Quantitative Trait Loci for Heat Stress Tolerance in Brassica rapa L. Are Distributed across the Genome and Occur in Diverse Genetic Groups, Flowering Phenologies and Morphotypes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Temperature Treatments
2.3. Phenotyping
2.4. Statistical Analysis of Phenotypic Traits
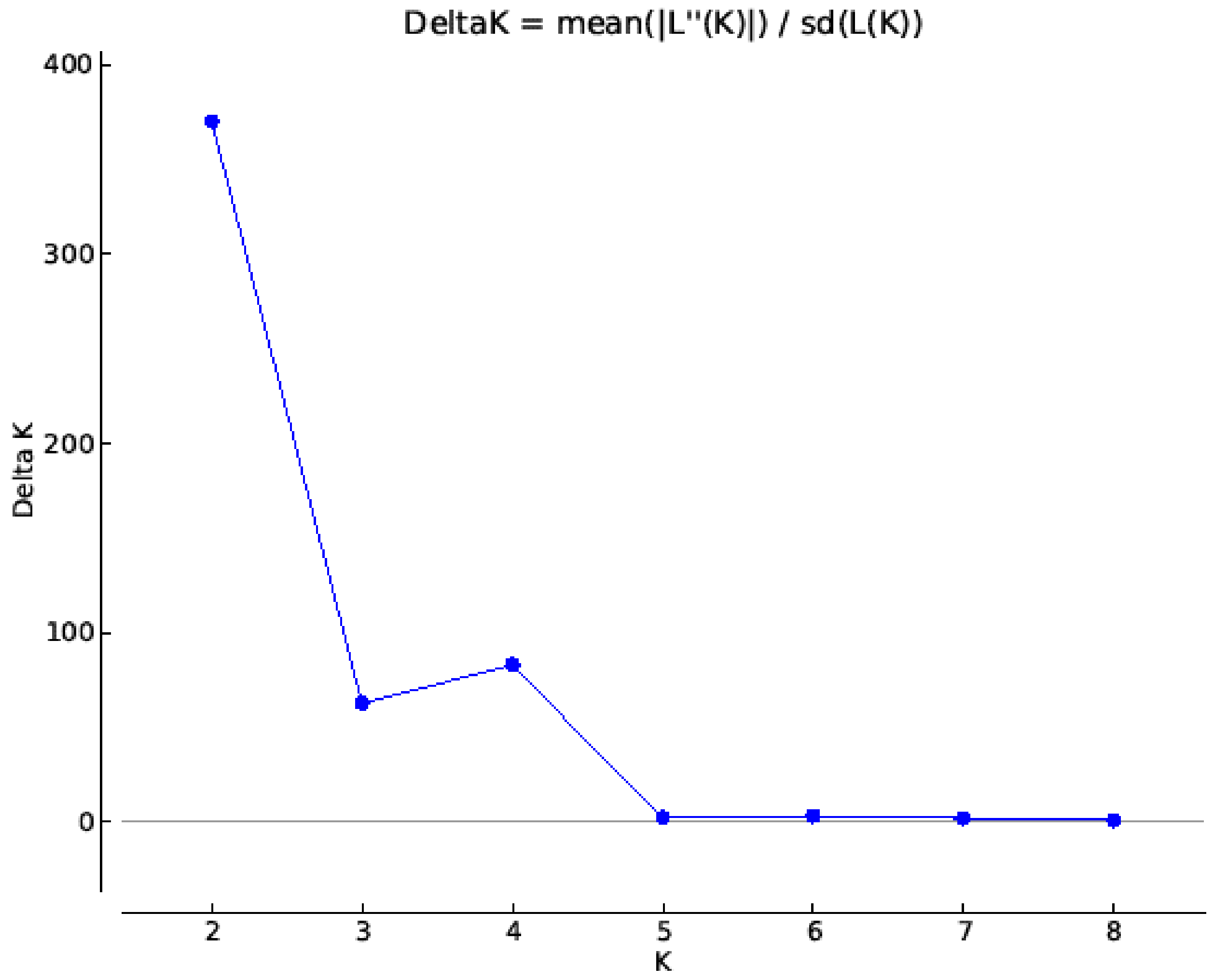
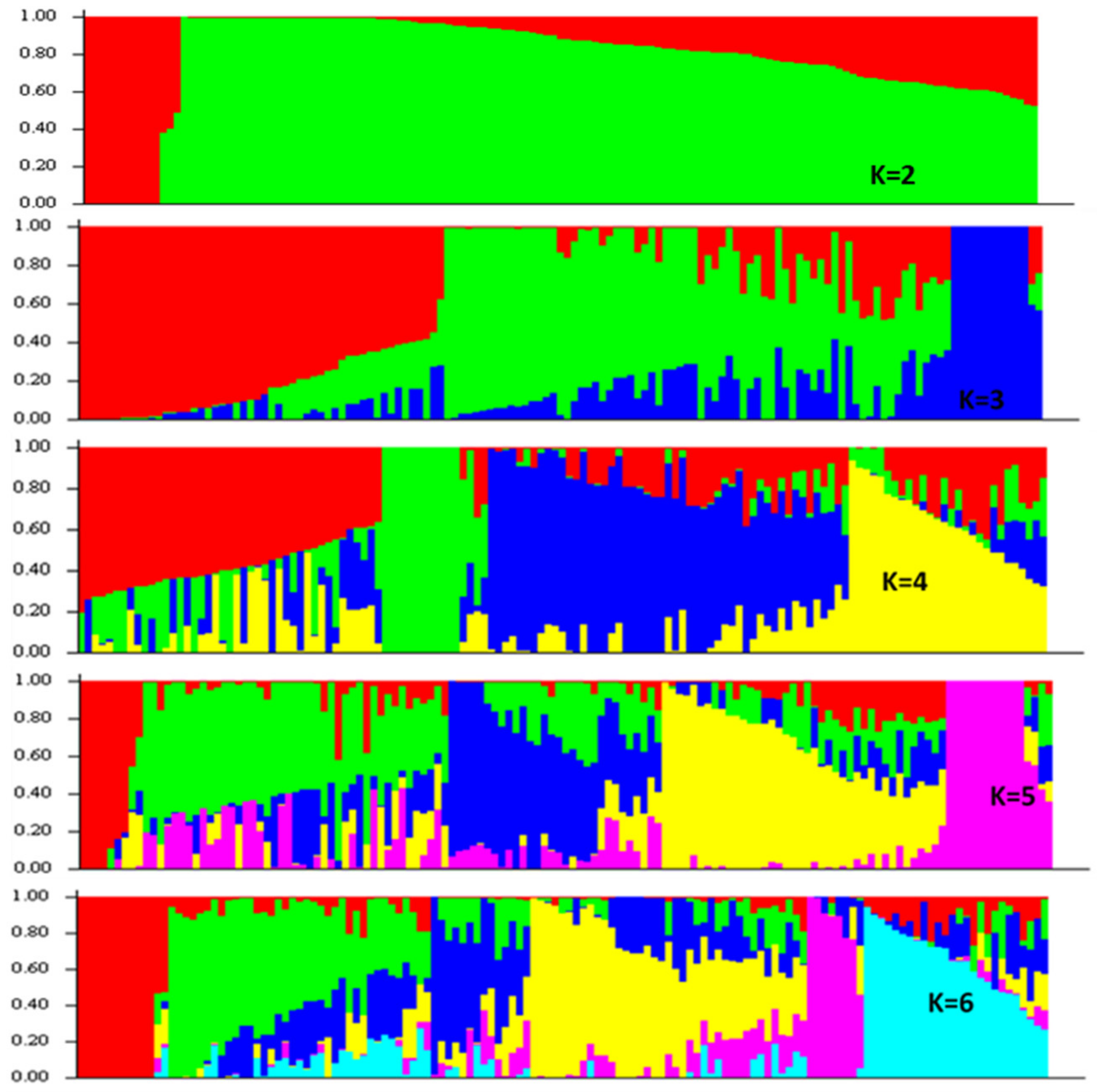
2.5. SNP Genotyping, Population Structure and Genetic Diversity Analysis
2.6. Genome-Wide Association Mapping
2.7. Candidate Gene Discovery
3. Results
3.1. Phenotypic Variation and Heat Stress Tolerance
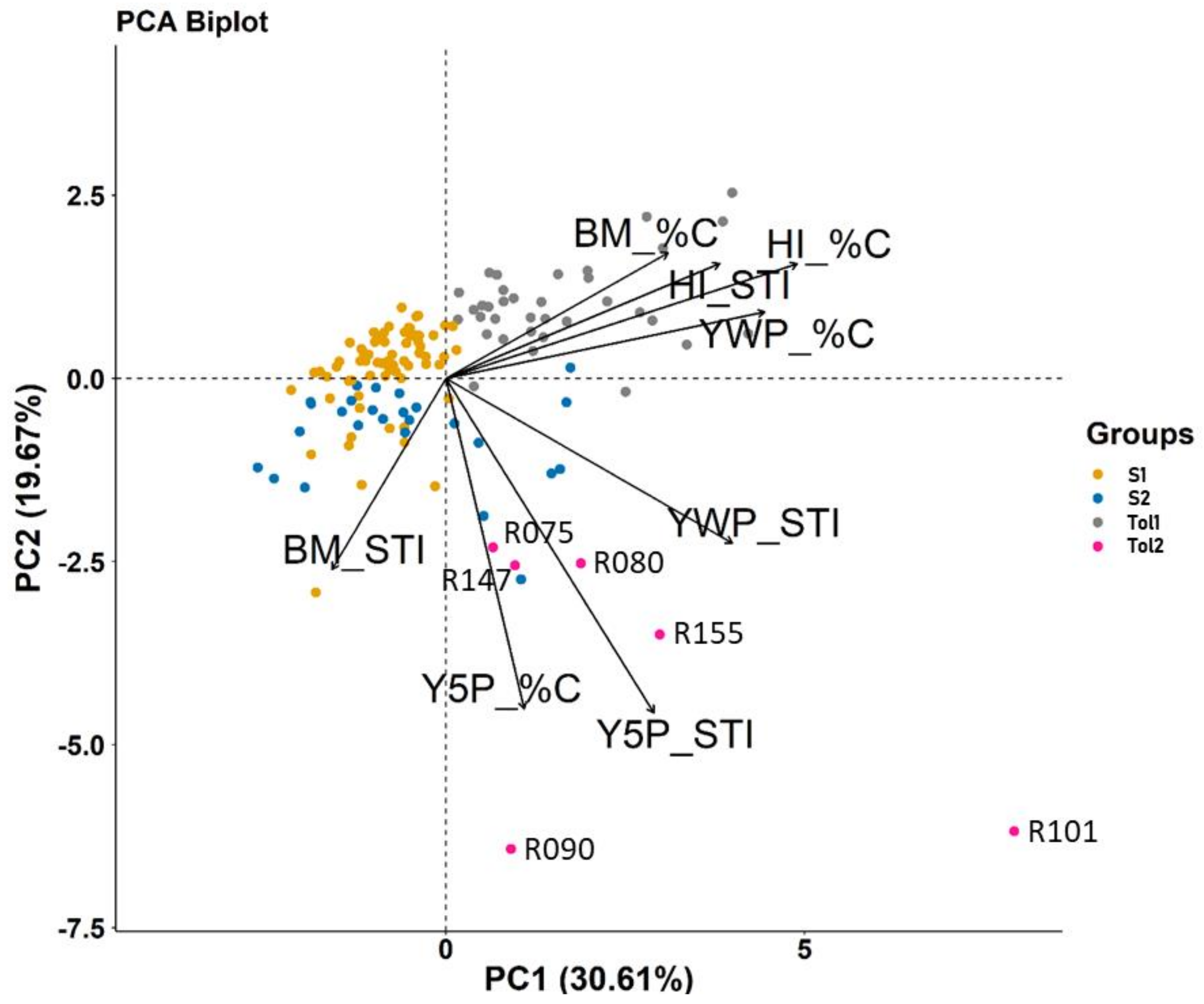
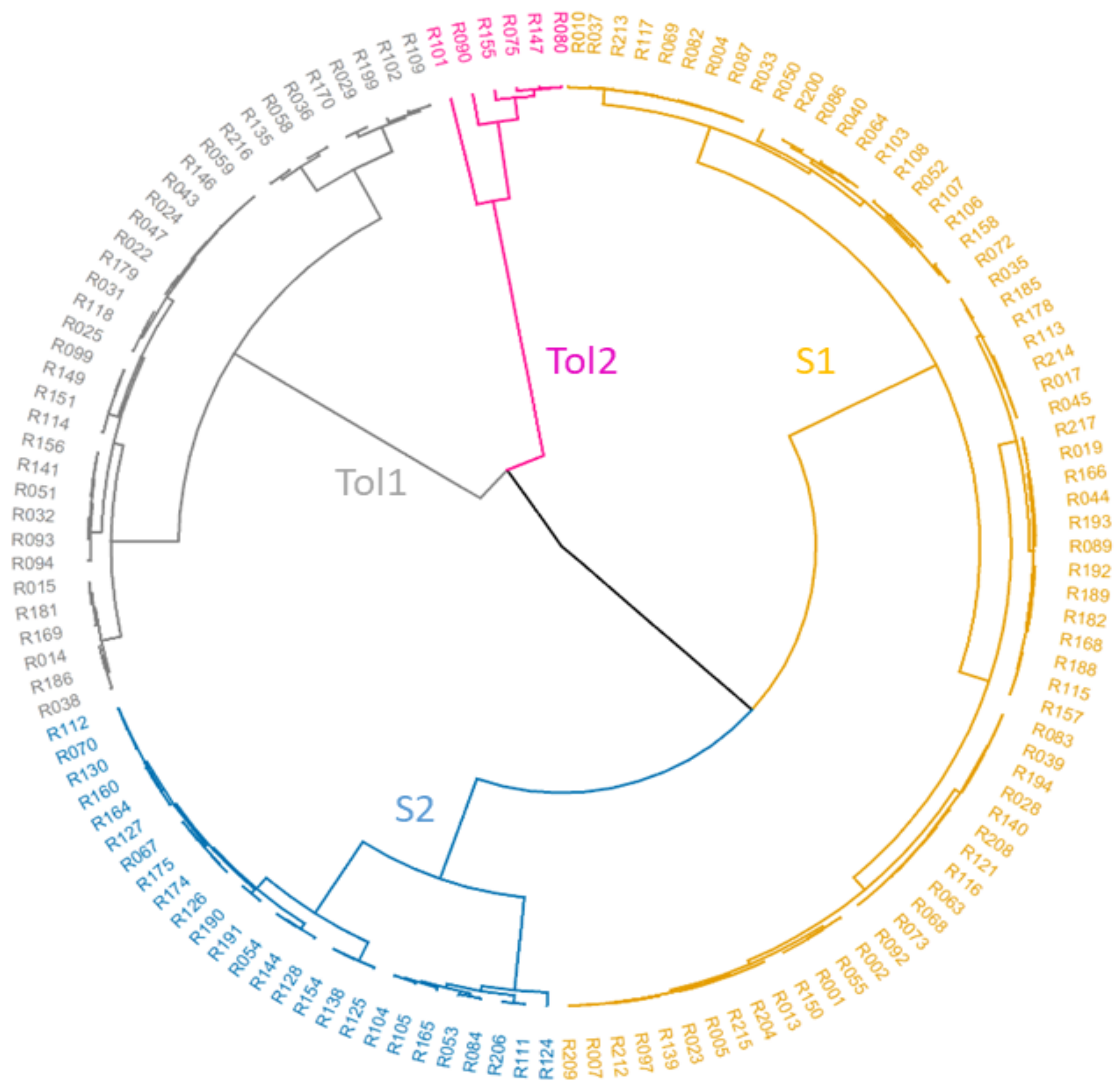
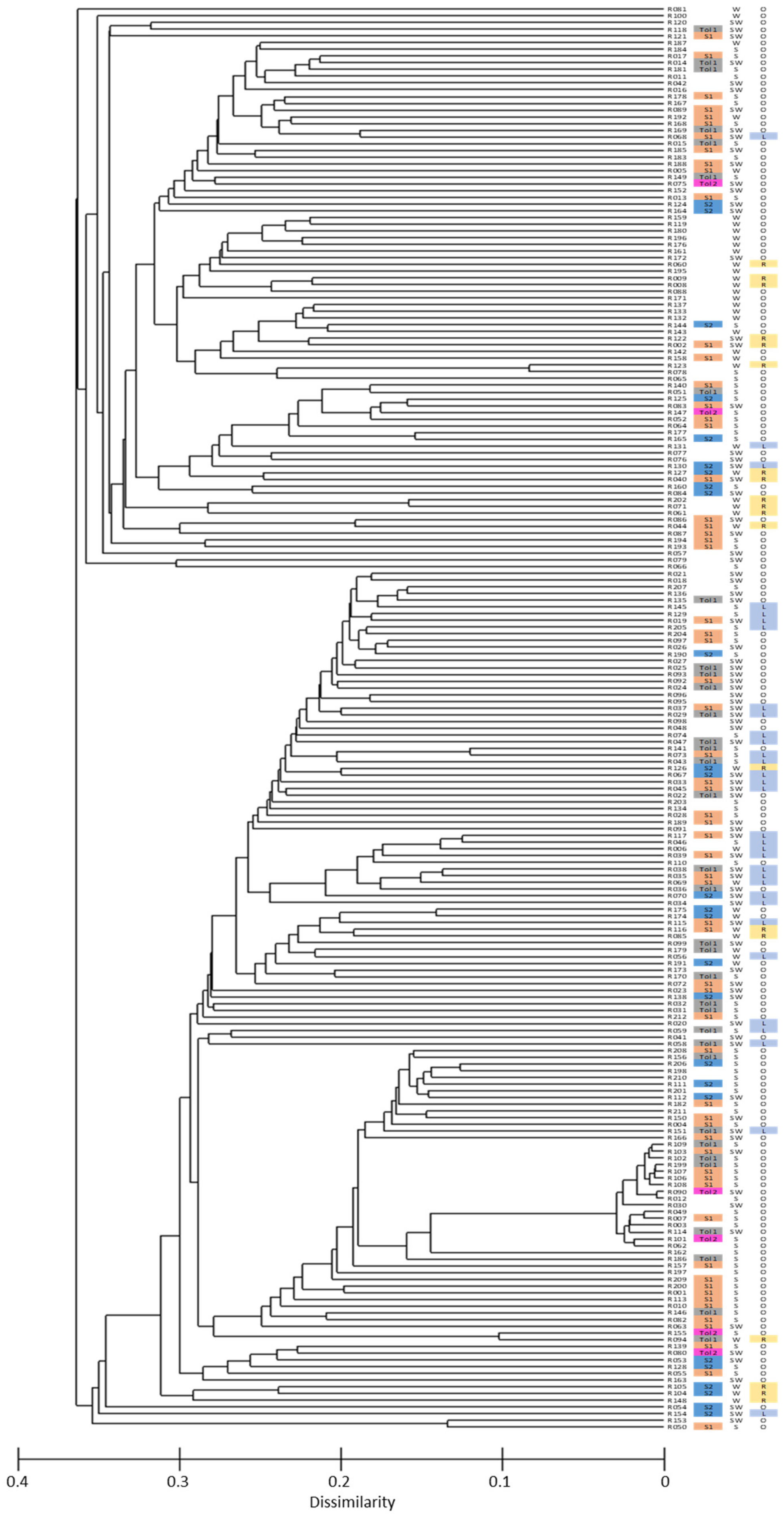
3.2. SNP Genetic Diversity and Genetic Population Structure versus Morphotype, Flowering Phenology and Heat Tolerance and Sensitivity
3.3. Genome-Wide Association Analysis and Candidate Genes Related to Heat Stress Tolerance
