Epigenetic and Physiological Responses to Varying Root-Zone Temperatures in Greenhouse Rocket
Abstract
:1. Introduction
2. Materials and Methods
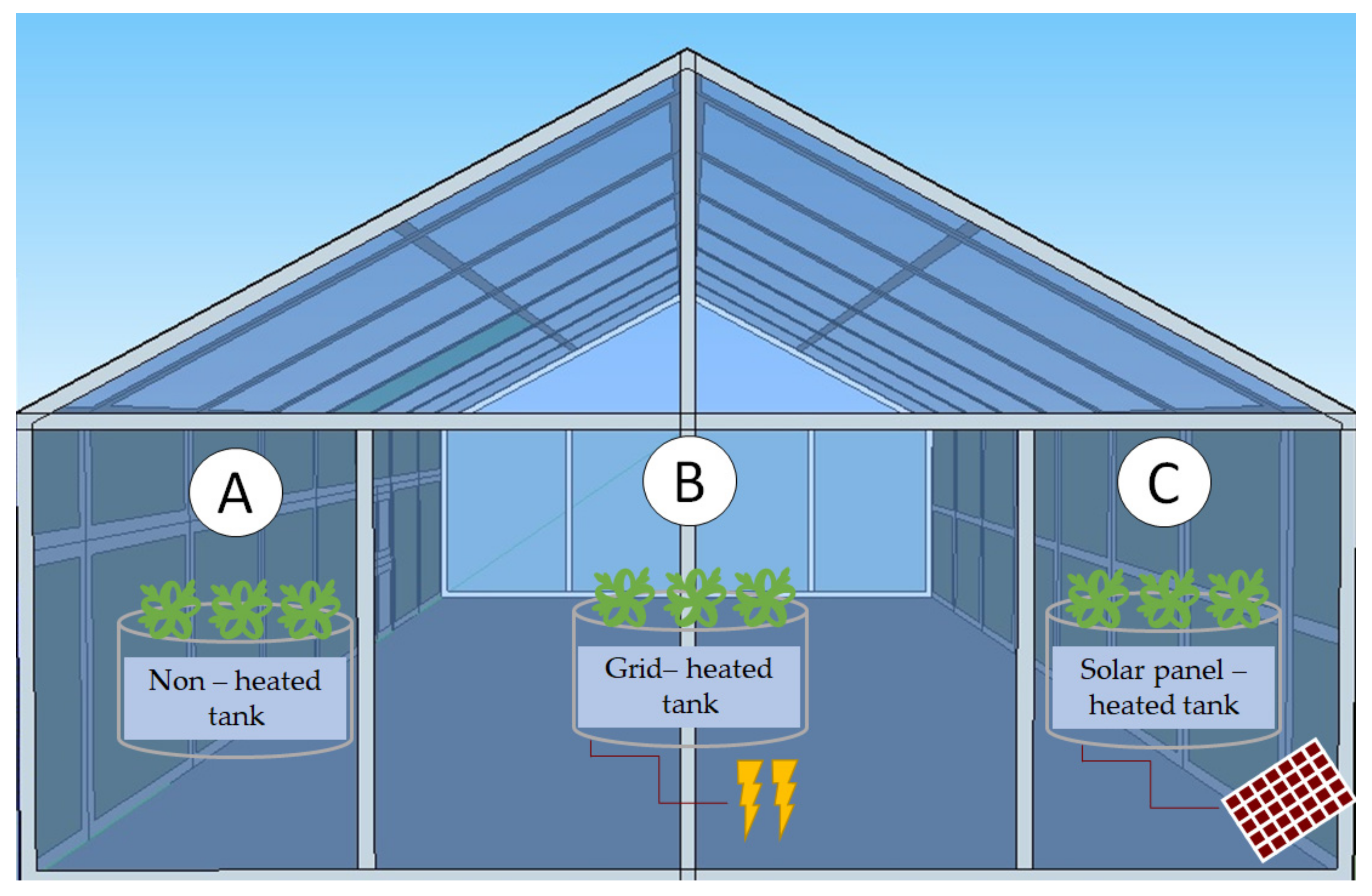
2.1. Experimental Set-Up and Plant Material
