Was the Last Bacterial Common Ancestor a Monoderm after All?
Abstract
1. Introduction
2. Materials and Methods
2.1. Dataset Assembly
2.1.1. Data Download
2.1.2. Genome Dereplication and Selection
2.1.3. Identification of Orthologous Groups
2.1.4. Database Creation
2.2. Evolution of the Bacterial Domain
2.2.1. Supermatrix Assembly
2.2.2. Phylogenomic Analyses
2.2.3. Congruence Tests
2.3. Evolution of the Cell-Wall
2.3.1. Cell-Wall Architecture of Extant Organisms
2.3.2. Correlation between Cell-Wall Traits
2.3.3. Ancestral State Reconstruction of Cell-Wall Traits
2.3.4. Comparison of the Selected Models
2.4. Evolution of the dcw Cluster
2.4.1. Synteny Analyses of Extant Genomes
2.4.2. Ancestral Gene Order Reconstruction
2.4.3. Phylogenetic Analyses
2.5. Evolution of the Genes Related to the Outer Membrane
2.5.1. Homology Searches in Complete Proteomes
2.5.2. Taxonomic and Phylogenetic Analyses
3. Results
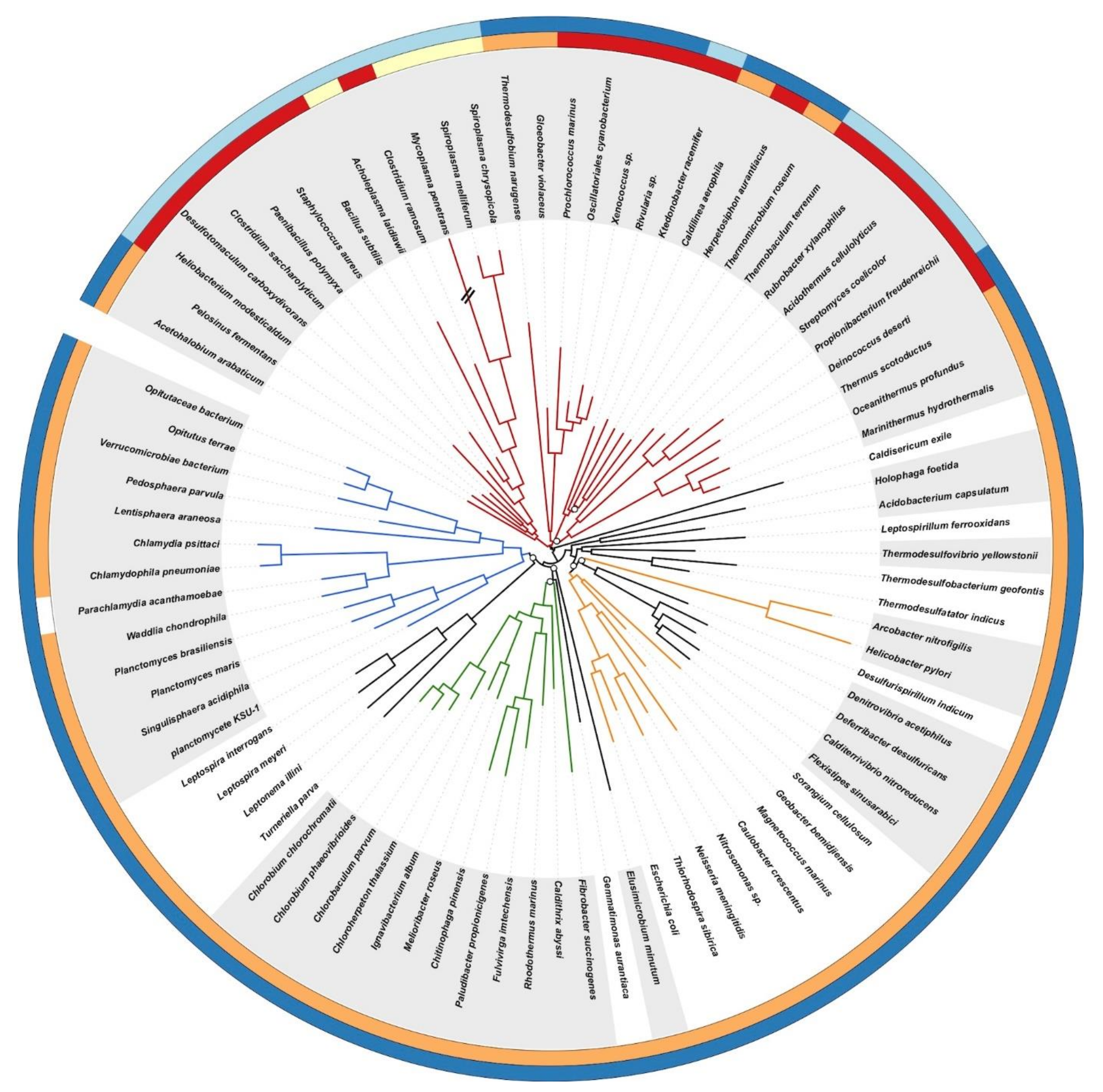
3.1. A Robust Tree of the Bacterial Domain
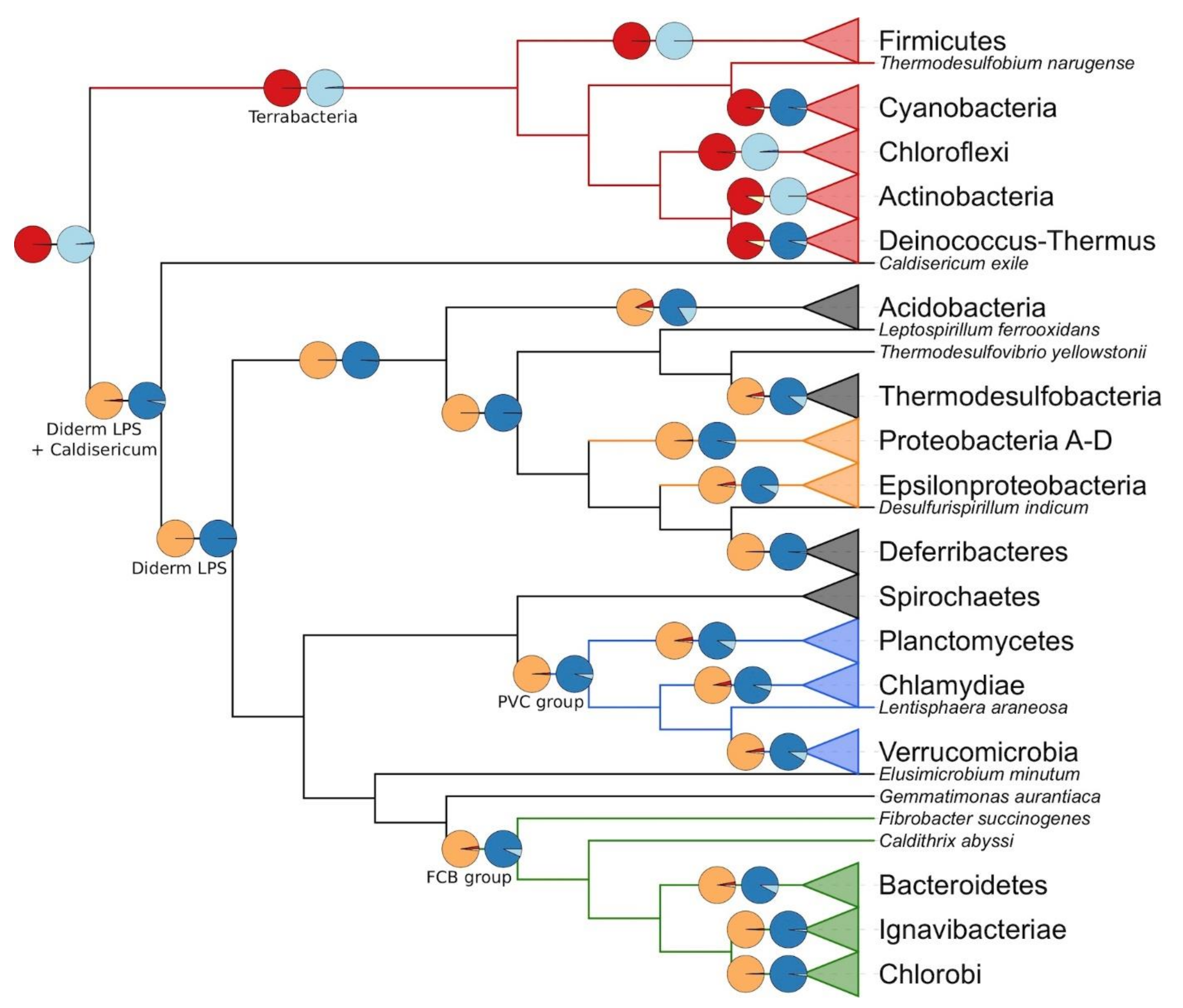
3.2. Evolution of the Cell-Wall Architecture
3.3. Evolution of the Gene Order within the dcw Cluster
3.4. Evolution of the Genes Related to the Outer Membrane
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schleifer, K.H.; Kandler, O. Peptidoglycan Types of Bacterial Cell Walls and Their Taxonomic Implications. Bacteriol. Rev. 1972, 36, 407–477. [Google Scholar] [CrossRef] [PubMed]
- Coico, R. Gram Staining. Curr. Protoc. Microbiol. 2006, A-3C. [Google Scholar]
- Silhavy, T.J.; Kahne, D.; Walker, S. The Bacterial Cell Envelope. Cold Spring Harb. Perspect. Biol. 2010, 2, a000414. [Google Scholar] [CrossRef] [PubMed]
- Woese, C.R. Bacterial Evolution. Microbiol. Rev. 1987, 51, 221–271. [Google Scholar] [CrossRef]
- Megrian, D.; Taib, N.; Witwinowski, J.; Beloin, C.; Gribaldo, S. One or Two Membranes? Diderm Firmicutes Challenge the Gram-Positive/Gram-Negative Divide. Mol. Microbiol. 2020, 113, 659–671. [Google Scholar] [CrossRef] [PubMed]
- Cavalier-Smith, T. The Origin of Eukaryote and Archaebacterial Cells. Ann. N. Y. Acad. Sci. 1987, 503, 17–54. [Google Scholar] [CrossRef] [PubMed]
- Cavalier-Smith, T. Deep Phylogeny, Ancestral Groups and the Four Ages of Life. Philos. Trans. R. Soc. B Biol. Sci. 2010, 365, 111–132. [Google Scholar] [CrossRef]
- Koch, A.L. Were Gram-Positive Rods the First Bacteria? TRENDS Microbiol. 2003, 11, 166–170. [Google Scholar] [CrossRef]
- Sutcliffe, I.C. A Phylum Level Perspective on Bacterial Cell Envelope Architecture. Trends Microbiol. 2010, 18, 464–470. [Google Scholar] [CrossRef]