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chalhoub, B.; Denoeud, F.; Liu, S.; Parkin, I.A.P.; Tang, H.; Wang, X.; Chiquet, J.; Belcram, H.; Tong, C.; Samans, B.; et al. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 2014, 345, 950–953. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Becker, H.C.; Engqvist, G.M.; Karlsson, B. Comparison of rapeseed cultivars and resynthesized lines based on allozyme and RFLP markers. Theor. Appl. Genet. 1995, 91, 62–67. [Google Scholar] [CrossRef] [PubMed]
- Cowling, W. Genetic diversity in Australian canola and implications for crop breeding for changing future environments. Field Crop. Res. 2007, 104, 103–111. [Google Scholar] [CrossRef]
- Annisa; Chen, S.; Turner, N.C.; Cowling, W.A. Genetic variation for heat tolerance during the reproductive phase in Brassica rapa. J. Agron. Crop Sci. 2013, 199, 424–435. [Google Scholar] [CrossRef]
- Liu, H. Rapeseed Genetics and Breeding; Shanghai Science and Technology Press: Shanghai, China, 1985. [Google Scholar]
- Dixon, G.R. Vegetable Brassicas and related crucifers. In Crop Production Science in Horticulture Series; Atherton, J., Rees, H., Eds.; CAB International: Oxfordshire, UK, 2007. [Google Scholar]
