A New Approach Using Targeted Sequence Capture for Phylogenomic Studies across Cactaceae
Abstract
:1. Introduction
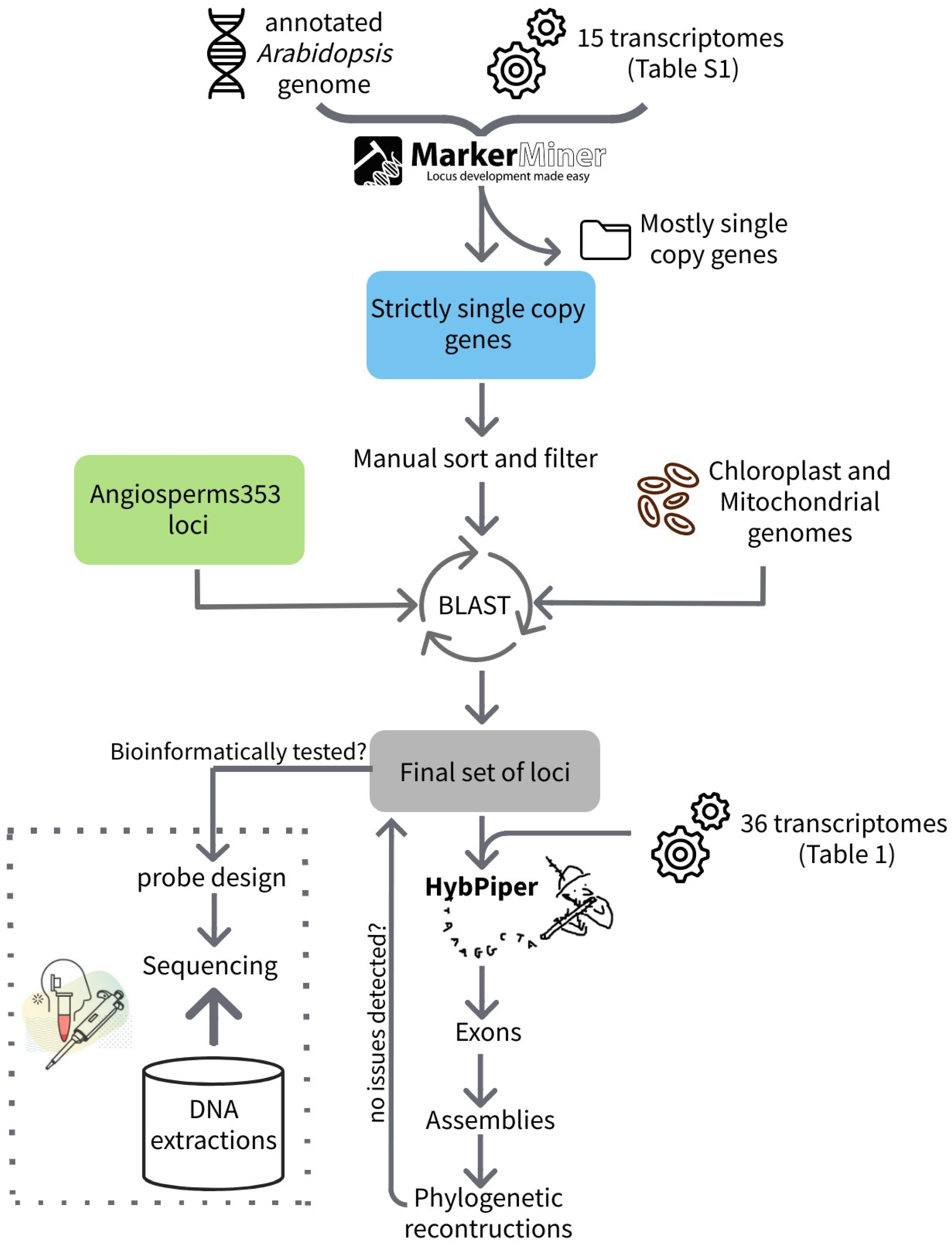
2. Materials and Methods
2.1. Probe Sets
2.1.1. Cactaceae120
2.1.2. Angiosperms353
2.2. Bioinformatic Evaluation
2.3. Phylogenetic Reconstruction
2.4. Experimental Evaluation
3. Results
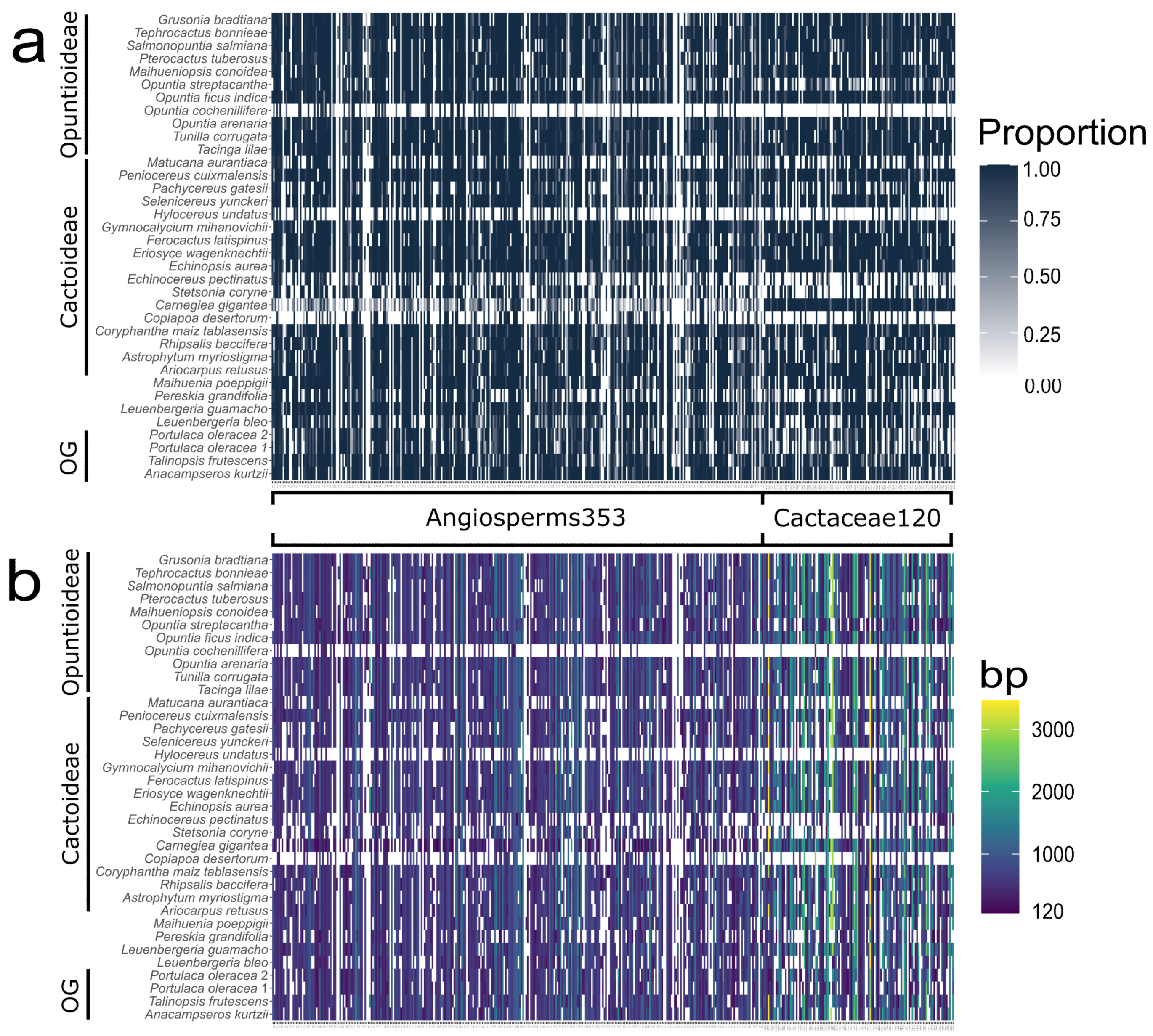
3.1. Cactaceae120 Loci
3.2. Angiosperms353
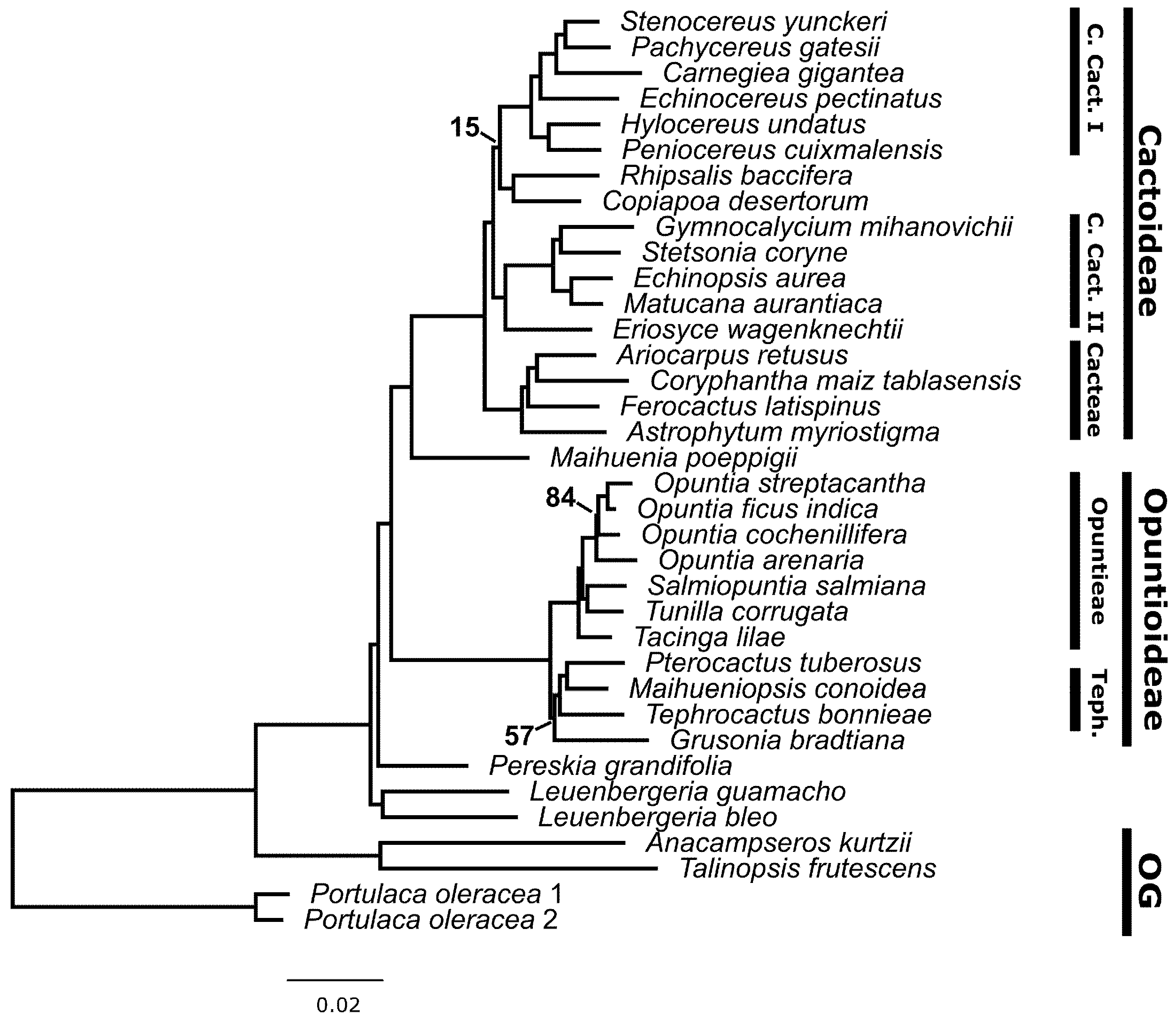
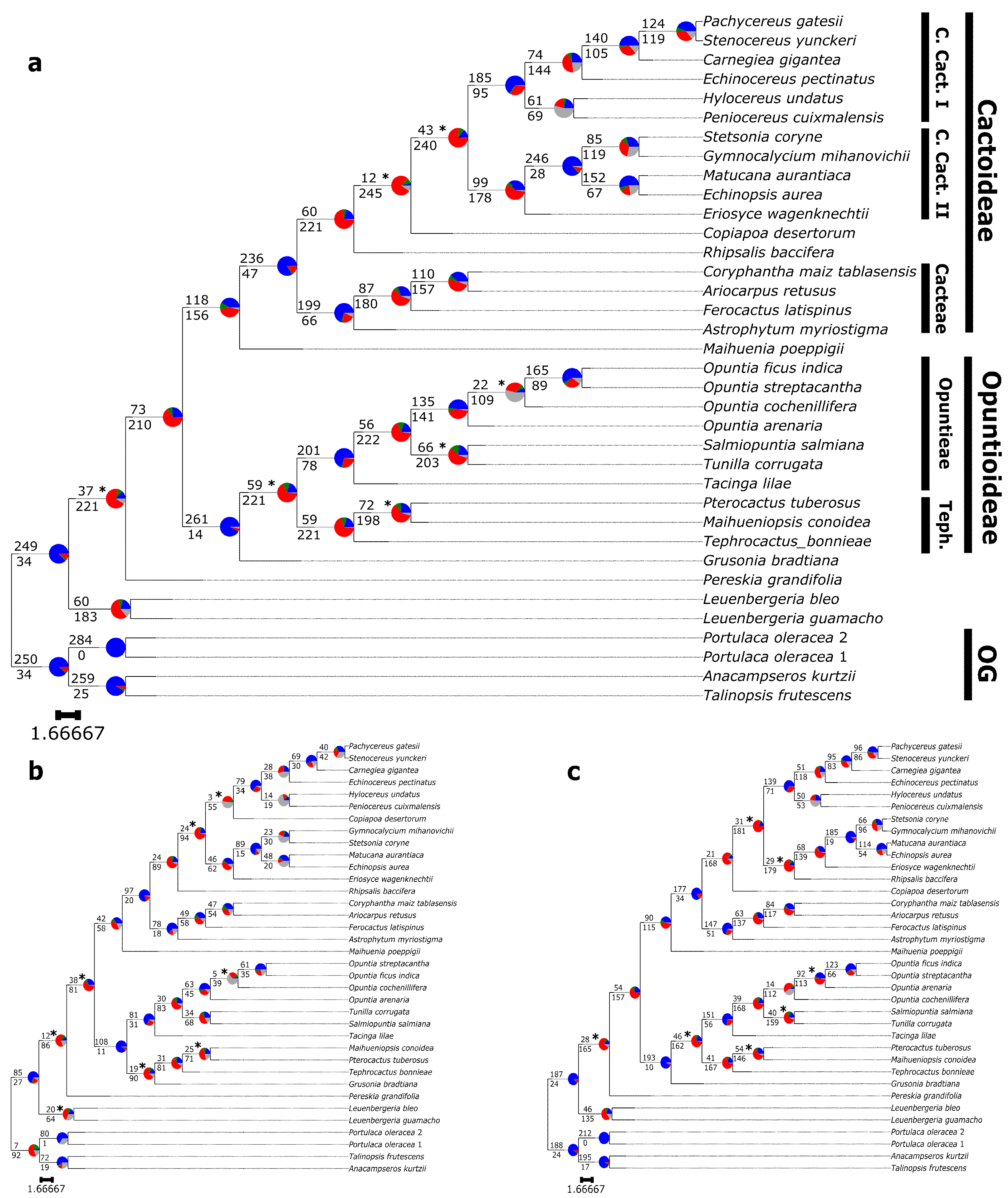
3.3. Phylogenetic Results
3.4. Experimental Evaluation
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Korotkova, N.; Aquino, D.; Arias, S.; Eggli, U.; Franck, A.; Gómez-Hinostrosa, C.; Guerrero, P.C.; Hernández, H.M.; Kohlbecker, A.; Köhler, M.; et al. Cactaceae at Caryophyllales.Org- A Dynamic Online Species-Level Taxonomic Backbone for the Family. Willdenowia 2021, 51, 251–270. [Google Scholar] [CrossRef]
- Anderson, E.F. The Cactus Family; Timber Press: Portland, OR, USA, 2001. [Google Scholar]
- Wallace, R.S.; Gibson, A.C. Evolution and Systematics. In Cacti: Biology and Uses; Nobel, P.S., Ed.; University of California Press: London, UK, 2002; pp. 1–21. [Google Scholar]
- Hershkovitz, M.A.; Zimmer, E.A. On the Evolutionary Origins of the Cacti. Taxon 1997, 46, 217–232. [Google Scholar] [CrossRef]
- Butterworth, C.A.; Wallace, R.S. Phylogenetic Studies of Mammillaria (Cactaceae)-Insights from Chloroplast Sequence Variation and Hypothesis Testing Using the Parametric Bootstrap. Am. J. Bot. 2004, 91, 1086–1098. [Google Scholar] [CrossRef]
- Griffith, M.P.; Porter, J.M. Phylogeny of Opuntioideae (Cactaceae). Int. J. Plant Sci. 2009, 170, 107–116. [Google Scholar] [CrossRef] [Green Version]
- Bárcenas, R.T.; Yesson, C.; Hawkins, J.A. Molecular Systematics of the Cactaceae. Cladistics 2011, 27, 470–489. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Hernández, T.; Hernández, H.M.; Arturo De-Nova, J.; Puente, R.; Eguiarte, L.E.; Magallón, S. Phylogenetic Relationships and Evolution of Growth Form in Cactaceae (Caryophyllales, Eudicotyledoneae). Am. J. Bot. 2011, 98, 44–61. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Majure, L.C.; Puente, R.; Patrick Griffith, M.; Judd, W.S.; Soltis, P.S.; Soltis, D.E. Phylogeny of Opuntia s.s. (Cactaceae): Clade Delineation, Geographic Origins, Reticulate Evolution. Am. J. Bot. 2012, 99, 847–864. [Google Scholar] [CrossRef] [Green Version]