- Gupta, R.S. Origin of Diderm (Gram-Negative) Bacteria: Antibiotic Selection Pressure Rather than Endosymbiosis Likely Led to the Evolution of Bacterial Cells with Two Membranes. Antonie Van Leeuwenhoek Int. J. Gen. Mol. Microbiol. 2011, 100, 171–182. [Google Scholar] [CrossRef]
- Errington, J. L-Form Bacteria, Cell Walls and the Origins of Life. Open Biol. 2013, 3, 120143. [Google Scholar] [CrossRef]
- Lake, J.A. Evidence for an Early Prokaryotic Endosymbiosis. Nature 2009, 460, 967–971. [Google Scholar] [CrossRef] [PubMed]
- Tocheva, E.I.; Matson, E.G.; Morris, D.M.; Moussavi, F.; Leadbetter, J.R.; Jensen, G.J. Peptidoglycan Remodeling and Conversion of an Inner Membrane into an Outer Membrane during Sporulation. Cell 2011, 146, 799–812. [Google Scholar] [CrossRef]
- Tocheva, E.I.; Ortega, D.R.; Jensen, G.J. Sporulation, Bacterial Cell Envelopes and the Origin of Life. Nat. Rev. Microbiol. 2016, 14, 535–542. [Google Scholar] [CrossRef]
- Vollmer, W. Bacterial Outer Membrane Evolution via Sporulation? Nat. Chem. Biol. 2011, 8, 14–18. [Google Scholar] [CrossRef]
- Antunes, L.C.S.; Poppleton, D.; Klingl, A.; Criscuolo, A.; Dupuy, B.; Brochier-Armanet, C.; Beloin, C.; Gribaldo, S. Phylogenomic Analysis Supports the Ancestral Presence of LPS-Outer Membranes in the Firmicutes. eLife 2016, 5, 1–21. [Google Scholar] [CrossRef]
- Taib, N.; Megrian, D.; Witwinowski, J.; Adam, P.; Poppleton, D.; Borrel, G.; Beloin, C.; Gribaldo, S. Genome-Wide Analysis of the Firmicutes Illuminates the Diderm/Monoderm Transition. Nat. Ecol. Evol. 2020, 4, 1661–1672. [Google Scholar] [CrossRef]
- Mingorance, J.; Tamames, J. The Bacterial Dcw Gene Cluster: An Island in the Genome? In Molecules in Time and Space; Springer: Boston, MA, USA, 2004; pp. 249–271. ISBN 978-0-306-48579-4. [Google Scholar]
- Tamames, J. Evolution of Gene Order Conservation in Prokaryotes. Genome Biol. 2001, 2, 1–11. [Google Scholar] [CrossRef]
- Real, G.; Henriques, A.O. Localization of the Bacillus Subtilis MurB Gene within the Dcw Cluster Is Important for Growth and Sporulation. J. Bacteriol. 2006, 188, 1721–1732. [Google Scholar] [CrossRef] [PubMed]
- Barloy-Hubler, F.; Lelaure, V.; Galibert, F. Ribosomal Protein Gene Cluster Analysis in Eubacterium Genomics: Homology between Sinorhizobium Meliloti Strain 1021 and Bacillus Subtilis. Nucleic Acids Res. 2001, 29, 2747–2756. [Google Scholar] [CrossRef] [PubMed]
- Nikolaichik, Y.A.; Donachie, W.D. Conservation of Gene Order amongst Cell Wall and Cell Division Genes in Eubacteria, and Ribosomal Genes in Eubacteria and Eukaryotic Organelles. Genetica 2000, 108, 1–7. [Google Scholar] [CrossRef]
