Impact of Variants in the ATIC and ARID5B Genes on Therapeutic Failure with Imatinib in Patients with Chronic Myeloid Leukemia
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics, Consent, and Permissions
2.2. Investigated Population
2.3. Selected Markers
2.4. DNA Extraction and Quantification
2.5. Genotyping
2.6. Genetic Ancestry
2.7. Statistical Analysis
3. Results
3.1. Genotype and Imatinib Response-Relative Risk Assessment (OR)
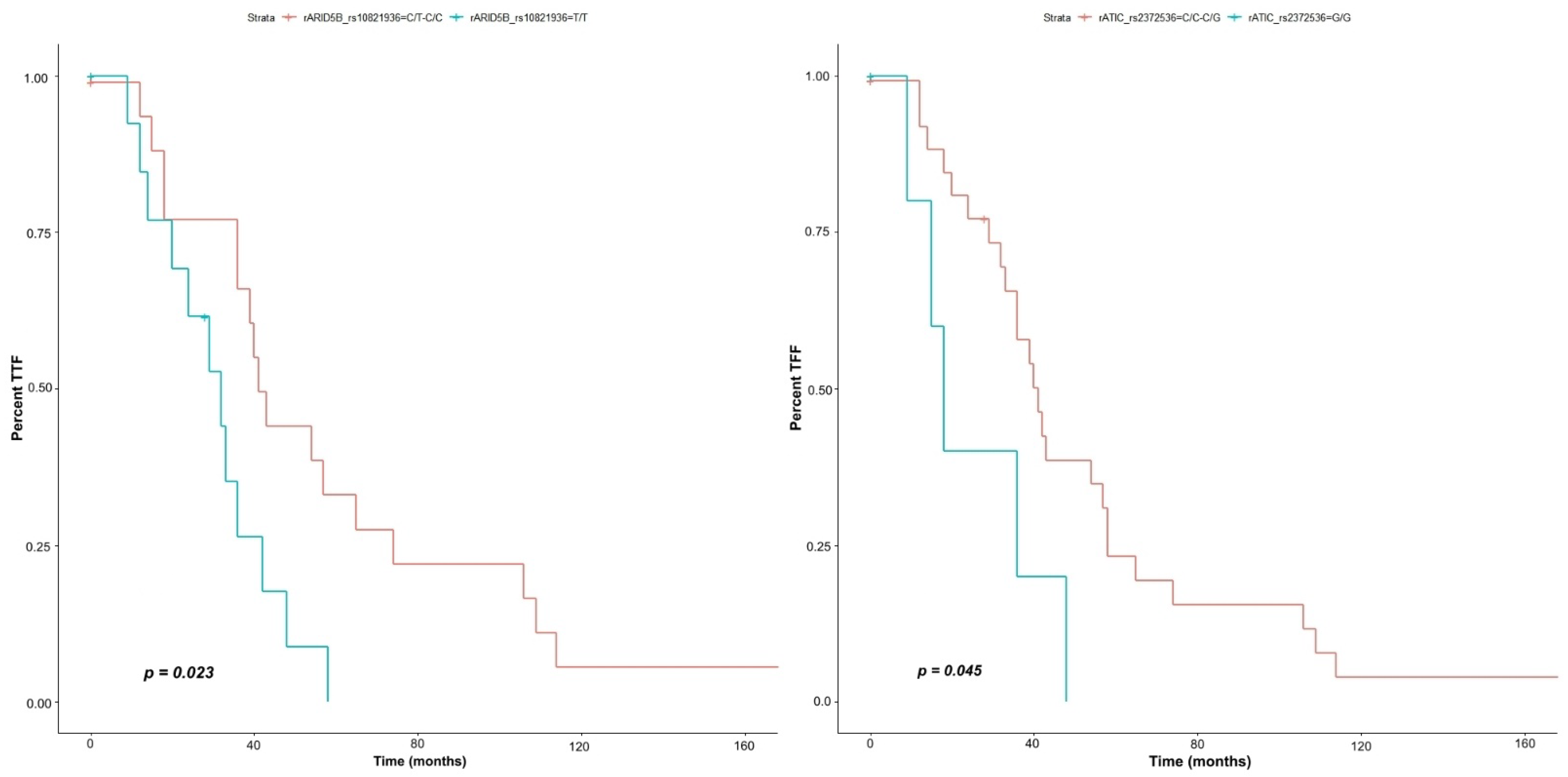
3.2. Time of Treatment Failure (TTF)/Risk Analysis over the Response Time (HR)
4. Discussion
4.1. ATIC
4.2. ARID5B
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Perrotti, D.; Jamieson, C.; Goldman, J.; Skorski, T. Chronic Myeloid Leukemia: Mechanisms of Blastic Transformation. J. Clin. Investig. 2010, 120, 2254–2264. [Google Scholar] [CrossRef]
- Apperley, J.F. CML and Tyrosine Kinase Inhibition: The Hope Becomes Reality. Lancet Haematol. 2015, 2, e176–e177. [Google Scholar] [CrossRef]
- Kang, Z.J.; Liu, Y.F.; Xu, L.Z.; Long, Z.J.; Huang, D.; Yang, Y.; Liu, B.; Feng, J.X.; Pan, Y.J.; Yan, J.S.; et al. The Philadelphia Chromosome in Leukemogenesis. Chin. J. Cancer 2016, 35, 48. [Google Scholar] [CrossRef]
- Ankathil, R.; Azlan, H.; Dzarr, A.A.; Baba, A.A. Pharmacogenetics and the Treatment of Chronic Myeloid Leukemia: How Relevant Clinically? An Update. Pharmacogenomics 2018, 19, 475–493. [Google Scholar] [CrossRef] [PubMed]
- Fausel, C. Targeted Chronic Myeloid Leukemia Therapy: Seeking a Cure. In Proceedings of the American Journal of Health-System Pharmacy, Oxford, UK, 15 December 2007; Volume 64. [Google Scholar]
- Bixby, D.; Talpaz, M. Mechanisms of Resistance to Tyrosine Kinase Inhibitors in Chronic Myeloid Leukemia and Recent Therapeutic Strategies to Overcome Resistance. Hematol. Am. Soc. Hematol. Educ. Program 2009, 461–476. [Google Scholar] [CrossRef] [PubMed]
- Milojkovic, D.; Apperley, J.F. Mechanisms of Resistance to Imatinib and Second-Generation Tyrosine Inhibitors in Chronic Myeloid Leukemia. Clin. Cancer Res. 2009, 15, 7519–7527. [Google Scholar] [CrossRef] [PubMed]
- Shinohara, Y.; Takahashi, N.; Nishiwaki, K.; Hino, M.; Kashimura, M.; Wakita, H.; Hatano, Y.; Hirasawa, A.; Nakagawa, Y.; Itoh, K.; et al. A Multicenter Clinical Study Evaluating the Confirmed Complete Molecular Response Rate in Imatinib-Treated Patients with Chronic Phase Chronic Myeloid Leukemia by Using the International Scale of Real-Time Quantitative Polymerase Chain Reaction. Haematologica 2013, 98, 1407–1413. [Google Scholar] [CrossRef] [PubMed]