- Arakaki, M.; Christin, P.A.; Nyffeler, R.; Lendel, A.; Eggli, U.; Ogburn, R.M.; Spriggs, E.; Moore, M.J.; Edwards, E.J. Contemporaneous and Recent Radiations of the World’s Major Succulent Plant Lineages. Proc. Natl. Acad. Sci. USA 2011, 108, 8379–8384. [Google Scholar] [CrossRef] [Green Version]
- Majure, L.C.; Baker, M.A.; Cloud-Hughes, M.; Salywon, A.; Neubig, K.M. Phylogenomics in Cactaceae: A Case Study Using the Chollas Sensu Lato (Cylindropuntieae, Opuntioideae) Reveals a Common Pattern out of the Chihuahuan and Sonoran Deserts. Am. J. Bot. 2019, 106, 1327–1345. [Google Scholar] [CrossRef]
- Majure, L.C.; Encarnación, Y.; Clase, T.; Peguero, B.; Ho, K.; Barrios, D. Phylogenetics of Leptocereus (Cactaceae) on Hispaniola: Clarifying Species Limits in the L. Weingartianus Complex and a New Species from the Sierra de Bahoruco. PhytoKeys 2021, 172, 17–37. [Google Scholar] [CrossRef]
- Majure, L.C.; Barrios, D.; Díaz, E.; Zumwalde, B.A.; Testo, W.; Negrón-Ortíz, V. Pleistocene Aridification Underlies the Evolutionary History of the Caribbean Endemic, Insular, Giant Consolea (Opuntioideae). Am. J. Bot. 2021, 108, 200–215. [Google Scholar] [CrossRef]
- Köhler, M.; Reginato, M.; Souza-Chies, T.T.; Majure, L.C. Insights into Chloroplast Genome Evolution Across Opuntioideae (Cactaceae) Reveals Robust Yet Sometimes Conflicting Phylogenetic Topologies. Front. Plant Sci. 2020, 11, 729. [Google Scholar] [CrossRef] [PubMed]
- Breslin, P.B.; Wojciechowski, M.F.; Majure, L.C. Molecular Phylogeny of the Mammilloid Clade (Cactaceae) Resolves the Monophyly of Mammillaria. Taxon 2021, 70, 308–323. [Google Scholar] [CrossRef]
- Johnson, M.G.; Pokorny, L.; Dodsworth, S.; Botigué, L.R.; Cowan, R.S.; Devault, A.; Eiserhardt, W.L.; Epitawalage, N.; Forest, F.; Kim, J.T.; et al. A Universal Probe Set for Targeted Sequencing of 353 Nuclear Genes from Any Flowering Plant Designed Using K-Medoids Clustering. Syst. Biol. 2019, 68, 594–606. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Breinholt, J.W.; Carey, S.B.; Tiley, G.P.; Davis, E.C.; Endara, L.; McDaniel, S.F.; Neves, L.G.; Sessa, E.B.; von Konrat, M.; Chantanaorrapint, S.; et al. A Target Enrichment Probe Set for Resolving the Flagellate Land Plant Tree of Life. Appl. Plant Sci. 2021, 9, e11406. [Google Scholar] [CrossRef] [PubMed]