- Eraso, J.M.; Markillie, L.M.; Mitchell, H.D.; Taylor, R.C.; Orr, G.; Margolin, W. The Highly Conserved MraZ Protein Is a Transcriptional Regulator in Escherichia Coli. J. Bacteriol. 2014, 196, 2053–2066. [Google Scholar] [CrossRef]
- Pilhofer, M.; Rappl, K.; Eckl, C.; Bauer, A.P.; Ludwig, W.; Schleifer, K.H.; Petroni, G. Characterization and Evolution of Cell Division and Cell Wall Synthesis Genes in the Bacterial Phyla Verrucomicrobia, Lentisphaerae, Chlamydiae, and Planctomycetes and Phylogenetic Comparison with RRNA Genes. J. Bacteriol. 2008, 190, 3192–3202. [Google Scholar] [CrossRef]
- Coleman, G.A.; Davín, A.A.; Mahendrarajah, T.A.; Szánthó, L.L.; Spang, A.; Hugenholtz, P.; Szöllősi, G.J.; Williams, T.A. A Rooted Phylogeny Resolves Early Bacterial Evolution. Science 2021, 372. [Google Scholar] [CrossRef] [PubMed]
- Kersey, P.J.; Allen, J.E.; Christensen, M.; Davis, P.; Falin, L.J.; Grabmueller, C.; Hughes, D.S.T.; Humphrey, J.; Kerhornou, A.; Khobova, J.; et al. Ensembl Genomes 2013: Scaling up Access to Genome-Wide Data. Nucleic Acids Res. 2014, 42, D546–D552. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Hagelsieb, G.; Wang, Z.; Walsh, S.; ElSherbiny, A. Phylogenomic Clustering for Selecting Non-Redundant Genomes for Comparative Genomics. Bioinformatics 2013, 29, 947–949. [Google Scholar] [CrossRef] [PubMed]
- Léonard, R.R.; Leleu, M.; Vlierberghe, M.V.; Cornet, L.; Kerff, F.; Baurain, D. ToRQuEMaDA: Tool for Retrieving Queried Eubacteria, Metadata and Dereplicating Assemblies. PeerJ 2021, 9, e11348. [Google Scholar] [CrossRef]
- Rice, P.; Longden, I.; Bleasby, A. EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet. 2000, 16, 276–277. [Google Scholar] [CrossRef]
- Sayers, E.W.; Barrett, T.; Benson, D.A.; Bolton, E.; Bryant, S.H.; Canese, K.; Chetvernin, V.; Church, D.M.; DiCuccio, M.; Federhen, S.; et al. Database Resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2011, 39, D38–D51. [Google Scholar] [CrossRef]
- R Core Team, Rf. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2013. [Google Scholar]
- Edgar, R.C. Search and Clustering Orders of Magnitude Faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef]
- Li, L.; Stoeckert, C.J.; Roos, D.S. OrthoMCL: Identification of Ortholog Groups for Eukaryotic Genomes. Genome Res. 2003, 13, 2178–2189. [Google Scholar] [CrossRef]