- Polillo, M.; Galimberti, S.; Baratè, C.; Petrini, M.; Danesi, R.; Di Paolo, A. Pharmacogenetics of BCR/ABL Inhibitors in Chronic Myeloid Leukemia. Int. J. Mol. Sci. 2015, 16, 22811–22829. [Google Scholar] [CrossRef]
- Guo, C.; Xie, X.; Li, J.; Huang, L.; Chen, S.; Li, X.; Yi, X.; Wu, Q.; Yang, G.; Zhou, H.; et al. Pharmacogenomics Guidelines: Current Status and Future Development. Clin. Exp. Pharmacol. Physiol. 2019, 46, 689–693. [Google Scholar] [CrossRef]
- Weinshilboum, R.M.; Wang, L. Pharmacogenetics and Pharmacogenomics: Development, Science, and Translation. Annu. Rev. Genom. Hum. Genet. 2006, 7, 223–245. [Google Scholar] [CrossRef]
- Rodrigues, J.C.G.; Fernandes, M.R.; Guerreiro, J.F.; Silva, A.L.D.C.D.; Ribeiro-dos-Santos, Â.; Santos, S.; Santos, N.P.C. Dos Polymorphisms of ADME-Related Genes and Their Implications for Drug Safety and Efficacy in Amazonian Amerindians. Sci. Rep. 2019, 9, 7201. [Google Scholar] [CrossRef] [PubMed]
- De Carvalho, D.C.; Wanderley, A.V.; dos Santos, A.M.R.; Moreira, F.C.; de Sá, R.B.A.; Fernandes, M.R.; Modesto, A.A.C.; de Souza, T.P.; Cohen-Paes, A.; Leitão, L.P.C.; et al. Characterization of Pharmacogenetic Markers Related to Acute Lymphoblastic Leukemia Toxicity in Amazonian Native Americans Population. Sci. Rep. 2020, 10, 10292. [Google Scholar] [CrossRef] [PubMed]
- Santos, N.P.C.; Ribeiro-Rodrigues, E.M.; Ribeiro-dos-Santos, Â.K.; Pereira, R.; Gusmão, L.; Amorim, A.; Guerreiro, J.F.; Zago, M.A.; Matte, C.; Hutz, M.H.; et al. Assessing Individual Interethnic Admixture and Population Substructure Using a 48-Insertion-Deletion (INSEL) Ancestry-Informative Marker (AIM) Panel. Hum. Mutat. 2010, 31, 184–190. [Google Scholar] [CrossRef]
- Suarez-Kurtz, G. Population Impact of Pharmacogenetic Tests in Admixed Populations across the Americas. Pharmacogenom. J. 2021, 21, 216–221. [Google Scholar] [CrossRef]
- Shah, N.P. NCCN Guidelines Updates: Discontinuing TKI Therapy in the Treatment of Chronic Myeloid Leukemia. J. Natl. Compr. Cancer Netw. 2019, 17, 611–613. [Google Scholar]
- Ramos, B.R.; Mendes, N.D.; Tanikawa, A.A.; Amador, M.A.; dos Santos, N.P.; dos Santos, S.E.; Castelli, E.C.; Witkin, S.S.; da Silva, M.G. Ancestry informative markers and selected single nucleotide polymorphisms in immunoregulatory genes on preterm labor and preterm premature rupture of membranes: A case control study. BMC Pregnancy Childbirth 2016, 16, 30. [Google Scholar] [CrossRef]