2.2. MSAP Procedure
2.3. Data Analysis
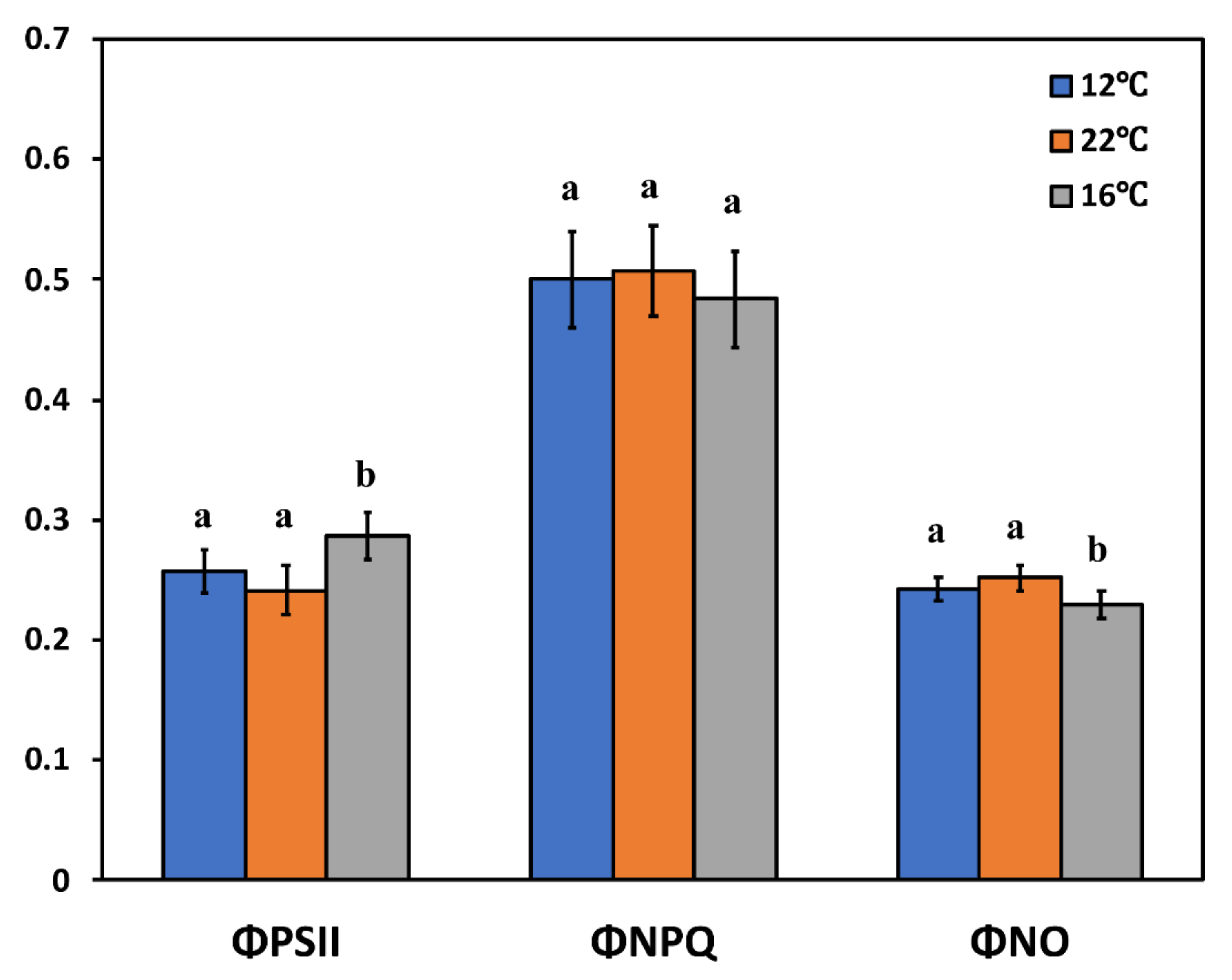
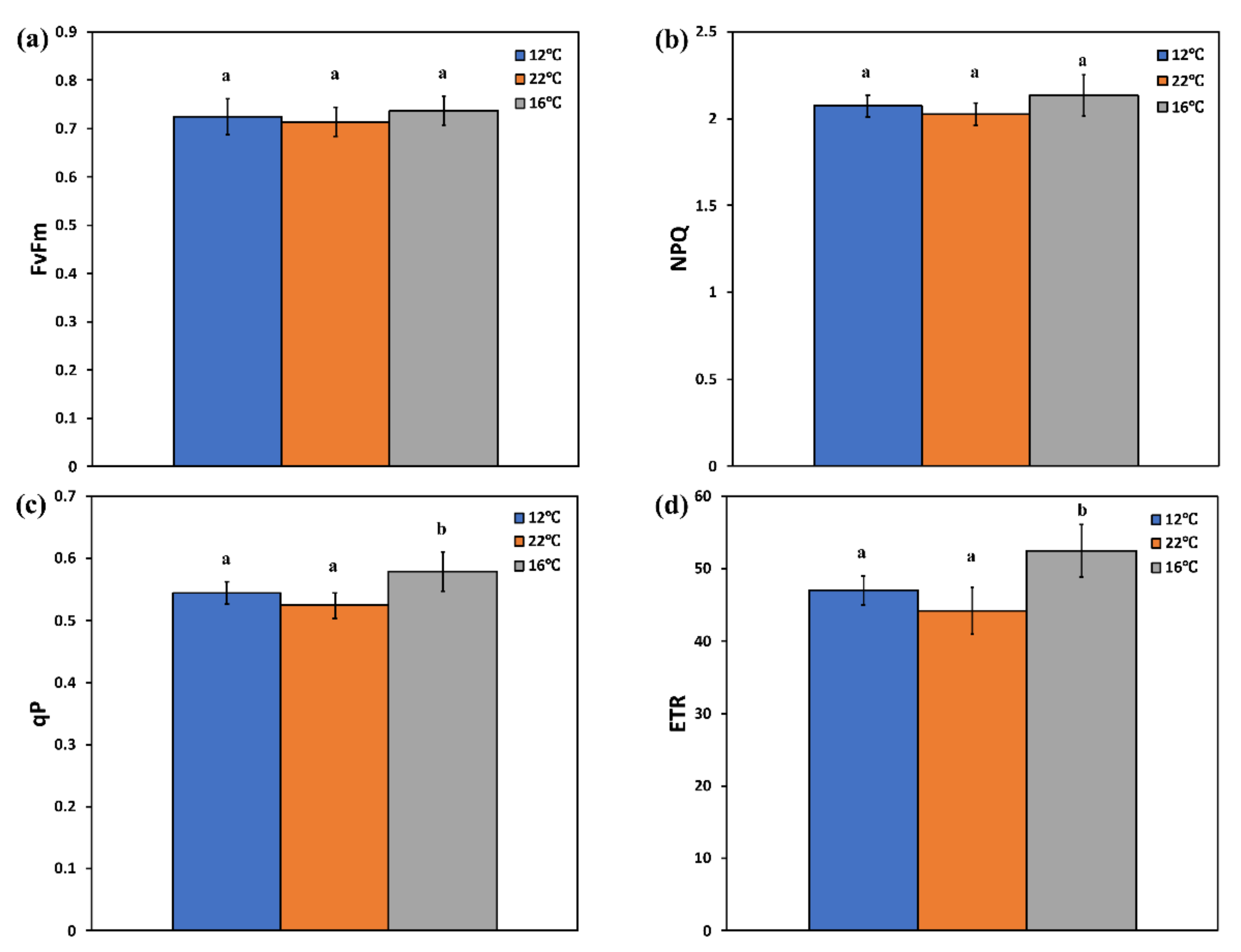
2.4. Chlorophyll Fluorescence Measurements
3. Results and Discussion
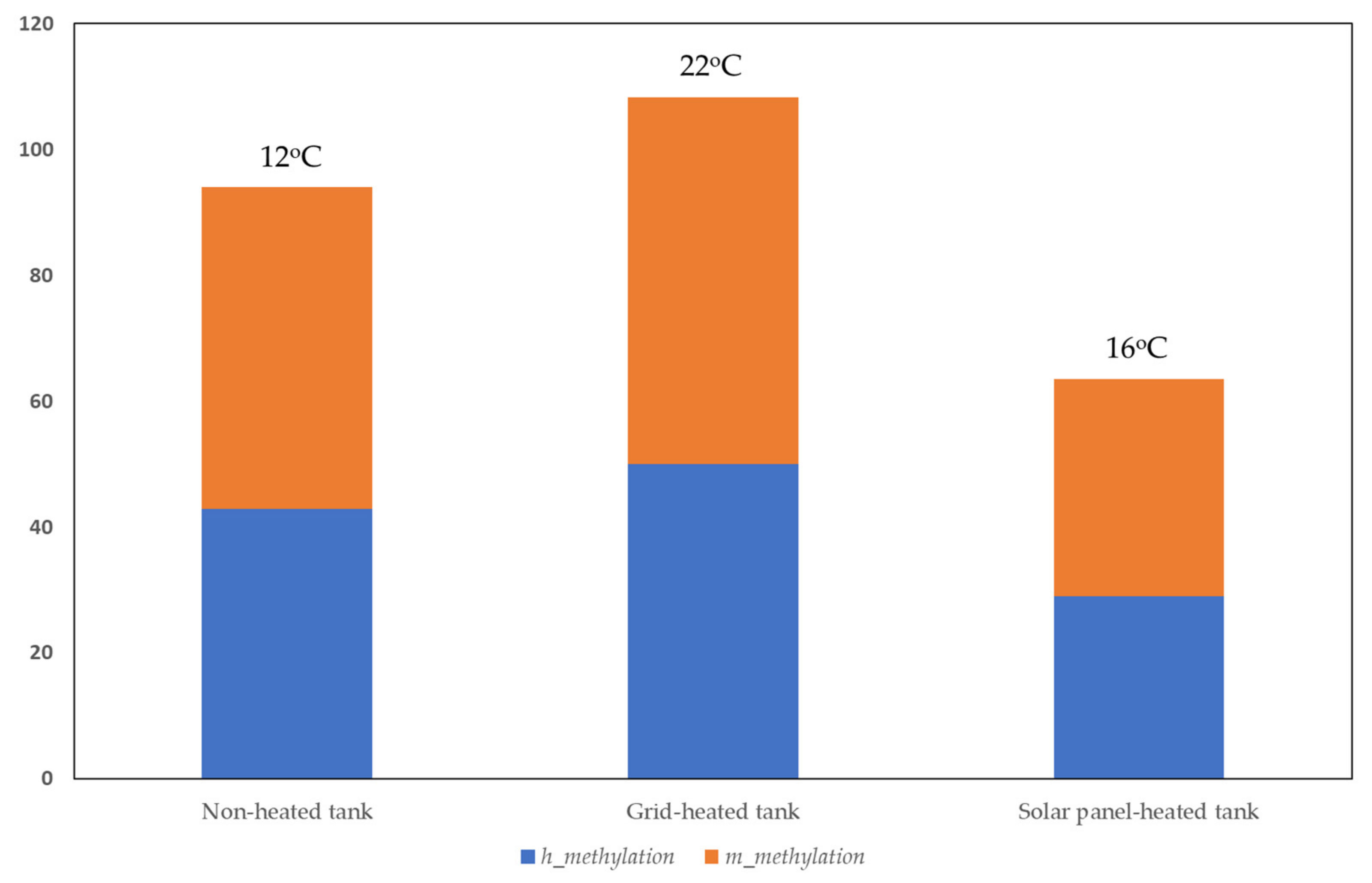
- (a)
- For h alleles, significant differences were found between plants grown in the non-heated control tank (12 °C) and the solar panel-heated tank (16 °C) and between the grid-heated tank (22 °C) and the solar panel-heated tank.