- Van Vlierberghe, M.; Philippe, H.; Baurain, D. Broadly Sampled Orthologous Groups of Eukaryotic Proteins for the Phylogenetic Study of Plastid-Bearing Lineages. BMC Res. Notes 2021, 14, 143. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Castresana, J. Selection of Conserved Blocks from Multiple Alignments for Their Use in Phylogenetic Analysis. Mol. Biol. Evol. 2000, 17, 540–552. [Google Scholar] [CrossRef] [PubMed]
- Roure, B.; Rodriguez-Ezpeleta, N.; Philippe, H. SCaFoS: A Tool for Selection, Concatenation and Fusion of Sequences for Phylogenomics. BMC Evol. Biol. 2007, 7 Suppl. S1, S2. [Google Scholar] [CrossRef]
- Lartillot, N.; Rodrigue, N.; Stubbs, D.; Richer, J. PhyloBayes MPI: Phylogenetic Reconstruction with Infinite Mixtures of Profiles in a Parallel Environment. Syst. Biol. 2013, 62, 611–615. [Google Scholar] [CrossRef]
- Lartillot, N.; Philippe, H. A Bayesian Mixture Model for Across-Site Heterogeneities in the Amino-Acid Replacement Process. Mol. Biol. Evol. 2004, 21, 1095–1109. [Google Scholar] [CrossRef]
- Lartillot, N.; Philippe, H. Computing Bayes Factors Using Thermodynamic Integration. Syst. Biol. 2006, 55, 195–207. [Google Scholar] [CrossRef]
- Lartillot, N.; Brinkmann, H.; Philippe, H. Suppression of Long-Branch Attraction Artefacts in the Animal Phylogeny Using a Site-Heterogeneous Model. BMC Evol. Biol. 2007, 7 Suppl. S1, S4. [Google Scholar] [CrossRef]
- Lartillot, N.; Lepage, T.; Blanquart, S. PhyloBayes 3: A Bayesian Software Package for Phylogenetic Reconstruction and Molecular Dating. Bioinformatics 2009, 25, 2286–2288. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree Of Life (ITOL) v5: An Online Tool for Phylogenetic Tree Display and Annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef] [PubMed]
- De Vienne, D.M.; Ollier, S.; Aguileta, G. Phylo-MCOA: A Fast and Efficient Method to Detect Outlier Genes and Species in Phylogenomics Using Multiple Co-Inertia Analysis. Mol. Biol. Evol. 2012, 29, 1587–1598. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A. RAxML Version 8: A Tool for Phylogenetic Analysis and Post-Analysis of Large Phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed]
- Pagel, M.; Meade, A.; Barker, D. Bayesian Estimation of Ancestral Character States on Phylogenies. Syst. Biol. 2004, 53, 673–684. [Google Scholar] [CrossRef] [PubMed]
- Pagel, M.; Meade, A. Bayesian Analysis of Correlated Evolution of Discrete Characters by Reversible-Jump Markov Chain Monte Carlo. Am. Nat. 2015, 167, 808–825. [Google Scholar] [CrossRef]
- Meade, A.; Pagel, M. BayesTraits V3. 0.1. 2017. Available online: http://www.evolution.rdg.ac.uk/BayesTraitsV3.0.1/BayesTraitsV3.0.1.html (accessed on 9 February 2022).