- Zuntini, A.R.; Frankel, L.P.; Pokorny, L.; Forest, F.; Baker, W.J. A Comprehensive Phylogenomic Study of the Monocot Order Commelinales, with a New Classification of Commelinaceae. Am. J. Bot. 2021, 108, 1066–1086. [Google Scholar] [CrossRef] [PubMed]
- Maurin, O.; Anest, A.; Bellot, S.; Biffin, E.; Brewer, G.; Charles-Dominique, T.; Cowan, R.S.; Dodsworth, S.; Epitawalage, N.; Gallego, B.; et al. A Nuclear Phylogenomic Study of the Angiosperm Order Myrtales, Exploring the Potential and Limitations of the Universal Angiosperms353 Probe Set. Am. J. Bot. 2021, 108, 1087–1111. [Google Scholar] [CrossRef]
- Thomas, S.K.; Liu, X.; Du, Z.Y.; Dong, Y.; Cummings, A.; Pokorny, L.; Xiang, Q.Y.; Leebens-Mack, J.H. Comprehending Cornales: Phylogenetic Reconstruction of the Order Using the Angiosperms353 Probe Set. Am. J. Bot. 2021, 108, 1112–1121. [Google Scholar] [CrossRef]
- Ufimov, R.; Zeisek, V.; Píšová, S.; Baker, W.J.; Fér, T.; van Loo, M.; Dobeš, C.; Schmickl, R. Relative Performance of Customized and Universal Probe Sets in Target Enrichment: A Case Study in Subtribe Malinae. Appl. Plant Sci. 2021, 9, e11442. [Google Scholar] [CrossRef]
- Siniscalchi, C.M.; Hidalgo, O.; Palazzesi, L.; Pellicer, J.; Pokorny, L.; Maurin, O.; Leitch, I.J.; Forest, F.; Baker, W.J.; Mandel, J.R. Lineage-Specific vs. Universal: A Comparison of the Compositae1061 and Angiosperms353 Enrichment Panels in the Sunflower Family. Appl. Plant Sci. 2021, 9. [Google Scholar] [CrossRef]
- Ogutcen, E.; Christe, C.; Nishii, K.; Salamin, N.; Möller, M.; Perret, M. Phylogenomics of Gesneriaceae Using Targeted Capture of Nuclear Genes. Mol. Phylogenet. Evol. 2021, 157, 107068. [Google Scholar] [CrossRef] [PubMed]
- Shah, T.; Schneider, J.V.; Zizka, G.; Maurin, O.; Baker, W.; Forest, F.; Brewer, G.E.; Savolainen, V.; Darbyshire, I.; Larridon, I. Joining Forces in Ochnaceae Phylogenomics: A Tale of Two Targeted Sequencing Probe Kits. Am. J. Bot. 2021, 108, 1201–1216. [Google Scholar] [CrossRef] [PubMed]
- Bagley, J.C.; Uribe-Convers, S.; Carlsen, M.M.; Muchhala, N. Utility of Targeted Sequence Capture for Phylogenomics in Rapid, Recent Angiosperm Radiations: Neotropical Burmeistera Bellflowers as a Case Study. Mol. Phylogenet. Evol. 2020, 152, 106769. [Google Scholar] [CrossRef] [PubMed]