- NOVARTIS Gleevec: Imatinib Mesylate Tablet. Available online: https://www.accessdata.fda.gov/scripts/cder/daf/index.cfm?event=overview.process&varApplNo=021588 (accessed on 25 November 2021).
- Yang, K.; Fu, L.W. Mechanisms of Resistance to BCR-ABL TKIs and the Therapeutic Strategies: A Review. Crit. Rev. Oncol. Hematol. 2015, 93, 277–292. [Google Scholar] [CrossRef] [PubMed]
- Wall, M.; Shim, J.H.; Benkovic, S.J. Human AICAR Transformylase: Role of the 4-Carboxamide of AICAR in Binding and Catalysis. Biochemistry 2000, 39, 11303–11311. [Google Scholar] [CrossRef]
- Martinez-Outschoorn, U.E.; Peiris-Pagés, M.; Pestell, R.G.; Sotgia, F.; Lisanti, M.P. Erratum: Cancer Metabolism: A Therapeutic Perspective. Nat. Rev. Clin. Oncol. 2017, 14, 113. [Google Scholar] [CrossRef]
- Yamaoka, T.; Kondo, M.; Honda, S.; Iwahana, H.; Moritani, M.; Ii, S.; Yoshimoto, K.; Itakura, M. Amidophosphoribosyltransferase Limits the Rate of Cell Growth-Linked de Novo Purine Biosynthesis in the Presence of Constant Capacity of Salvage Purine Biosynthesis. J. Biol. Chem. 1997, 272, 17719–17725. [Google Scholar] [CrossRef]
- Chan, C.Y.; Zhao, H.; Pugh, R.J.; Pedley, A.M.; French, J.; Jones, S.A.; Zhuang, X.; Jinnah, H.; Huan, T.J.; Benkovic, S.J. Purinosome Formation as a Function of the Cell Cycle. Proc. Natl. Acad. Sci. USA 2015, 112, 1368–1373. [Google Scholar] [CrossRef]
- Liu, X.; Chhipa, R.R.; Pooya, S.; Wortman, M.; Yachyshin, S.; Chow, L.M.L.; Kumar, A.; Zhou, X.; Sun, Y.; Quinn, B.; et al. Discrete Mechanisms of MTOR and Cell Cycle Regulation by AMPK Agonists Independent of AMPK. Proc. Natl. Acad. Sci. USA 2014, 111, E435–E444. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Jin, C.; Xu, M.; Zhou, L.; Li, D.; Yin, Y. Bifunctional Enzyme ATIC Promotes Propagation of Hepatocellular Carcinoma by Regulating AMPK-MTOR-S6 K1 Signaling. Cell Commun. Signal. 2017, 15, 52. [Google Scholar] [CrossRef]
- Huo, X.; Qi, J.; Huang, K.; Bu, S.; Yao, W.; Chen, Y.; Nie, J. Identification of an Autophagy-Related Gene Signature That Can Improve Prognosis of Hepatocellular Carcinoma Patients. BMC Cancer 2020, 20, 771. [Google Scholar] [CrossRef]
- Zhu, J.; Wang, M.; Hu, D. Development of an Autophagy-Related Gene Prognostic Signature in Lung Adenocarcinoma and Lung Squamous Cell Carcinoma. PeerJ 2020, 2020, e8288. [Google Scholar] [CrossRef]