- McAlvay, A.C.; Ragsdale, A.P.; Mabry, M.E.; Qi, X.; Bird, K.A.; Velasco, P.; An, H.; Pires, J.C.; Emshwiller, E. Brassica rapa domestication: Untangling wild and feral forms and convergence of crop morphotypes. Mol. Biol. Evol. 2021, 38, 3358–3372. [Google Scholar] [CrossRef] [PubMed]
- Warwick, S.I.; James, T.; Falk, K.C. AFLP-based molecular characterization of Brassica rapa and diversity in Canadian spring turnip rape cultivars. Plant Genet. Resour. 2008, 6, 11–21. [Google Scholar] [CrossRef]
- McGrath, J.M.; Quiros, C.F. Genetic diversity at isozyme and RFLP loci in Brassica campestris as related to crop type and geographical origin. Theor. Appl. Genet. 1992, 83, 783–790. [Google Scholar] [CrossRef]
- Annisa; Chen, S.; Cowling, W.A. Global genetic diversity in oilseed Brassica rapa. Crop. Pasture Sci. 2013, 64, 993–1007. [Google Scholar] [CrossRef]
- Zhao, J.; Wang, X.; Deng, B.; Lou, P.; Wu, J.; Sun, R.; Xu, Z.; Vromans, J.; Koornneef, M.; Bonnema, G. Genetic relationships within Brassica rapa as inferred from AFLP fingerprints. Theor. Appl. Genet. 2005, 110, 1301–1314. [Google Scholar] [CrossRef]