- Couvreur, T.L.P.; Helmstetter, A.J.; Koenen, E.J.M.; Bethune, K.; Brandão, R.D.; Little, S.A.; Sauquet, H.; Erkens, R.H.J. Phylogenomics of the Major Tropical Plant Family Annonaceae Using Targeted Enrichment of Nuclear Genes. Front. Plant Sci. 2019, 9, 1941. [Google Scholar] [CrossRef] [Green Version]
- Carlsen, M.M.; Fér, T.; Schmickl, R.; Leong-Škorničková, J.; Newman, M.; Kress, W.J. Resolving the Rapid Plant Radiation of Early Diverging Lineages in the Tropical Zingiberales: Pushing the Limits of Genomic Data. Mol. Phylogenet. Evol. 2018, 128, 55–68. [Google Scholar] [CrossRef]
- Jantzen, J.R.; Amarasinghe, P.; Folk, R.A.; Reginato, M.; Michelangeli, F.A.; Soltis, D.E.; Cellinese, N.; Soltis, P.S. A Two-Tier Bioinformatic Pipeline to Develop Probes for Target Capture of Nuclear Loci with Applications in Melastomataceae. Appl. Plant Sci. 2020, 8, e11345. [Google Scholar] [CrossRef]
- Chamala, S.; García, N.; Godden, G.T.; Krishnakumar, V.; Jordon-Thaden, I.E.; de Smet, R.; Barbazuk, W.B.; Soltis, D.E.; Soltis, P.S. MarkerMiner 1.0: A New Application for Phylogenetic Marker Development Using Angiosperm Transcriptomes. Appl. Plant Sci. 2015, 3, 1400115. [Google Scholar] [CrossRef] [Green Version]
- Lamesch, P.; Berardini, T.Z.; Li, D.; Swarbreck, D.; Wilks, C.; Sasidharan, R.; Muller, R.; Dreher, K.; Alexander, D.L.; Garcia-Hernandez, M.; et al. The Arabidopsis Information Resource (TAIR): Improved Gene Annotation and New Tools. Nucleic Acids Res. 2012, 40, D1202–D1210. [Google Scholar] [CrossRef]
- Kubo, T.; Nishizawa, S.; Sugawara, A.; Itchoda, N.; Estiati, A.; Mikami, T. The Complete Nucleotide Sequence of the Mitochondrial Genome of Sugar Beet (B. vulgaris L.) Reveals a Novel Gene for TRNA Cys (GCA). Nucleic Acids Res. 2000, 28, 2571–2576. [Google Scholar] [CrossRef] [Green Version]
- Cheng, C.Y.; Krishnakumar, V.; Chan, A.P.; Thibaud-Nissen, F.; Schobel, S.; Town, C.D. Araport11: A Complete Reannotation of the Arabidopsis Thaliana Reference Genome. Plant J. 2017, 89, 789–804. [Google Scholar] [CrossRef] [Green Version]