- Li, R.; Chen, G.; Dang, Y.; He, R.; Liu, A.; Ma, J.; Wang, C. Upregulation of ATIC in Multiple Myeloma Tissues Based on Tissue Microarray and Gene Microarrays. Int. J. Lab. Hematol. 2021, 43, 409–417. [Google Scholar] [CrossRef] [PubMed]
- Trinei, M.; Lanfrancone, L.; Campo, E.; Pulford, K.; Mason, D.Y.; Pelicci, P.G.; Falini, B. A New Variant Anaplastic Lymphoma Kinase (ALK)-Fusion Protein (ATIC-ALK) in a Case of ALK-Positive Anaplastic Large Cell Lymphoma. Cancer Res. 2000, 60, 793–798. [Google Scholar] [PubMed]
- Van Der Krogt, J.A.; Bempt, M.V.; Ferreiro, J.F.; Mentens, N.; Jacobs, K.; Pluys, U.; Doms, K.; Geerdens, E.; Uyttebroeck, A.; Pierre, P.; et al. Anaplastic Lymphoma Kinase-Positive Anaplastic Large Cell Lymphoma with the Variant RNF213-, ATIC- and TPM3-ALK Fusions Is Characterized by Copy Number Gain of the Rearranged ALK Gene. Haematologica 2017, 102, 1605–1616. [Google Scholar] [CrossRef]
- Boccalatte, F.E.; Voena, C.; Riganti, C.; Bosia, A.; D’Amico, L.; Riera, L.; Cheng, M.; Ruggeri, B.; Jensen, O.N.; Goss, V.L.; et al. The Enzymatic Activity of 5-Aminoimidazole-4-Carboxamide Ribonucleotide Formyltransferase/IMP Cyclohydrolase Is Enhanced by NPM-ALK: New Insights in ALK-Mediated Pathogenesis and the Treatment of ALCL. Blood 2009, 113, 2776–2790. [Google Scholar] [CrossRef]
- Su, W.J.; Lu, P.Z.; Wu, Y.; Kalpana, K.; Yang, C.K.; Lu, G.D. Identification of Key Genes in Purine Metabolism as Prognostic Biomarker for Hepatocellular Carcinoma. Front. Oncol. 2021, 10, 3057. [Google Scholar] [CrossRef]
- Zhang, K.; Jiang, K.; Hong, R.; Xu, F.; Xia, W.; Qin, G.; Lee, K.; Zheng, Q.; Lu, Q.; Zhai, Q.; et al. Identification and Characterization of Critical Genes Associated with Tamoxifen Resistance in Breast Cancer. PeerJ 2020, 8, e10468. [Google Scholar] [CrossRef]
- Visser, S.; Koolen, S.; Van Donk, N.; Van Walree, N.; Van Der Leest, C.; Cornelissen, R.; Van Schaik, R.; Mathijssen, R.; Aerts, J.; Stricker, B.H. Genetic Polymorphism in ATIC Is Associated with Effectiveness and Toxicity of Pemetrexed in Non-Small-Cell Lung Cancer. Thorax 2021, 76, 1150–1153. [Google Scholar] [CrossRef] [PubMed]
- Kurzawski, M.; Malinowski, D.; Szarmach, N.; Nowak, A.; Goryniak, A.; Pawlik, A.; Droździk, M. ATIC Missense Variant Affects Response to Methotrexate Treatment in Rheumatoid Arthritis Patients. Pharmacogenomics 2016, 17, 1971–1978. [Google Scholar] [CrossRef]
- Park, J.A.; Shin, H.Y. ATIC Gene Polymorphism and Histologic Response to Chemotherapy in Pediatric Osteosarcoma. J. Pediatr. Hematol. Oncol. 2017, 39, e270–e274. [Google Scholar] [CrossRef]
