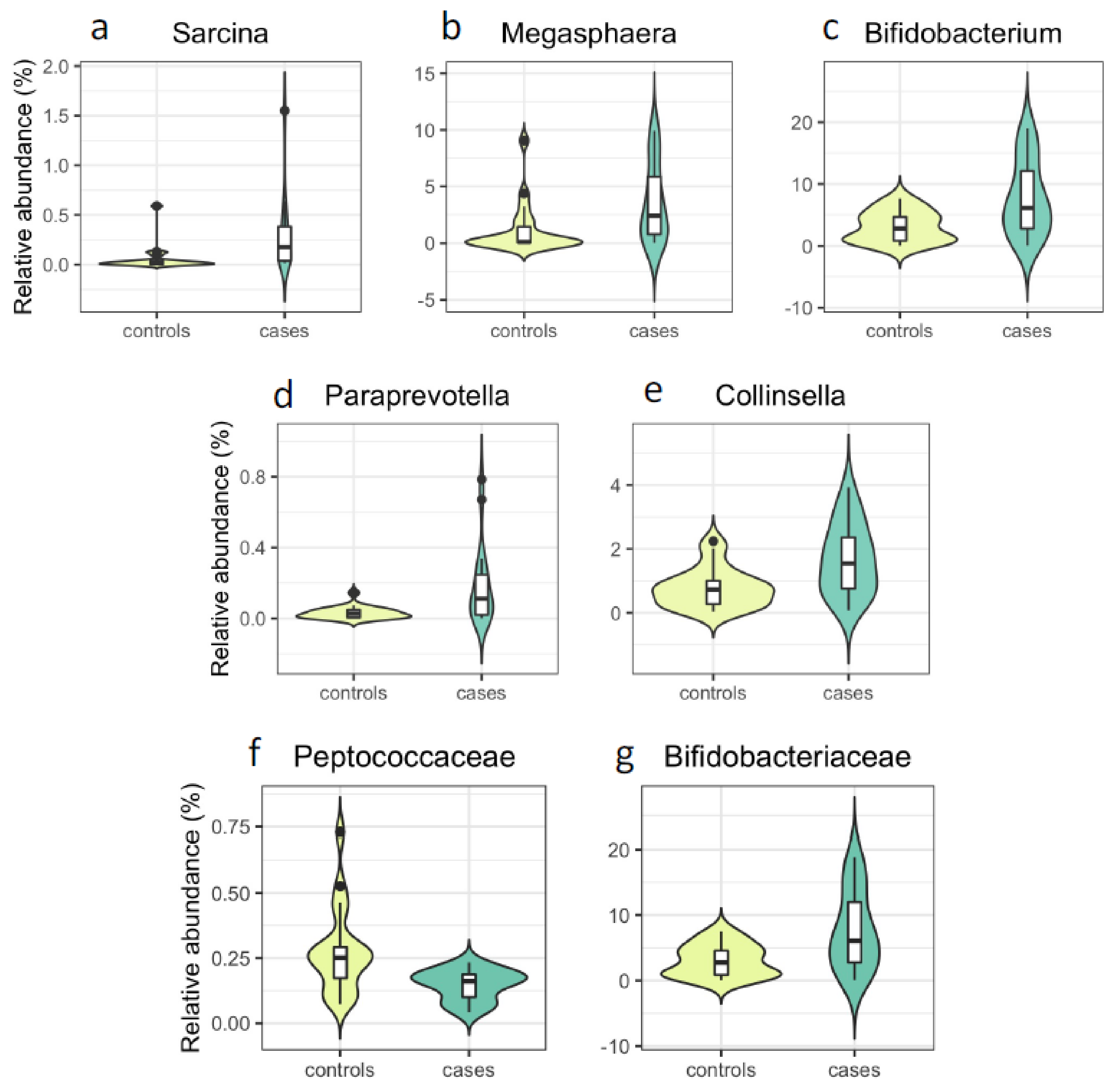
| Sarcina | 0.28 | 0.059 | 3.4 × 10−4 | 36 | 0.023 | 0.001 | 0.025 | 0.023 | 0.001 | 0.066 | 0.024 | 0.001 | 0.127 |
| Megasphaera | 3.62 | 1.17 | 2.1 × 10−3 | 39 | 0.093 | 0.003 | 0.048 | 0.099 | 0.003 | 0.104 | 0.098 | 0.005 | 0.190 |
| Bifidobacterium | 7.72 | 3.12 | 7.5 × 10−3 | 39 | 0.096 | 0.006 | 0.048 | 0.100 | 0.004 | 0.115 | 0.103 | 0.005 | 0.190 |
| Paraprevotella | 0.18 | 0.032 | 7.3 × 10−3 | 37 | 0.014 | 0.004 | 0.048 | 0.014 | 0.001 | 0.066 | 0.012 | 0.006 | 0.192 |
| Collinsella | 1.68 | 0.78 | 7.5 × 10−3 | 39 | 0.041 | 0.004 | 0.048 | 0.043 | 0.002 | 0.101 | 0.043 | 0.004 | 0.179 |
| Erysipelothrix | 1.52 | 0.29 | 7.5 × 10−3 | 39 | 0.017 | 0.036 | 0.219 | 0.018 | 0.022 | 0.342 | 0.016 | 0.058 | 0.475 |
| Lactobacillus | 5.30 | 1.90 | 0.012 | 39 | 0.086 | 0.006 | 0.048 | 0.085 | 0.008 | 0.207 | 0.084 | 0.014 | 0.304 |
| Dysgonomonas | 0.66 | 0.18 | 0.013 | 39 | 0.016 | 0.035 | 0.219 | 0.016 | 0.047 | 0.481 | 0.016 | 0.063 | 0.501 |
| Oscillospira | 2.22 | 1.68 | 0.018 | 39 | 0.033 | 0.168 | 0.454 | 0.039 | 0.06 | 0.487 | 0.036 | 0.104 | 0.574 |
| Natronincola | 0.35 | 0.31 | 0.030 | 38 | 0.016 | 0.049 | 0.256 | 0.017 | 0.016 | 0.298 | 0.016 | 0.031 | 0.456 |
| Alkalibacterium | 0.089 | 0.32 | 0.049 | 35 | 0.007 | 0.001 | 0.025 | 0.007 | 0.000 | 0.066 | 0.006 | 0.002 | 0.145 |
| Atopobium | 0.27 | 0.08 | 0.066 | 36 | 0.020 | 0.038 | 0.219 | 0.019 | 0.042 | 0.481 | 0.019 | 0.057 | 0.475 |
| Feature | Mann–Whitney U-Test | MaAsLin | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean (%) in Cases (n = 19) | Mean (%) in Controls (n = 20) | p-Value | Unadjusted | Model 1 | Model 2 | ||||||||
| n | Coefficient | p-Value | Q-Value | Coefficient | p-Value | Q-Value | Coefficient | p-Value | Q-Value | ||||
| Peptococcaceae | 0.15 | 0.27 | 1.5 × 10−3 | 39 | −0.011 | 0.003 | 0.055 | −0.010 | 0.005 | 0.246 | −0.010 | 0.005 | 0.233 |