- Andermann, T.; Cano, Á.; Zizka, A.; Bacon, C.; Antonelli, A. SECAPR-A Bioinformatics Pipeline for the Rapid and User-Friendly Processing of Targeted Enriched Illumina Sequences, from Raw Reads to Alignments. PeerJ 2018, 6, e5175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnson, M.G.; Gardner, E.M.; Liu, Y.; Medina, R.; Goffinet, B.; Shaw, A.J.; Zerega, N.J.C.; Wickett, N.J. HybPiper: Extracting Coding Sequence and Introns for Phylogenetics from High-Throughput Sequencing Reads Using Target Enrichment. Appl. Plant Sci. 2016, 4, 1600016. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.; Durbin, R. Fast and Accurate Long-Read Alignment with Burrows-Wheeler Transform. Bioinformatics 2010, 26, 589–595. [Google Scholar] [CrossRef] [Green Version]
- Wickham, H. Ggplot2; Springer: New York, NY, USA, 2009; ISBN 978-0-387-98140-6. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing 2020; R Foundation for Statistical Computing: Vienna, Austria, 2020. [Google Scholar]
- Edwards, E.J.; Nyffeler, R.; Donoghue, M.J. Basal Cactus Phylogeny: Implications of Pereskia (Cactaceae) Paraphyly for the Transition to the Cactus Life Form. Am. J. Bot. 2005, 92, 1177–1188. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walker, J.F.; Yang, Y.; Feng, T.; Timoneda, A.; Mikenas, J.; Hutchison, V.; Edwards, C.; Wang, N.; Ahluwalia, S.; Olivieri, J.; et al. From Cacti to Carnivores: Improved Phylotranscriptomic Sampling and Hierarchical Homology Inference Provide Further Insight into the Evolution of Caryophyllales. Am. J. Bot. 2018, 105, 446–462. [Google Scholar] [CrossRef] [Green Version]
- Guerrero, P.C.; Majure, L.C.; Cornejo-Romero, A.; Hernández-Hernández, T. Phylogenetic Relationships and Evolutionary Trends in the Cactus Family. J. Hered. 2019, 110, 4–21. [Google Scholar] [CrossRef]
- Majure Lucas, C.; Acha, S.; Baker Marc, A.; Puente-Martínez; Matias, K.; Shannon, F. Phylogenomics of One of the World’s Most Intriguing Groups of CAM Plants, The Opuntioids (Opuntioideae: Cactaceae): Adaptation to Tropical Dry Forests Helped Drive Prominent Morphological Features in the Clade. University of Florida: Gainesville, FL, USA, 2022; (manuscript in preparation). [Google Scholar]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [Green Version]