- Adam de Beaumais, T.; Jacqz-Aigrain, E. Intracellular Disposition of Methotrexate in Acute Lymphoblastic Leukemia in Children. Curr. Drug Metab. 2012, 13, 822–834. [Google Scholar] [CrossRef] [PubMed]
- Hinks, A.; Moncrieffe, H.; Martin, P.; Ursu, S.; Lal, S.; Kassoumeri, L.; Weiler, T.; Glass, D.N.; Thompson, S.D.; Wedderburn, L.R.; et al. Association of the 5-Aminoimidazole-4-Carboxamide Ribonucleotide Transformylase Gene with Response to Methotrexate in Juvenile Idiopathic Arthritis. Ann. Rheum. Dis. 2011, 70, 1395–1400. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.H.; Bae, S.C. Association of the ATIC 347 C/G Polymorphism with Responsiveness to and Toxicity of Methotrexate in Rheumatoid Arthritis: A Meta-Analysis. Rheumatol. Int. 2016, 36, 1591–1599. [Google Scholar] [CrossRef]
- Eektimmerman, F.; Swen, J.J.; Madhar, M.B.; Allaart, C.F.; Guchelaar, H.J. Predictive Genetic Biomarkers for the Efficacy of Methotrexate in Rheumatoid Arthritis: A Systematic Review. Pharmacogenom. J. 2020, 20, 159–168. [Google Scholar] [CrossRef]
- Lahoud, M.H.; Ristevski, S.; Venter, D.J.; Jermiin, L.S.; Bertoncello, I.; Zavarsek, S.; Hasthorpe, S.; Drago, J.; de Kretser, D.; Hertzog, P.J.; et al. Gene targeting of Desrt, a novel ARID class DNA-binding protein, causes growth retardation and abnormal development of reproductive organs. Genome Res. 2001, 11, 1327–1334. [Google Scholar] [CrossRef]
- Lin, C.; Song, W.; Bi, X.; Zhao, J.; Huang, Z.; Li, Z.; Zhou, J.; Cai, J.; Zhao, H. Recent advances in the ARID family: Focusing on roles in human cancer. OncoTargets Ther. 2014, 7, 315–324. [Google Scholar] [CrossRef]
- Baba, A.; Ohtake, F.; Okuno, Y.; Yokota, K.; Okada, M.; Imai, Y.; Ni, M.; Meyer, C.A.; Igarashi, K.; Kanno, J.; et al. PKA-dependent regulation of the histone lysine demethylase complex PHF2-ARID5B. Nat. Cell Biol. 2011, 13, 668–675. [Google Scholar] [CrossRef]
- Orsi, L.; Rudant, J.; Bonaventure, A.; Goujon-Bellec, S.; Corda, E.; Evans, T.J.; Petit, A.; Bertrand, Y.; Nelken, B.; Robert, A.; et al. Genetic polymorphisms and childhood acute lymphoblastic leukemia: GWAS of the ESCALE study (SFCE). Leukemia 2012, 26, 2561–2564. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Vijayakrishnan, J.; Qian, M.; Studd, J.B.; Yang, W.; Kinnersley, B.; Law, P.J.; Broderick, P.; Raetz, E.A.; Allan, J.; Pui, C.H.; et al. Identification of four novel associations for B-cell acute lymphoblastic leukaemia risk. Nat. Commun. 2019, 10, 5348. [Google Scholar] [CrossRef]