| Bifidobacteriaceae | 7.63 | 3.07 | 7.5 × 10−3 | 39 | 0.096 | 0.005 | 0.055 | 0.100 | 0.003 | 0.233 | 0.103 | 0.005 | 0.233 |
| Lactobacillaceae | 5.26 | 1.89 | 0.012 | 39 | 0.086 | 0.005 | 0.055 | 0.085 | 0.008 | 0.307 | 0.083 | 0.014 | 0.349 |
| Erysipelotrichaceae | 2.50 | 0.91 | 0.026 | 39 | 0.030 | 0.034 | 0.254 | 0.029 | 0.045 | 0.457 | 0.022 | 0.124 | 0.680 |
| Porphyromonadaceae | 0.88 | 0.43 | 0.047 | 39 | 0.015 | 0.058 | 0.254 | 0.016 | 0.051 | 0.461 | 0.015 | 0.094 | 0.680 |
| Aerococcaceae | 0.08 | 0.31 | 0.052 | 35 | 0.007 | 0.001 | 0.037 | 0.007 | 0.000 | 0.082 | 0.006 | 0.002 | 0.233 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hassan, S.; Kaakinen, M.A.; Draisma, H.; Zudina, L.; Ganie, M.A.; Rashid, A.; Balkhiyarova, Z.; Kiran, G.S.; Vogazianos, P.; Shammas, C.; et al. Bifidobacterium Is Enriched in Gut Microbiome of Kashmiri Women with Polycystic Ovary Syndrome. Genes 2022, 13, 379. https://doi.org/10.3390/genes13020379
Hassan S, Kaakinen MA, Draisma H, Zudina L, Ganie MA, Rashid A, Balkhiyarova Z, Kiran GS, Vogazianos P, Shammas C, et al. Bifidobacterium Is Enriched in Gut Microbiome of Kashmiri Women with Polycystic Ovary Syndrome. Genes. 2022; 13(2):379. https://doi.org/10.3390/genes13020379
Chicago/Turabian StyleHassan, Saqib, Marika A. Kaakinen, Harmen Draisma, Liudmila Zudina, Mohd A. Ganie, Aafia Rashid, Zhanna Balkhiyarova, George S. Kiran, Paris Vogazianos, Christos Shammas, and et al. 2022. "Bifidobacterium Is Enriched in Gut Microbiome of Kashmiri Women with Polycystic Ovary Syndrome" Genes 13, no. 2: 379. https://doi.org/10.3390/genes13020379
APA StyleHassan, S., Kaakinen, M. A., Draisma, H., Zudina, L., Ganie, M. A., Rashid, A., Balkhiyarova, Z., Kiran, G. S., Vogazianos, P., Shammas, C., Selvin, J., Antoniades, A., Demirkan, A., & Prokopenko, I. (2022). Bifidobacterium Is Enriched in Gut Microbiome of Kashmiri Women with Polycystic Ovary Syndrome. Genes, 13(2), 379. https://doi.org/10.3390/genes13020379