- Eddy, S.R. Accelerated Profile HMM Searches. PLoS Comput. Biol. 2011, 7, e1002195. [Google Scholar] [CrossRef]
- Mistry, J.; Finn, R.D.; Eddy, S.R.; Bateman, A.; Punta, M. Challenges in Homology Search: HMMER3 and Convergent Evolution of Coiled-Coil Regions. Nucleic Acids Res. 2013, 41, e121-e121. [Google Scholar] [CrossRef]
- Perrin, A.; Varré, J.-S.; Blanquart, S.; Ouangraoua, A. ProCARs: Progressive Reconstruction of Ancestral Gene Orders. BMC Genomics 2015, 16 Suppl 5, S6. [Google Scholar] [CrossRef][Green Version]
- Jones, P.; Binns, D.; Chang, H.-Y.; Fraser, M.; Li, W.; McAnulla, C.; McWilliam, H.; Maslen, J.; Mitchell, A.; Nuka, G.; et al. InterProScan 5: Genome-Scale Protein Function Classification. Bioinformatics 2014, 30, 1236–1240. [Google Scholar] [CrossRef]
- Sánchez-Pulido, L.; Devos, D.; Genevrois, S.; Vicente, M.; Valencia, A. POTRA: A Conserved Domain in the FtsQ Family and a Class of β-Barrel Outer Membrane Proteins. Trends Biochem. Sci. 2003, 28, 523–526. [Google Scholar] [CrossRef] [PubMed]
- Heinz, E.; Lithgow, T. A Comprehensive Analysis of the Omp85/TpsB Protein Superfamily Structural Diversity, Taxonomic Occurrence, and Evolution. Front. Microbiol. 2014, 5, 370. [Google Scholar] [CrossRef]
- Delsuc, F.; Brinkmann, H.; Philippe, H. Phylogenomics and the Reconstruction of the Tree of Life. Nat. Rev. Genet. 2005, 6, 361–375. [Google Scholar] [CrossRef]
- Philippe, H.; de Vienne, D.M.; Ranwez, V.; Roure, B.; Baurain, D.; Delsuc, F. Pitfalls in Supermatrix Phylogenomics. Eur. J. Taxon. 2017, 283, 1–25. [Google Scholar] [CrossRef]
- Liu, L.; Anderson, C.; Pearl, D.; Edwards, S.V. Modern Phylogenomics: Building Phylogenetic Trees Using the Multispecies Coalescent Model. In Evolutionary Genomics: Statistical and Computational Methods; Anisimova, M., Ed.; Humana: New York, NY, USA, 2019; pp. 211–239. [Google Scholar]
- Battistuzzi, F.U.; Hedges, S.B. A Major Clade of Prokaryotes with Ancient Adaptations to Life on Land. Mol. Biol. Evol. 2009, 26, 335–343. [Google Scholar] [CrossRef]
- Wu, D.; Hugenholtz, P.; Mavromatis, K.; Pukall, R.; Dalin, E.; Ivanova, N.N.; Kunin, V.; Goodwin, L.; Wu, M.; Tindall, B.J.; et al. A Phylogeny-Driven Genomic Encyclopaedia of Bacteria and Archaea. Nature 2009, 462, 1056–1060. [Google Scholar] [CrossRef] [PubMed]
- Yutin, N.; Puigbo, P.; Koonin, E.V.; Wolf, Y.I. Phylogenomics of Prokaryotic Ribosomal Proteins. Curr. Sci. 2012, 101, 1435–1439. [Google Scholar] [CrossRef] [PubMed]
- Lasek-nesselquist, E.; Gogarten, J.P. The Effects of Model Choice and Mitigating Bias on the Ribosomal Tree of Life. Mol. Phylogenet. Evol. 2013, 69, 17–38. [Google Scholar] [CrossRef]
- Rinke, C.; Schwientek, P.; Sczyrba, A.; Ivanova, N.N.; Anderson, I.J.; Cheng, J.-F.; Darling, A.E.; Malfatti, S.; Swan, B.K.; Gies, E.A.; et al. Insights into the Phylogeny and Coding Potential of Microbial Dark Matter. Nature 2013, 499, 431–437. [Google Scholar] [CrossRef] [PubMed]
- Raymann, K.; Brochier-Armanet, C.; Gribaldo, S. The Two-Domain Tree of Life Is Linked to a New Root for the Archaea. Proc. Natl. Acad. Sci. USA 2015, 112, 6670–6675. [Google Scholar] [CrossRef] [PubMed]
- Hug, L.A.; Baker, B.J.; Anantharaman, K.; Brown, C.T.; Probst, A.J.; Castelle, C.J.; Butterfield, C.N.; Hernsdorf, A.W.; Amano, Y.; Kotaro, I.; et al. A New View of the Tree of Life. Nat. Microbiol. 2016, 1, 16048. [Google Scholar] [CrossRef]
- Castelle, C.J.; Banfield, J.F. Major New Microbial Groups Expand Diversity and Alter Our Understanding of the Tree of Life. Cell 2018, 172, 1181–1197. [Google Scholar] [CrossRef]
- Parks, D.H.; Chuvochina, M.; Waite, D.W.; Rinke, C.; Skarshewski, A.; Chaumeil, P.-A.; Hugenholtz, P. A Standardized Bacterial Taxonomy Based on Genome Phylogeny Substantially Revises the Tree of Life. Nat. Biotechnol. 2018, 36, 996–1004. [Google Scholar] [CrossRef]
- Zhu, Q.; Mai, U.; Pfeiffer, W.; Janssen, S.; Asnicar, F.; Sanders, J.G.; Belda-ferre, P.; Al-ghalith, G.A.; Kopylova, E.; Mcdonald, D.; et al. Phylogenomics of 10,575 Genomes Reveals Evolutionary Proximity between Domains Bacteria and Archaea. Nat. Commun. 2019, 10, 1–14. [Google Scholar] [CrossRef]