- (b)
- For m alleles, significant differences were found between plants grown in the non-heated control tank and the solar panel-heated tank and between the grid-heated tank and the solar panel-heated tank.
- (c)
- For total methylation (h + m), significant differences were found between plants grown in the non-heated control tank and the solar panel-heated tank and between plants grown in the grid-heated tank and the solar panel-heated tank.
- (d)
- For u alleles, significant differences were found between the non-heated control tank and the grid-heated tank and between the control and the solar panel-heated tank. U alleles are the unmethylated alleles.
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ntinas, G.K.; Kadoglidou, K.; Tsivelika, N.; Krommydas, K.; Kalivas, A.; Ralli, P.; Irakli, M. Performance and Hydroponic Tomato Crop Quality Characteristics in a Novel Greenhouse Using Dye-Sensitized Solar Cell Technology for Covering Material. Horticulturae 2019, 5, 42. [Google Scholar] [CrossRef] [Green Version]
- Savvas, D.; Gruda, N. Application of Soilless Culture Technologies in the Modern Greenhouse Industry—A Review. Eur. J. Hortic. Sci. 2018, 83, 280–293. [Google Scholar] [CrossRef]
- Pardossi, A.; Malorgio, F.; Incrocci, L.; Campiotti, C.A.; Tognoni, F. A comparison between two methods to control nutrient delivery to greenhouse melons grown in recirculating nutrient solution culture. Sci. Hortic. 2002, 92, 89–95. [Google Scholar] [CrossRef]
- Tomasi, N.; Pinton, R.; Costa, L.D.; Cortella, G.; Terzano, R.; Mimmo, T.; Scampicchio, M.; Cesco, S. New ‘solutions’ for floating cultivation system of ready-to-eat salad: A review. Trends Food Sci. Technol. 2015, 46, 267–276. [Google Scholar] [CrossRef]
- Dalla Costa, L.; Tomasi, N.; Gottardi, S.; Iacuzzo, F.; Cortella, G.; Manzocco, L.; Pinton, R.; Mimmo, T.; Cesco, S. The Effect of Growth Medium Temperature on Corn Salad [Valerianella Locusta (L.) Laterr] Baby Leaf Yield and Quality. HortScience 2011, 46, 1619–1625. [Google Scholar] [CrossRef]
- Ashraf, M.; Harris, P.J.C. Photosynthesis under Stressful Environments: An Overview. Photosynthetica 2013, 51, 163–190. [Google Scholar] [CrossRef]
- Velitchkova, M.; Popova, A.V.; Faik, A.; Gerganova, M.; Ivanov, A.G. Low Temperature and High Light Dependent Dynamic Photoprotective Strategies in Arabidopsis thaliana. Physiol. Plant. 2020, 170, 93–108. [Google Scholar] [CrossRef]
- Lourkisti, R.; Froelicher, Y.; Herbette, S.; Morillon, R.; Tomi, F.; Gibernau, M.; Giannettini, J.; Berti, L.; Santini, J. Triploid Citrus Genotypes Have a Better Tolerance to Natural Chilling Conditions of Photosynthetic Capacities and Specific Leaf Volatile Organic Compounds. Front. Plant Sci. 2020, 11, 330. [Google Scholar] [CrossRef] [Green Version]
- Hussain, H.A.; Hussain, S.; Khaliq, A.; Ashraf, U.; Anjum, S.A.; Men, S.; Wang, L. Chilling and Drought Stresses in Crop Plants: Implications, Cross Talk, and Potential Management Opportunities. Front. Plant Sci. 2018, 9, 393. [Google Scholar] [CrossRef]
- Jurczyk, B.; Grzesiak, M.; Pociecha, E.; Wlazło, M.; Rapacz, M. Diverse Stomatal Behaviors Mediating Photosynthetic Acclimation to Low Temperatures in Hordeum Vulgare. Front. Plant Sci. 2019, 9, 1963. [Google Scholar] [CrossRef]
- Sperdouli, I.; Moustaka, J.; Antonoglou, O.; Adamakis, I.-D.S.; Dendrinou-Samara, C.; Moustakas, M. Leaf Age-Dependent Effects of Foliar-Sprayed CuZn Nanoparticles on Photosynthetic Efficiency and ROS Generation in Arabidopsis thaliana. Materials 2019, 12, 2498. [Google Scholar] [CrossRef] [Green Version]