- Capella-Gutiérrez, S.; Silla-Martínez, J.M.; Gabaldón, T. TrimAl: A Tool for Automated Alignment Trimming in Large-Scale Phylogenetic Analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML Version 8: A Tool for Phylogenetic Analysis and Post-Analysis of Large Phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Morel, B.; Kozlov, A.M.; Stamatakis, A. ParGenes: A Tool for Massively Parallel Model Selection and Phylogenetic Tree Inference on Thousands of Genes. Bioinformatics 2019, 35, 1771–1773. [Google Scholar] [CrossRef] [Green Version]
- Darriba, D.; Posada, D.; Kozlov, A.M.; Stamatakis, A.; Morel, B.; Flouri, T. ModelTest-NG: A New and Scalable Tool for the Selection of DNA and Protein Evolutionary Models. Mol. Biol. Evol. 2020, 37, 291–294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kozlov, A.M.; Darriba, D.; Flouri, T.; Morel, B.; Stamatakis, A. RAxML-NG: A Fast, Scalable and User-Friendly Tool for Maximum Likelihood Phylogenetic Inference. Bioinformatics 2019, 35, 4453–4455. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, C.; Rabiee, M.; Sayyari, E.; Mirarab, S. ASTRAL-III: Polynomial Time Species Tree Reconstruction from Partially Resolved Gene Trees. BMC Bioinform. 2018, 19, 15–30. [Google Scholar] [CrossRef] [Green Version]
- Smith, S.A.; Moore, M.J.; Brown, J.W.; Yang, Y. Analysis of Phylogenomic Datasets Reveals Conflict, Concordance, and Gene Duplications with Examples from Animals and Plants. BMC Evol. Biol. 2015, 15, 150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neubig, K.M.; Whitten, W.M.; Abbott, J.R.; Elliott, S.; Soltis, D.E.; Soltis, P.S. Variables Affecting DNA Preservation in Archival Plant Specimens. In Proceedings of the DNA Banking for the 21st Century: Proceedings of the US Workshop on DNA Banking; Applequist, W.L., Campbell, L.M., Eds.; William L. Brown Center: St. Louis, MO, USA, 2013; pp. 81–112. [Google Scholar]
- Castro, J.P.; Moraes, A.P.; Chase, M.W.; Santos, A.M.S.; Batista, F.R.C.; Felix, L.P. Karyotype Characterization and Evolution of Chromosome Number in Cactaceae with Special Emphasis on Subfamily Cactoideae. Acta Bot. Bras. 2020, 34, 135–148. [Google Scholar] [CrossRef]
- Wang, N.; Yang, Y.; Moore, M.J.; Brockington, S.F.; Walker, J.F.; Brown, J.W.; Liang, B.; Feng, T.; Edwards, C.; Mikenas, J.; et al. Evolution of Portulacineae Marked by Gene Tree Conflict and Gene Family Expansion Associated with Adaptation to Harsh Environments. Mol. Biol. Evol. 2019, 36, 112–126. [Google Scholar] [CrossRef] [PubMed]