- Cavalier-Smith, T.; Ema, E.; Chao, Y. Multidomain Ribosomal Protein Trees and the Planctobacterial Origin of Neomura (Eukaryotes, Archaebacteria). Protoplasma 2020, 1–133. [Google Scholar] [CrossRef] [PubMed]
- Cavalier-Smith, T. Rooting the Tree of Life by Transition Analyses. Biol. Direct 2006, 1, 19. [Google Scholar] [CrossRef][Green Version]
- Zhaxybayeva, O.; Swithers, K.S.; Lapierre, P.; Fournier, G.P.; Bickhart, D.M.; DeBoy, R.T.; Nelson, K.E.; Nesbø, C.L.; Doolittle, W.F.; Gogarten, J.P.; et al. On the Chimeric Nature, Thermophilic Origin, and Phylogenetic Placement of the Thermotogales. Proc. Natl. Acad. Sci. USA 2009, 106, 5865–5870. [Google Scholar] [CrossRef]
- Bhandari, V.; Naushad, H.S.; Gupta, R.S. Protein Based Molecular Markers Provide Reliable Means to Understand Prokaryotic Phylogeny and Support Darwinian Mode of Evolution. Front. Cell. Infect. Microbiol. 2012, 2, 1–14. [Google Scholar] [CrossRef]
- Eveleigh, R.J.M.; Meehan, C.J.; Archibald, J.M.; Beiko, R.G. Being Aquifex Aeolicus: Untangling a Hyperthermophile’s Checkered Past. Genome Biol. Evol. 2013, 5, 2478–2497. [Google Scholar] [CrossRef] [PubMed]
- Rachel, R.; Wildhaber, I.; Stetter, K.O.; Baumeister, W. The Structure of the Surface Protein of Thermotoga Maritima. In Crystalline Bacterial Cell Surface Layers; Springer: Berlin/Heidelberg, Germany, 1988; pp. 83–86. [Google Scholar]
- Rachel, R.; Engel, A.M.; Huber, R.; Stetter, K.-O.; Baumeister, W. A Porin-Type Protein Is the Main Constituent of the Cell Envelope of the Ancestral Eubacterium Thermotoga Maritima. FEBS Lett. 1990, 262, 64–68. [Google Scholar] [CrossRef]
- Bapteste, E.; Boucher, Y.; Leigh, J.; Doolittle, W.F. Phylogenetic Reconstruction and Lateral Gene Transfer. Trends Microbiol. 2004, 12, 406–411. [Google Scholar] [CrossRef]
- Mira, A.; Pushker, R.; Legault, B.A.; Moreira, D.; Rodríguez-Valera, F. Evolutionary Relationships of Fusobacterium Nucleatum Based on Phylogenetic Analysis and Comparative Genomics. BMC Evol. Biol. 2004, 4, 50. [Google Scholar] [CrossRef]
- Beiko, R.G.; Harlow, T.J.; Ragan, M. a Highways of Gene Sharing in Prokaryotes. Proc. Natl. Acad. Sci. USA 2005, 102, 14332–14337. [Google Scholar] [CrossRef]
- Koonin, E.V. Orthologs, Paralogs, and Evolutionary Genomics. Annu. Rev. Genet. 2005, 39, 309–338. [Google Scholar] [CrossRef]
- Koonin, E.V. Horizontal Gene Transfer: Essentiality and Evolvability in Prokaryotes, and Roles in Evolutionary Transitions. F1000Research 2016, 5, F1000 Faculty Rev-1805. [Google Scholar] [CrossRef]
- Boussau, B.; Guéguen, L.; Gouy, M. Accounting for Horizontal Gene Transfers Explains Conflicting Hypotheses Regarding the Position of Aquificales in the Phylogeny of Bacteria. BMC Evol. Biol. 2008, 8, 1–18. [Google Scholar] [CrossRef]
- Philippe, H.; Brinkmann, H.; Lavrov, D.V.; Littlewood, D.T.J.; Manuel, M.; Wörheide, G.; Baurain, D. Resolving Difficult Phylogenetic Questions: Why More Sequences Are Not Enough. PLoS Biol. 2011, 9. [Google Scholar] [CrossRef] [PubMed]
- Gouy, R.; Baurain, D.; Philippe, H. Rooting the Tree of Life: The Phylogenetic Jury Is Still Out. Philos. Trans. R. Soc. B Biol. Sci. 2015, 370, 20140329. [Google Scholar] [CrossRef] [PubMed]
- Rivas-Marín, E.; Canosa, I.; Devos, D.P. Evolutionary Cell Biology of Division Mode in the Bacterial Planctomycetes-Verrucomicrobia- Chlamydiae Superphylum. Front. Microbiol. 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Cruz-Morales, P.; Orellana, C.A.; Moutafis, G.; Moonen, G.; Rincon, G.; Nielsen, L.K.; Marcellin, E. Revisiting the Evolution and Taxonomy of Clostridia, a Phylogenomic Update. Genome Biol. Evol. 2019, 11, 2035–2044. [Google Scholar] [CrossRef]
- Mavromatis, K.; Ivanova, N.; Anderson, I.; Lykidis, A.; Hooper, S.D.; Sun, H.; Kunin, V.; Lapidus, A.; Hugenholtz, P.; Patel, B.; et al. Genome Analysis of the Anaerobic Thermohalophilic Bacterium Halothermothrix Orenii. PLoS ONE 2009, 4, e4192. [Google Scholar] [CrossRef]
- Kivistö, A.T.; Karp, M.T. Halophilic Anaerobic Fermentative Bacteria. J. Biotechnol. 2011, 152, 114–124. [Google Scholar] [CrossRef] [PubMed]
- Sutcliffe, I.C. Cell Envelope Architecture in the Chloroflexi: A Shifting Frontline in a Phylogenetic Turf War. Environ. Microbiol. 2011, 13, 279–282. [Google Scholar] [CrossRef] [PubMed]