- Yang, J.L.; Liu, Y.N.; Bi, Y.Y.; Wang, H. ARID5B gene polymorphisms and the risk of childhood acute lymphoblastic leukemia: A meta-analysis. Int. J. Hematol. 2019, 110, 272–284. [Google Scholar] [CrossRef]
- Tamai, M.; Huang, M.; Kagami, K.; Abe, M.; Somazu, S.; Shinohara, T.; Harama, D.; Watanabe, A.; Akahane, K.; Goi, K.; et al. Association of relapse-linked ARID5B single nucleotide polymorphisms with drug resistance in B-cell precursor acute lymphoblastic leukemia cell lines. Cancer Cell Int. 2020, 20, 434. [Google Scholar] [CrossRef]
- Studd, J.B.; Vijayakrishnan, J.; Yang, M.; Migliorini, G.; Paulsson, K.; Houlston, R.S. Genetic and regulatory mechanism of susceptibility to high-hyperdiploid acute lymphoblastic leukaemia at 10p21.2. Nat. Commun. 2017, 8, 14616. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Zhao, X.; Bhojwani, D.; E, S.; Goodings, C.; Zhang, H.; Seibel, N.L.; Yang, W.; Li, C.; Carroll, W.L.; et al. ARID5B Influences Antimetabolite Drug Sensitivity and Prognosis of Acute Lymphoblastic Leukemia. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2020, 26, 256–264. [Google Scholar] [CrossRef]
- Sharma, B.; Angurana, S.; Shah, R.; Verma, S.; Bhat, A.; Bhat, G.R.; Bakshi, D.; Jamwal, R.S.; Tanwar, M.; Singh, S.; et al. Genetic association of ARID5B with the risk of colorectal cancer within Jammu and Kashmir. India. Genes Genet. Syst. 2021, 96, 187–191. [Google Scholar] [CrossRef] [PubMed]
- Singh, B.; Kinne, H.E.; Milligan, R.D.; Washburn, L.J.; Olsen, M.; Lucci, A. Important role of FTO in the survival of rare panresistant triple-negative inflammatory breast cancer cells facing a severe metabolic challenge. PLoS ONE 2016, 11, e0159072. [Google Scholar] [CrossRef]
- Davalieva, K.; Kostovska, I.M.; Kiprijanovska, S.; Markoska, K.; Kubelka-Sabit, K.; Filipovski, V.; Polenakovic, M. Proteomics analysis of malignant and benign prostate tissue by 2D DIGE/MS reveals new insights into proteins involved in prostate cancer. Prostate 2015, 75, 1586–1600. [Google Scholar] [CrossRef]
| Variable | Responders | No Responders | p-Value |
|---|---|---|---|
| 103 (63.1%) | 62 (36.9%) | ||
| Age (years) | 48.50 ± 15.07 | 46.76 ± 15.00 | 0.451 |
| Sex (%) | 0.078 | ||
| Men | 64 (62.1) | 29 (46.7) | |
| Women | 39 (37.9) | 33 (53.3) | |
| Ancestry Mean | |||
| European | 0.468 ± 0.147 | 0.458 ± 0.147 | 0.797 |
| Amerindian | 0.299 ± 0.132 | 0.319 ± 0.133 | 0.604 |
| African | 0.233 ± 0.101 | 0.223 ± 0.101 | 0.686 |