- Moustaka, J.; Panteris, E.; Adamakis, I.-D.S.; Tanou, G.; Giannakoula, A.; Eleftheriou, E.P.; Moustakas, M. High Anthocyanin Accumulation in Poinsettia Leaves Is Accompanied by Thylakoid Membrane Unstacking, acting as a Photoprotective Mechanism, to Prevent ROS Formation. Environ. Exp. Bot. 2018, 154, 44–55. [Google Scholar] [CrossRef]
- Sperdouli, I.; Moustakas, M. Differential Blockage of Photosynthetic Electron Flow in Young and Mature Leaves of Arabidopsis thaliana by Exogenous Proline. Photosynthetica 2015, 53, 471–477. [Google Scholar] [CrossRef]
- Zhang, H.; Lang, Z.; Zhu, J.-K. Dynamics and Function of DNA Methylation in Plants. Nat. Rev. Mol. Cell Biol. 2018, 19, 489–506. [Google Scholar] [CrossRef] [PubMed]
- Taudt, A.; Colomé-Tatché, M.; Johannes, F. Genetic Sources of Population Epigenomic Variation. Nat. Rev. Genet. 2016, 17, 319. [Google Scholar] [CrossRef] [PubMed]
- Lauria, M.; Rossi, V. Epigenetic Control of Gene Regulation in Plants. Biochim. Biophys. Acta (BBA) Gene Regul. Mech. 2011, 1809, 369–378. [Google Scholar] [CrossRef]
- Liu, R.; Lang, Z. The Mechanism and Function of Active DNA Demethylation in Plants. J. Integr. Plant Biol. 2020, 62, 148–159. [Google Scholar] [CrossRef] [Green Version]
- Karan, R.; DeLeon, T.; Biradar, H.; Subudhi, P.K. Salt Stress Induced Variation in DNA Methylation Pattern and Its Influence on Gene Expression in Contrasting Rice Genotypes. PLoS ONE 2012, 7, e40203. [Google Scholar] [CrossRef]
- Joel, A.J. Epigenetic Responses to Drought Stress in Rice (Oryza sativa L.). Physiol. Mol. Biol. Plants 2013, 19, 379–387. [Google Scholar]
- Shan, X.; Wang, X.; Yang, G.; Wu, Y.; Su, S.; Li, S.; Liu, H.; Yuan, Y. Analysis of the DNA Methylation of Maize (Zea mays L.) in Response to Cold Stress Based on Methylation-Sensitive Amplified Polymorphisms. J. Plant Biol. 2013, 56, 32–38. [Google Scholar] [CrossRef]
- Song, Y.; Liu, L.; Feng, Y.; Wei, Y.; Yue, X.; He, W.; Zhang, H.; An, L. Chilling- and Freezing- Induced Alterations in Cytosine Methylation and Its Association with the Cold Tolerance of an Alpine Subnival Plant, Chorispora bungeana. PLoS ONE 2015, 10, e0135485. [Google Scholar] [CrossRef] [PubMed]
- Forestan, C.; Farinati, S.; Rouster, J.; Lassagne, H.; Lauria, M.; Dal Ferro, N.; Varotto, S. Control of Maize Vegetative and Reproductive Development, Fertility, and RRNAs Silencing by HISTONE DEACETYLASE 108. Genetics 2018, 208, 1443–1466. [Google Scholar] [CrossRef] [Green Version]
- Varotto, S.; Tani, E.; Abraham, E.; Krugman, T.; Kapazoglou, A.; Melzer, R.; Radanović, A.; Miladinović, D. Epigenetics: Possible Applications in Climate-Smart Crop Breeding. J. Exp. Bot. 2020, 71, 5223–5236. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Li, Y.; Duan, W.; Huang, F.; Hou, X. Cold Acclimation Alters DNA Methylation Patterns and Confers Tolerance to Heat and Increases Growth Rate in Brassica rapa. J. Exp. Bot. 2017, 68, 1213–1224. [Google Scholar] [CrossRef] [Green Version]
- Tang, X.; Wang, Q.; Yuan, H.; Huang, X. Chilling-Induced DNA Demethylation Is Associated with the Cold Tolerance of Hevea brasiliensis. BMC Plant Biol. 2018, 18, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Boyko, A.; Blevins, T.; Yao, Y.; Golubov, A.; Bilichak, A.; Ilnytskyy, Y.; Hollander, J.; Meins, F., Jr.; Kovalchuk, I. Transgenerational Adaptation of Arabidopsis to Stress Requires DNA Methylation and the Function of Dicer-Like Proteins. PLoS ONE 2010, 5, e9514. [Google Scholar] [CrossRef]