- Hendriks, K.P.; Mandáková, T.; Hay, N.M.; Ly, E.; Hooft van Huysduynen, A.; Tamrakar, R.; Thomas, S.K.; Toro-Núñez, O.; Pires, J.C.; Nikolov, L.A.; et al. The Best of Both Worlds: Combining Lineage-Specific and Universal Bait Sets in Target-Enrichment Hybridization Reactions. Appl. Plant Sci. 2021, 9. [Google Scholar] [CrossRef]
- Mclay, T.G.B.; Birch, J.L.; Gunn, B.F.; Ning, W.; Tate, J.A.; Nauheimer, L.; Joyce, E.M.; Simpson, L.; Weigner, N.; Schmidt-Lebuhn, A.N.; et al. New Targets Acquired: Improving Locus Recovery from the Angiosperms353 Probe Set. Appl. Plant Sci. 2021, 9. [Google Scholar] [CrossRef]
- McDonnell, A.J.; Baker, W.J.; Dodsworth, S.; Forest, F.; Graham, S.W.; Johnson, M.G.; Pokorny, L.; Tate, J.; Wicke, S.; Wickett, N.J. Exploring Angiosperms353: Developing and Applying a Universal Toolkit for Flowering Plant Phylogenomics. Appl. Plant Sci. 2021, 9. [Google Scholar] [CrossRef]
| Species | Clade | NCBI SRA # |
|---|---|---|
| Anacampseros kurtzii | Outgroup | SRR6435355 (Bioproject: PRJNA428216) |
| Ariocarpus retusus | Cactoideae | SRR7905834 |
| Astrophytum myriostigma | Cactoideae | SRR7905836 |
| Carnegiea gigantea | Cactoideae | SRR5036296 |
| Copiapoa desertorum | Cactoideae | SRR7905838 (Bioproject: PRJNA493215) |
| Coryphantha maiz tablasensis | Cactoideae | SRR7905839 (Bioproject: PRJNA493215) |
| Echinocereus pectinatus | Cactoideae | SRR1698109 (Bioproject: PRJNA269655) |
| Echinopsis aurea | Cactoideae | SRR7905840 (Bioproject: PRJNA493215) |
| Eriosyce wagenknechtii | Cactoideae | SRR7905831 (Bioproject: PRJNA493215) |
| Ferocactus latispinus | Cactoideae | SRR7905830 (Bioproject: PRJNA493215) |
| Grusonia bradtiana | Opuntioideae | SRR7905852 (Bioproject: PRJNA493215) |
| Gymnocalycium mihanovichii | Cactoideae | SRR7905853 (Bioproject: PRJNA493215) |
| Hylocereus undatus | Cactoideae | SRR11603181 |
| Leuenbergeria bleo | Leuenbergeria + Pereskia | SRR1698112 |
| Leuenbergeria guamacho | Leuenbergeria + Pereskia | SRR7905854 (Bioproject: PRJNA493215) |
| Maihuenia poeppigii | Maihuenia | SRR7905849 (Bioproject: PRJNA493215) |
| Maihueniopsis conoidea | Opuntioideae | SRR7905848 (Bioproject: PRJNA493215) |
| Matucana aurantiaca | Cactoideae | SRR7905855 (Bioproject: PRJNA493215) |
| Opuntia arenaria | Opuntioideae | SRR7905850 (Bioproject: PRJNA493215) |
| O. cochenillifera | Opuntioideae | SRR1698108 |