| Genotype | HR (95% CI) | Lower | Upper | p-Value |
|---|---|---|---|---|
| ATIC rs2372536 | ||||
| CC/CG vs. GG 1 | 2.726 | 0.9986 | 7.441 | 0.04 |
| CC vs. CG + GG 2 | 1.484 | 0.7235 | 3.046 | 0.3 |
| ARID5B rs10821936 | ||||
| TT vs. CT + CC 1 | 0.4053 | 0.1802 | 0.911 | 0.02 |
| CC vs. TT + CT 2 | 0.6114 | 0.2436 | 1.535 | 0.3 |
| TPMT rs1142345 | ||||
| TT + CT vs. CC 1 | 0.711 | 0.165 | 3.063 | 0.6 |
| TT vs. CT + CC 2 | 0.9895 | 0.4341 | 2.255 | 1.00 |
| TPMT rs12201199 | ||||
| NA 1,3 | ||||
| AA vs. AT + TT 2 | 0.6824 | 0.2561 | 1.818 | 0.4 |
| SLC01B1 rs4149056 | ||||
| NA 1,3 | ||||
| TT vs. CT + CC 2 | 1.109 | 0.3502 | 2.323 | 0.8 |
| ABCC2 rs717620 | ||||
| NA 1,3 | ||||
| CC vs. CT + TT 2 | 1.909 | 0.1807 | 1.518 | 0.2 |
| ABCC3 rs9895420 | ||||
| NA 1,3 | ||||
| TT vs. AT + AA 2 | 1.738 | 0.1983 | 1.67 | 0.3 |
| GGH rs11545078 | ||||
| NA 1,3 | ||||
| GG vs. AG + AA 2 | 1.769 | 0.81 | 3.862 | 0.1 |
| GGH rs3758149 | ||||
| GG + AG vs. AA 1 | 0.9296 | 0.2162 | 3.997 | 0.9 |
| GG vs. AG + AA 2 | 1.148 | 0.5384 | 2.446 | 0.7 |
| ATIC rs4673993 | ||||
| TT + CT vs. CC 1 | 2.561 | 0.3905 | 0.9388 | 0.06 |
| TT vs. CT + CC 2 | 1.633 | 0.7888 | 3.382 | 0.2 |
| AMPD1 rs17602729 | ||||
| NA 1,3 | ||||
| GG vs. AG + AA 2 | 0.7357 | 0.2756 | 1.964 | 0.5 |
| CCND1 rs9344 | ||||
| GG + AG vs. AA 1 | 1.536 | 0.5155 | 4.576 | 0.4 |
| GG vs. AG + AA 2 | 1.174 | 0.5439 | 2.532 | 0.7 |
| IKZF1 rs4132601 | ||||
| TT + GT vs. GG 1 | 0.7569 | 0.1013 | 5.657 | 0.8 |
| TT vs. GT + GG 2 | 1.164 | 0.5258 | 2.579 | 0.7 |
| ITPA rs1127354 | ||||
| CC vs. AC 1 | 1.957 | 0.4465 | 8.575 | 0.4 |
| MTRR rs1801394 | ||||
| AA + AG vs. GG 1 | 0.8794 | 0.328 | 2.358 | 0.8 |
| AA vs. AG + GG 2 | 0.9615 | 0.4536 | 2.038 | 0.9 |
| MTHFD1 rs2236225 | ||||
| GG + AG vs. AA 1 | 0.6535 | 0.2462 | 1.735 | 0.4 |
| GG vs. AG + AA 2 | 0.7404 | 0.3318 | 1.652 | 0.5 |
| NOS3 rs1799983 | ||||
| NA 1,3 | ||||
| GG vs. GT 2 | 1.052 | 0.2408 | 4.592 | 0.9 |
| MTHFR rs1801133 | ||||
| GG + GA vs. AA 1 | 1.445 | 0.4198 | 4.97 | 0.6 |
| GG vs. GA + AA 2 | 0.9837 | 0.4521 | 2.141 | 1.00 |
| TLR4 rs4986790 | ||||
| NA 1,3 | ||||
| AA vs. AG 2 | 1.545 | 0.6471 | 0.2034 | 0.7 |
| TPMT rs1800460 | ||||
| CC vs. CT + TT 1 | 0.6339 | 0.218 | 1.843 | 0.4 |
| NA2 | ||||
| SLCO1B1 rs4149015 | ||||
| GG vs. AG + AA 1 | 0.8043 | 0.2271 | 2.849 | 0.7 |
| AA vs. GG + AG 2 | 3.472 | 0.288 | 0.4051 | 0.2 |
| GGH rs1800909 | ||||
| AA+AG vs. GG 1 | 0.8698 | 0.2995 | 2.526 | 0.8 |