- Guo, Y.; Chen, S.; Li, Z.; Cowling, W.A. Center of origin and centers of civersity in an ancient crop, Brassica rapa (turnip rape). J. Hered. 2014, 105, 555–565. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.; Guo, Y.; Sirault, X.; Stefanova, K.; Saradadevi, R.; Turner, N.C.; Nelson, M.N.; Furbank, R.T.; Siddique, K.H.M.; Cowling, W.A. Nondestructive phenomic tools for the prediction of heat and drought tolerance at anthesis in Brassica species. Plant Phenomics 2019, 2019, 1–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, S.; Stefanova, K.; Siddique, K.H.M.; Cowling, W.A. Transient daily heat stress during the early reproductive phase disrupts pod and seed development in Brassica napus L. Food Energy Secur. 2020, 10, e262. [Google Scholar] [CrossRef]
- Morrison, M.J.; Stewart, D.W. Heat stress during flowering in summer Brassica. Crop Sci. 2002, 3, 797–803. [Google Scholar] [CrossRef]
- Young, L.W.; Wilen, R.W.; Bonham-Smith, P.C. High temperature stress of Brassica napus during flowering reduces micro- and megagametophyte fertility, induces fruit abortion, and disrupts seed production. J. Exp. Bot. 2004, 55, 485–495. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.; Saradadevi, R.; Vidotti, M.S.; Frische-Neto, R.; Crossa, J.; Siddique, K.H.M.; Crowling, W.A. Female reproductive organs of Brassica napus are more sensitive than male to transient heat stress. Euphytica 2021, 6, 117. [Google Scholar] [CrossRef]
- Fernandez, G.C.J. Effective selection criteria for assessing plant stress tolerance. Proceedings Of The International Symposium On Adaptation Of Vegetables And Other Food Crops In Temperature And Water Stress, Shanhua, Taiwan, 13–16 August 1992. [Google Scholar]
- Butler, D.G.; Cullis, B.R.; Gilmour, A.R.; Gogel, B.J.; Thompson, R. ASReml-R Reference Manual Version 4; VSN International Ltd.: Hemel Hempstead, UK, 2017. [Google Scholar]
- Dalton-Morgan, J.; Hayward, A.; Alamery, S.; Tollenaere, R.; Mason, A.S.; Campbell, E.; Patel, D.; Lorenc, M.T.; Yi, B.; Long, Y.; et al. A high-throughput SNP array in the amphidiploid species Brassica napus. Funct. Integr. Genom. 2014, 4, 643–655. [Google Scholar] [CrossRef]