- Zhang, P.Y.; Wang, J.G.; Geng, Y.P.; Dai, J.R.; Zhong, Y.; Chen, Z.Z.; Zhu, K.; Wang, X.Z.; Chen, S.Y. MSAP-Based Analysis of DNA Methylation Diversity in Tobacco Exposed to Different Environments and at Different Development Phases. Biochem. Syst. Ecol. 2015, 62, 249–260. [Google Scholar] [CrossRef]
- Tang, X.-M.; Tao, X.; Wang, Y.; Ma, D.-W.; Li, D.; Yang, H.; Ma, X.-R. Analysis of DNA Methylation of Perennial Ryegrass under Drought Using the Methylation-Sensitive Amplification Polymorphism (MSAP) Technique. Mol. Genet. Genom. 2014, 289, 1075–1084. [Google Scholar] [CrossRef]
- Ventouris, Y.E.; Tani, E.; Avramidou, E.V.; Abraham, E.M.; Chorianopoulou, S.N.; Vlachostergios, D.N.; Papadopoulos, G.; Kapazoglou, A. Recurrent Water Deficit and Epigenetic Memory in Medicago sativa L. Varieties. Appl. Sci. 2020, 10, 3110. [Google Scholar] [CrossRef]
- Wang, W.; Huang, F.; Qin, Q.; Zhao, X.; Li, Z.; Fu, B. Comparative Analysis of DNA Methylation Changes in Two Rice Genotypes under Salt Stress and Subsequent Recovery. Biochem. Biophys. Res. Commun. 2015, 465, 790–796. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.; Yu, S.; Xing, C.; Fan, S.; Song, M. Analysis of DNA Methylation in Cotton Hybrids and Their Parents. Mol. Biol. 2008, 42, 169. [Google Scholar] [CrossRef]
- Wu, S.; Xue, J.; Zhang, H.; Xu, P.; Wu, X. Special DNA Methylated Sites between Haploid of Twin-Seedling and Its Hybrids in Rice (Oryza sativa). Rice Sci. 2012, 19, 94–99. [Google Scholar] [CrossRef]
- Lauria, M.; Rupe, M.; Guo, M.; Kranz, E.; Pirona, R.; Viotti, A.; Lund, G. Extensive Maternal DNA Hypomethylation in the Endosperm of Zea mays. Plant Cell 2004, 16, 510–522. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, B.; Saltveit, M.E.; Beckles, D.M. Chilling-Stress Modifies DNA Methylation Level in Cucumber (Cucumis sativus L.) Seedling Radicle to Regulate Elongation Rate. Sci. Hortic. 2019, 252, 14–19. [Google Scholar] [CrossRef]
- Marfil, C.; Ibañez, V.; Alonso, R.; Varela, A.; Bottini, R.; Masuelli, R.; Fontana, A.; Berli, F. Changes in Grapevine DNA Methylation and Polyphenols Content Induced by Solar Ultraviolet-B Radiation, Water Deficit and Abscisic Acid Spray Treatments. Plant Physiol. Biochem. 2019, 135, 287–294. [Google Scholar] [CrossRef] [PubMed]
- Marconi, G.; Pace, R.; Traini, A.; Raggi, L.; Lutts, S.; Chiusano, M.; Guiducci, M.; Falcinelli, M.; Benincasa, P.; Albertini, E. Use of MSAP Markers to Analyse the Effects of Salt Stress on DNA Methylation in Rapeseed (Brassica napus Var. Oleifera). PLoS ONE 2013, 8, e75597. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fulneček, J.; Kovařík, A. How to Interpret Methylation Sensitive Amplified Polymorphism (MSAP) Profiles? BMC Genet. 2014, 15, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bednarek, P.T.; Orłowska, R.; Niedziela, A. A Relative Quantitative Methylation-Sensitive Amplified Polymorphism (MSAP) Method for the Analysis of Abiotic Stress. BMC Plant Biol. 2017, 17, 79. [Google Scholar] [CrossRef] [Green Version]
- Abid, G.; Mingeot, D.; Muhovski, Y.; Mergeai, G.; Aouida, M.; Abdelkarim, S.; Aroua, I.; El Ayed, M.; M’hamdi, M.; Sassi, K.; et al. Analysis of DNA Methylation Patterns Associated with Drought Stress Response in Faba Bean (Vicia faba L.) Using Methylation-Sensitive Amplification Polymorphism (MSAP). Environ. Exp. Bot. 2017, 142, 34–44. [Google Scholar] [CrossRef]
- Resh, H.M. Hydroponic Food Production: A Definitive Guidebook of Soilless Food-Growing Methods; Woodbridge Press Publishing Company: Santa Barbara, CA, USA, 1995; ISBN 0880072121. [Google Scholar]