- Gaisin, V.A.; Kooger, R.; Grouzdev, D.S.; Gorlenko, V.M.; Pilhofer, M. Cryo-Electron Tomography Reveals the Complex Ultrastructural Organization of Multicellular Filamentous Chloroflexota (Chloroflexi) Bacteria. Front. Microbiol. 2020, 11, 1373. [Google Scholar] [CrossRef] [PubMed]
- Gilks, W.R.; Richardson, S.; Spiegelhalter, D. Markov Chain Monte Carlo in Practice; CRC Press: Boca Raton, FL, USA, 1995; ISBN 978-1-4822-1497-0. [Google Scholar]
- Yutin, N.; Galperin, M.Y. A Genomic Update on Clostridial Phylogeny: Gram-Negative Spore Formers and Other Misplaced Clostridia. Environ. Microbiol. 2013, 15, 2631–2641. [Google Scholar] [CrossRef] [PubMed]
- Hagan, C.L.; Silhavy, T.J.; Kahne, D. β-Barrel Membrane Protein Assembly by the Bam Complex. Annu. Rev. Biochem. 2011, 80, 189–210. [Google Scholar] [CrossRef]
- Bowyer, A.; Baardsnes, J.; Ajamian, E.; Zhang, L.; Cygler, M. Characterization of Interactions between LPS Transport Proteins of the Lpt System. Biochem. Biophys. Res. Commun. 2011, 404, 1093–1098. [Google Scholar] [CrossRef]
- Walburger, A.; Lazdunski, C.; Corda, Y. The Tol/Pal System Function Requires an Interaction between the C-Terminal Domain of TolA and the N-Terminal Domain of TolB. Mol. Microbiol. 2002, 44, 695–708. [Google Scholar] [CrossRef]
- Ranjit, C.; Noll, K.M. Distension of the Toga of Thermotoga Maritima Involves Continued Growth of the Outer Envelope as Cells Enter the Stationary Phase. FEMS Microbiol. Lett. 2016, 363. [Google Scholar]
- Bernard, G.; Chan, C.X.; Ragan, M.A. Alignment-Free Microbial Phylogenomics under Scenarios of Sequence Divergence, Genome Rearrangement and Lateral Genetic Transfer. Sci. Rep. 2016, 6, 1–12. [Google Scholar] [CrossRef]
- Yu, J.; Lu, L. BamA Is a Pivotal Protein in Cell Envelope Synthesis and Cell Division in Deinococcus Radiodurans. Biochim. Biophys. Acta BBA-Biomembr. 2019, 1861, 1365–1374. [Google Scholar] [CrossRef]
- Voulhoux, R.; Bos, M.P.; Geurtsen, J.; Mols, M.; Tommassen, J. Role of a Highly Conserved Bacterial Protein in Outer Membrane Protein Assembly. Science 2003, 299, 262–265. [Google Scholar] [CrossRef]
- Gully, D.; Bouveret, E. A Protein Network for Phospholipid Synthesis Uncovered by a Variant of the Tandem Affinity Purification Method in Escherichia Coli. Proteomics 2006, 6, 282–293. [Google Scholar] [CrossRef]
- Narita, S.; Tokuda, H. Biochemical Characterization of an ABC Transporter LptBFGC Complex Required for the Outer Membrane Sorting of Lipopolysaccharides. FEBS Lett. 2009, 583, 2160–2164. [Google Scholar] [CrossRef] [PubMed]
- Cornet, L.; Meunier, L.; Van Vlierberghe, M.; Léonard, R.R.; Durieu, B.; Lara, Y.; Misztak, A.; Sirjacobs, D.; Javaux, E.J.; Philippe, H.; et al. Consensus Assessment of the Contamination Level of Publicly Available Cyanobacterial Genomes. PLoS ONE 2018, 13, e0200323. [Google Scholar] [CrossRef]
- Wang, Z.; Xiang, Q.; Zhu, X.; Dong, H.; He, C.; Wang, H.; Zhang, Y.; Wang, W.; Dong, C. Structural and Functional Studies of Conserved Nucleotide-Binding Protein LptB in Lipopolysaccharide Transport. Biochem. Biophys. Res. Commun. 2014, 452, 443–449. [Google Scholar] [CrossRef] [PubMed]
- Rigal, A.; Bouveret, E.; Lloubes, R.; Lazdunski, C.; Benedetti, H. The TolB Protein Interacts with the Porins of Escherichia Coli. J. Bacteriol. 1997, 179, 7274–7279. [Google Scholar] [CrossRef]
- Ray, M.-C.; Germon, P.; Vianney, A.; Portalier, R.; Lazzaroni, J.C. Identification by Genetic Suppression OfEscherichia Coli TolB Residues Important for TolB-Pal Interaction. J. Bacteriol. 2000, 182, 821–824. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Demoulin, C.F.; Lara, Y.J.; Cornet, L.; François, C.; Baurain, D.; Wilmotte, A.; Javaux, E.J. Cyanobacteria Evolution: Insight from the Fossil Record. Free Radic. Biol. Med. 2019, 140, 206–223. [Google Scholar] [CrossRef]
- Xavier, J.C.; Gerhards, R.E.; Wimmer, J.L.; Brueckner, J.; Tria, F.D.; Martin, W.F. The Metabolic Network of the Last Bacterial Common Ancestor. Commun. Biol. 2021, 4, 1–10. [Google Scholar] [CrossRef]