| NA 2 | ||||
| NALCN rs7992226 | ||||
| AA+AG vs. GG 1 | 0.03471 | 0.42 | 2.552 | 0.9 |
| AA vs. AG + GG 2 | 1.199 | 0.348 | 4.132 | 0.8 |
| SHMT1 rs1979277 | ||||
| AA+AG vs. GG 1 | 0.873 | 0.3515 | 2.168 | 0.8 |
| GG vs. AA + AG 2 | 3.361 | 0.6504 | 1.447 | 0.1 |
| SLCO1B1 rs2306283 | ||||
| GG+AG vs. AA 1 | 1.733 | 0.6368 | 4.718 | 0.3 |
| GG vs. AG + AA 2 | 1.276 | 0.7837 | 0.4663 | |
| CEBPE rs2239633 | ||||
| GG+AG vs. AA 1 | 1.899 | 0.8027 | 4.493 | 0.1 |
| GG vs. AG + AA 2 | 2.229 | 0.4485 | 0.506 | 0.3 |
| TNFAIP3 rs6920220 | ||||
| GG+AG vs. AA 1 | 0.6463 | 0.2426 | 1.722 | 0.4 |
| NA 2 | ||||
| PIP4K2A rs7088318 | ||||
| AA vs. AC + CC 1 | 0.5205 | 1.921 | 0.1135 | 0.4 |
| NA 2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cereja Pantoja, K.B.C.; Azevedo, T.C.d.B.; Carvalho, D.C.d.; Monte, N.; Cohen Paes, A.d.N.; Barros, M.C.d.C.; Vinagre, L.W.M.S.; Freitas, A.R.S.d.; Burbano, R.M.R.; Assumpção, P.P.d.; et al. Impact of Variants in the ATIC and ARID5B Genes on Therapeutic Failure with Imatinib in Patients with Chronic Myeloid Leukemia. Genes 2022, 13, 330. https://doi.org/10.3390/genes13020330
Cereja Pantoja KBC, Azevedo TCdB, Carvalho DCd, Monte N, Cohen Paes AdN, Barros MCdC, Vinagre LWMS, Freitas ARSd, Burbano RMR, Assumpção PPd, et al. Impact of Variants in the ATIC and ARID5B Genes on Therapeutic Failure with Imatinib in Patients with Chronic Myeloid Leukemia. Genes. 2022; 13(2):330. https://doi.org/10.3390/genes13020330
Chicago/Turabian StyleCereja Pantoja, Karla Beatriz Cardias, Tereza Cristina de Brito Azevedo, Darlen Cardoso de Carvalho, Natasha Monte, Amanda de Nazaré Cohen Paes, Maria Clara da Costa Barros, Lui Wallacy Morikawa Souza Vinagre, Ana Rosa Sales de Freitas, Rommel Mario Rodríguez Burbano, Paulo Pimentel de Assumpção, and et al. 2022. "Impact of Variants in the ATIC and ARID5B Genes on Therapeutic Failure with Imatinib in Patients with Chronic Myeloid Leukemia" Genes 13, no. 2: 330. https://doi.org/10.3390/genes13020330
APA StyleCereja Pantoja, K. B. C., Azevedo, T. C. d. B., Carvalho, D. C. d., Monte, N., Cohen Paes, A. d. N., Barros, M. C. d. C., Vinagre, L. W. M. S., Freitas, A. R. S. d., Burbano, R. M. R., Assumpção, P. P. d., Santos, S. E. B. d., Fernandes, M. R., & Santos, N. P. C. d. (2022). Impact of Variants in the ATIC and ARID5B Genes on Therapeutic Failure with Imatinib in Patients with Chronic Myeloid Leukemia. Genes, 13(2), 330. https://doi.org/10.3390/genes13020330