- He, J.; See, X.E.; Qin, L.; Choong, T.W. Effects of Root-Zone Temperature on Photosynthesis, Productivity and Nutritional Quality of Aeroponically Grown Salad Rocket (Eruca sativa) Vegetable. Am. J. Plant Sci. 2016, 7, 1993–2005. [Google Scholar] [CrossRef] [Green Version]
- Karnoutsos, P.; Karagiovanidis, M.; Bantis, F.; Chatzistathis, T.; Koukounaras, A.; Ntinas, G.K. Controlled Root-zone Temperature Effect on Baby Leaf Vegetables Yield and Quality in a Floating System under Mild and Extreme Weather Conditions. J. Sci. Food Agric. 2020, 101, 3933–3941. [Google Scholar] [CrossRef]
- Ruelland, E.; Zachowski, A. How Plants Sense Temperature. Environ. Exp. Bot. 2010, 69, 225–232. [Google Scholar] [CrossRef]
- Vekemans, X.; Beauwens, T.; Lemaire, M.; Roldán-Ruiz, I. Data from Amplified Fragment Length Polymorphism (AFLP) Markers Show Indication of Size Homoplasy and of a Relationship between Degree of Homoplasy and Fragment Size. Mol. Ecol. 2002, 11, 139–151. [Google Scholar] [CrossRef] [PubMed]
- Schulz, B.; Eckstein, R.L.; Durka, W. Scoring and Analysis of Methylation-sensitive Amplification Polymorphisms for Epigenetic Population Studies. Mol. Ecol. Resour. 2013, 13, 642–653. [Google Scholar] [CrossRef] [Green Version]
- Peakall, R.O.D.; Smouse, P.E. GENALEX 6: Genetic Analysis in Excel. Population Genetic Software for Teaching and Research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Moustakas, M.; Bayçu, G.; Sperdouli, I.; Eroğlu, H.; Eleftheriou, E.P. Arbuscular Mycorrhizal Symbiosis Enhances Photosynthesis in the Medicinal Herb Salvia fruticosa by Improving Photosystem II Photochemistry. Plants 2020, 9, 962. [Google Scholar] [CrossRef]
- Dobrikova, A.; Apostolova, E.; Hanć, A.; Yotsova, E.; Borisova, P.; Sperdouli, I.; Adamakis, I.D.S.; Moustakas, M. Tolerance Mechanisms of the Aromatic and Medicinal Plant Salvia sclarea L. To Excess Zinc. Plants 2021, 10, 16. [Google Scholar] [CrossRef]
- Pan, Y.; Wang, W.; Zhao, X.; Zhu, L.; Fu, B.; Li, Z. DNA Methylation Alterations of Rice in Response to Cold Stress. Plant Omics J. 2011, 4, 364–369. [Google Scholar]
- Banerjee, A.; Roychoudhury, A. Cold Stress and Photosynthesis. In Photosynthesis, Productivity and Environmental Stress; Ahmad, P., Ahanger, M.A., Alyemeni, M.N., Alam, P., Eds.; John Wiley & Sons: Hoboken, NJ, USA, 2019; pp. 23–37. [Google Scholar] [CrossRef]
- Moustaka, J.; Moustakas, M. Photoprotective Mechanism of the Non-Target Organism Arabidopsis thaliana to Paraquat Exposure. Pestic. Biochem. Physiol. 2014, 111, 6. [Google Scholar] [CrossRef]
- Genty, B.; Briantais, J.-M.; Baker, N.R. The Relationship between the Quantum Yield of Photosynthetic Electron Transport and Quenching of Chlorophyll Fluorescence. Biochim. Et Biophys. Acta (BBA)-Gen. Subj. 1989, 990, 87–92. [Google Scholar] [CrossRef]
- Zhang, C.; Wang, H.; Xu, Y.; Zhang, S.; Wang, J.; Hu, B.; Hou, X.; Li, Y.; Liu, T. Enhanced Relative Electron Transport Rate Contributes to Increased Photosynthetic Capacity in Autotetraploid Pak Choi. Plant Cell Physiol. 2020, 61, 761–774. [Google Scholar] [CrossRef] [PubMed]
- Ma, K.; Song, Y.; Jiang, X.; Zhang, Z.; Li, B.; Zhang, D. Photosynthetic Response to Genome Methylation Affects the Growth of Chinese White Poplar. Tree Genet. Genomes 2012, 8, 1407–1421. [Google Scholar] [CrossRef]
- Krieger-Liszkay, A.; Fufezan, C.; Trebst, A. Singlet Oxygen Production in Photosystem II and Related Protection Mechanism. Photosynth. Res. 2008, 98, 551–564. [Google Scholar] [CrossRef] [PubMed]