- Kuhn, A. The Bacterial Cell Wall and Membrane—A Treasure Chest for Antibiotic Targets. In Bacterial Cell Walls and Membranes; Kuhn, A., Ed.; Subcellular Biochemistry; Springer International Publishing: Cham, Switzerland, 2019; pp. 1–5. ISBN 978-3-030-18768-2. [Google Scholar]
- Melville, S.; Craig, L. Type IV Pili in Gram-Positive Bacteria. Microbiol. Mol. Biol. Rev. 2013, 77, 323–341. [Google Scholar] [CrossRef]
- Baurain, D.; Wilmotte, A.; Frère, J.-M. Gram-Negative Bacteria:" Inner" vs." Cytoplasmic" or" Plasma Membrane": A Question of Clarity Rather than Vocabulary. J. Microb. Biochem. Technol. 2016, 8, 325–326. [Google Scholar] [CrossRef]
- Verma, M.; Lal, D.; Kaur, J.; Saxena, A.; Kaur, J.; Anand, S.; Lal, R. Phylogenetic Analyses of Phylum Actinobacteria Based on Whole Genome Sequences. Res. Microbiol. 2013, 164, 718–728. [Google Scholar] [CrossRef]
- Yakhnina, A.A.; Bernhardt, T.G. The Tol-Pal System Is Required for Peptidoglycan-Cleaving Enzymes to Complete Bacterial Cell Division. Proc. Natl. Acad. Sci. USA 2020, 117, 6777–6783. [Google Scholar] [CrossRef]
- Parks, D.H.; Rinke, C.; Chuvochina, M.; Chaumeil, P.; Woodcroft, B.J.; Evans, P.N.; Hugenholtz, P.; Tyson, G.W. Recovery of Nearly 8,000 Metagenome-Assembled Genomes Substantially Expands the Tree of Life. Nat. Microbiol. 2017, 2. [Google Scholar] [CrossRef]
- Luef, B.; Frischkorn, K.R.; Wrighton, K.C.; Holman, H.-Y.N.; Birarda, G.; Thomas, B.C.; Singh, A.; Williams, K.H.; Siegerist, C.E.; Tringe, S.G.; et al. Diverse Uncultivated Ultra-Small Bacterial Cells in Groundwater. Nat. Commun. 2015, 6, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Gutierrez, C.A.; Aylward, F.O. Phylogenetic Signal, Congruence, and Uncertainty across Bacteria and Archaea. Mol. Biol. Evol. 2021, 38, 5514–5527. [Google Scholar] [CrossRef] [PubMed]
- Dawes, I.W. Sporulation in Evolution. Mol. Cell. Asp. Microb. Evol. Camb. Univ. Press N. Y. 1981, 85–130. [Google Scholar]

| Node | Trait | Statistic | E | H1 | H2 | R1 | R2 | H Biased |
|---|---|---|---|---|---|---|---|---|
| LBCA | MBN | mean q01 | 2.495 | 2.352 | 2.477 | 2.288 | 2.411 | 1.431 |
| LBCA | MBN | mean q10 | 0.132 | 0.113 | 0.121 | 0.012 | 0.008 | 0.210 |
| LBCA | MBN | mean P(0) | 94.951 | 94.204 | 95.375 | 97.134 | 98.161 | 67.092 |
| LBCA | PG | mean P(0) | 22.068 | 4.022 | 38.604 | 0.397 | 0.594 | N/A |
| LBCA | PG | mean P(2) | 76.497 | 94.622 | 60.147 | 99.535 | 99.358 | N/A |
| LBCA | PG | mean q01 | 4.626 | 1.634 | 7.317 | 0.798 | 0.827 | N/A |
| LBCA | PG | mean q02 | 6.935 | 2.020 | 20.967 | 0.953 | 1.041 | N/A |
| LBCA | PG | mean q10 | 0.166 | 0.102 | 0.187 | 0.000 | 0.000 | N/A |
| LBCA | PG | mean q12 | 0.128 | 0.109 | 0.118 | 0.001 | 0.000 | N/A |
| LBCA | PG | mean q20 | 2.088 | 0.937 | 4.941 | 1.347 | 1.413 | N/A |
| LBCA | PG | mean q21 | 1.890 | 2.165 | 1.600 | 1.398 | 1.419 | N/A |
| Firmicutes | PG | mean P(0) | 17.631 | 3.936 | 30.120 | 0.611 | 0.738 | N/A |
| Firmicutes | PG | mean P(2) | 81.891 | 95.648 | 69.435 | 99.378 | 99.237 | N/A |
| Complex | Simple | MBN | PG |
|---|---|---|---|
| R1 | H2 | 7.41 | 22.86 |
| E | 5.95 | 17.47 | |
| H1 | 2.69 | 8.38 | |
| R2 | 2.42 | 1.91 | |
| R2 | H2 | 4.99 | 20.95 |
| E | 3.53 | 15.56 | |
| H1 | 0.27 | 6.47 | |
| H1 | H2 | 4.71 | 14.47 |
| E | 3.25 | 9.09 | |
| E | H2 | 1.46 | 5.39 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Léonard, R.R.; Sauvage, E.; Lupo, V.; Perrin, A.; Sirjacobs, D.; Charlier, P.; Kerff, F.; Baurain, D. Was the Last Bacterial Common Ancestor a Monoderm after All? Genes 2022, 13, 376. https://doi.org/10.3390/genes13020376
Léonard RR, Sauvage E, Lupo V, Perrin A, Sirjacobs D, Charlier P, Kerff F, Baurain D. Was the Last Bacterial Common Ancestor a Monoderm after All? Genes. 2022; 13(2):376. https://doi.org/10.3390/genes13020376
Chicago/Turabian StyleLéonard, Raphaël R., Eric Sauvage, Valérian Lupo, Amandine Perrin, Damien Sirjacobs, Paulette Charlier, Frédéric Kerff, and Denis Baurain. 2022. "Was the Last Bacterial Common Ancestor a Monoderm after All?" Genes 13, no. 2: 376. https://doi.org/10.3390/genes13020376
APA StyleLéonard, R. R., Sauvage, E., Lupo, V., Perrin, A., Sirjacobs, D., Charlier, P., Kerff, F., & Baurain, D. (2022). Was the Last Bacterial Common Ancestor a Monoderm after All? Genes, 13(2), 376. https://doi.org/10.3390/genes13020376