| O. ficus indica | Opuntioideae | SRR3567682 |
| O. streptacantha | Opuntioideae | SRR3478181 |
| Pachycereus gatesii | Cactoideae | SRR7905847 (Bioproject: PRJNA493215) |
| Peniocereus cuixmalensis | Cactoideae | SRR7905861 (Bioproject: PRJNA493215) |
| Pereskia grandifolia | Leuenbergeria + Pereskia | SRR1698106 |
| Portulaca oleracea 1 | Outgroup | SRR10247085 |
| P. oleracea 2 | Outgroup | SRR10247116 |
| Pterocactus tuberosus | Opuntioideae | SRR7905860 (Bioproject: PRJNA493215) |
| Rhipsalis baccifera | Cactoideae | SRR7905851 (Bioproject: PRJNA493215) |
| Salmiopuntia salmiana | Opuntioideae | SRR7905862 (Bioproject: PRJNA493215) |
| Stenocereus yunckeri | Cactoideae | SRR7905856 (Bioproject: PRJNA493215) |
| Stetsonia coryne | Cactoideae | SRR7905865(Bioproject: PRJNA493215) |
| Tacinga lilae | Opuntioideae | SRR7905864 (Bioproject: PRJNA493215) |
| Talinopsis frutescens | Outgroup | SRR6435354 (Bioproject: PRJNA428216) |
| Tephrocactus bonnieae | Opuntioideae | SRR7905863 (Bioproject: PRJNA493215) |
| Tunilla corrugata | Opuntioideae | SRR7905866 (Bioproject: PRJNA493215) |
| Angiosperms353 | ||||||
|---|---|---|---|---|---|---|
| Cactoideae | Leuenbergeria + Pereskia | Maihuenia | Opuntioideae | Outgroup | ||
| Cactaceae120 | Cactoideae | 303 120 | 272 | 252 | 288 | 288 |
| Leuenbergeria + Pereskia | 109 | 273 109 | 246 | 267 | 269 | |
| Maihuenia | 92 | 91 | 254 92 | 249 | 253 | |
| Opuntioideae | 119 | 109 | 92 | 295 119 | 281 | |
| Outgroup | 117 | 108 | 92 | 116 | 292 117 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Acha, S.; Majure, L.C. A New Approach Using Targeted Sequence Capture for Phylogenomic Studies across Cactaceae. Genes 2022, 13, 350. https://doi.org/10.3390/genes13020350
Acha S, Majure LC. A New Approach Using Targeted Sequence Capture for Phylogenomic Studies across Cactaceae. Genes. 2022; 13(2):350. https://doi.org/10.3390/genes13020350
Chicago/Turabian StyleAcha, Serena, and Lucas C. Majure. 2022. "A New Approach Using Targeted Sequence Capture for Phylogenomic Studies across Cactaceae" Genes 13, no. 2: 350. https://doi.org/10.3390/genes13020350
APA StyleAcha, S., & Majure, L. C. (2022). A New Approach Using Targeted Sequence Capture for Phylogenomic Studies across Cactaceae. Genes, 13(2), 350. https://doi.org/10.3390/genes13020350