- Antonoglou, O.; Moustaka, J.; Adamakis, I.-D.S.; Sperdouli, I.; Pantazaki, A.A.; Moustakas, M.; Dendrinou-Samara, C. Nanobrass CuZn Nanoparticles as Foliar Spray Nonphytotoxic Fungicides. ACS Appl. Mater. Interfaces 2018, 10, 4450–4461. [Google Scholar] [CrossRef]
- Gallusci, P.; Dai, Z.; Génard, M.; Gauffretau, A.; Leblanc-Fournier, N.; Richard-Molard, C.; Vile, D.; Brunel-Muguet, S. Epigenetics for Plant Improvement: Current Knowledge and Modeling Avenues. Trends Plant Sci. 2017, 22, 610–623. [Google Scholar] [CrossRef]
- Iwasaki, M.; Paszkowski, J. Epigenetic Memory in Plants. EMBO J. 2014, 33, 1987–1998. [Google Scholar] [CrossRef] [Green Version]
- Parrilla-Doblas, J.T.; Roldán-Arjona, T.; Ariza, R.R.; Córdoba-Cañero, D. Active DNA Demethylation in Plants. Int. J. Mol. Sci. 2019, 20, 4683. [Google Scholar] [CrossRef] [Green Version]
| 5′ to 3′ Sequence | |
|---|---|
| EcoRI adapter | CTCGTAGACTGCGTACC AATTGGTACGCAGTC |
| HpaII/MspI adapter | GACGATGAGTCTCGAT CGATCGAGACTCAT |
| Pre-selective EcoRI primer | GACTGCGTACCAATTC−A |
| Pre-selective HpaII/MspI primer | ATGAGTCTCGATCGG−T |
| Selective EcoRI primers | GACTGCGTACCAATTC+ATG GACTGCGTACCAATTC+ACT GACTGCGTACCAATTC+AAC GACTGCGTACCAATTC+AAG |
| Selective HpaII/MspI primer | ATGAGTCTCGATCGG+TCA ATGAGTCTCGATCGG+ACT ATGAGTCTCGATCGG+AAT |
| h Alleles | u Alleles | m Alleles | Total Methylation (h + m) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Iepi | Hepi | Iepi | Hepi | Iepi | Hepi | Iepi | Hepi | ||
| Tank A * | Mean | 0.221 a | 0.143 a | 0.188 c,d | 0.125 c,d | 0.282 e | 0.188 e | 0.253 g | 0.167 g |
| SE | 0.017 | 0.011 | 0.016 | 0.011 | 0.018 | 0.012 | 0.012 | 0.008 | |
| Tank B | Mean | 0.254 b | 0.164 b | 0.252 c | 0.165 c | 0.321 f | 0.214 f | 0.289 h | 0.190 h |
| SE | 0.017 | 0.011 | 0.017 | 0.011 | 0.017 | 0.012 | 0.012 | 0.008 | |
| Tank C | Mean | 0.145 a,b | 0.093 a,b | 0.277 d | 0.186 d | 0.175 e,f | 0.113 e,f | 0.161 g,h | 0.104 g,h |
| SE | 0.015 | 0.010 | 0.018 | 0.012 | 0.015 | 0.010 | 0.011 | 0.007 | |
| h Methylation | m Methylation | Yield | TSS | ΦPSII | ΦNPQ | |
|---|---|---|---|---|---|---|
| h methylation | ||||||
| m methylation | 0.999 * | |||||
| Yield | 0.314 | 0.273 | ||||
| TSS | −0.761 | −0.732 | −0.855 | |||
| ΦPSII | −1.000 * | −0.998 * | −0.337 | 0.776 | ||
| ΦNPQ | 1.000 * | 1.000 * | 0.293 | −0.746 | −0.999 * | |
| ΦNO | 0.998 * | 0.995 | 0.368 | −0.797 | −0.999 * | 0.997 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tsaballa, A.; Sperdouli, I.; Avramidou, E.V.; Ganopoulos, I.; Koukounaras, A.; Ntinas, G.K. Epigenetic and Physiological Responses to Varying Root-Zone Temperatures in Greenhouse Rocket. Genes 2022, 13, 364. https://doi.org/10.3390/genes13020364
Tsaballa A, Sperdouli I, Avramidou EV, Ganopoulos I, Koukounaras A, Ntinas GK. Epigenetic and Physiological Responses to Varying Root-Zone Temperatures in Greenhouse Rocket. Genes. 2022; 13(2):364. https://doi.org/10.3390/genes13020364
Chicago/Turabian StyleTsaballa, Aphrodite, Ilektra Sperdouli, Evangelia V. Avramidou, Ioannis Ganopoulos, Athanasios Koukounaras, and Georgios K. Ntinas. 2022. "Epigenetic and Physiological Responses to Varying Root-Zone Temperatures in Greenhouse Rocket" Genes 13, no. 2: 364. https://doi.org/10.3390/genes13020364
APA StyleTsaballa, A., Sperdouli, I., Avramidou, E. V., Ganopoulos, I., Koukounaras, A., & Ntinas, G. K. (2022). Epigenetic and Physiological Responses to Varying Root-Zone Temperatures in Greenhouse Rocket. Genes, 13(2), 364. https://doi.org/10.3390/genes13020364