- Wang, X.; Wang, H.; Wang, J.; Sun, R.; Wu, J.; Liu, S.; Bai, Y.; Mun, J.-H.; Bancroft, I.; Cheng, F.; et al. The genome of the mesopolyploid crop species Brassica rapa. Nat Genet. 2011, 10, 1035–1039. [Google Scholar] [CrossRef] [Green Version]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef]
- Falush, D.; Stephens, M.; Pritchard, J.K. Inference of population structure using multilocus genotype data: Linked loci and correlated allele frequencies. Genetics 2003, 164, 1567–1587. [Google Scholar] [CrossRef]
- Loiselle, B.A.; Sork, V.L.; Nason, J.; Graham, C. Spatial genetic structure of a tropical understory shrub, Psychotria officinalis (Rubiaceae). Am. J. Bot. 1995, 11, 1420–1425. [Google Scholar] [CrossRef]
- Hardy, O.J.; Vekemans, X. Spagedi: A versatile computer program to analyse spatial genetic structure at the individual or population levels. Mol. Ecol. Notes 2002, 2, 618–620. [Google Scholar] [CrossRef] [Green Version]
- Clarke, K.R.; Gorley, R.N. PRIMER v6: User Manual/Tutorial; PRIMER-E Ltd.: Plymouth, UK, 2006. [Google Scholar]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef] [PubMed]
- Stich, B.; Möhring, J.; Piepho, H.-P.; Heckenberger, M.; Buckler, E.S.; Melchinger, A.E. Comparison of mixed-model approaches for association mapping. Genetics 2008, 178, 1745–1754. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, J.; Pressoir, G.; Briggs, W.H.; Bi, I.V.; Yamasaki, M.; Doebley, J.F.; McMullen, M.D.; Gaut, B.S.; Nielsen, D.M.; Holland, J.B.; et al. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat. Genet. 2005, 38, 203–208. [Google Scholar] [CrossRef]
- Kaler, A.S.; Gillman, J.D.; Beissinger, T.; Purcell, L.C. Comparing different statistical models and multiple testing corrections for association mapping in soybean and maize. Front. Plant Sci. 2020, 10, 1794. [Google Scholar] [CrossRef] [PubMed]
- Sun, Q.; Miao, C.; Hanel, M.; Borthwick, A.G.; Duan, Q.; Ji, D.; Li, H. Global heat stress on health, wildfires, and agricultural crops under different levels of climate warming. Environ. Int. 2019, 128, 125–136. [Google Scholar] [CrossRef]
- Bird, K.A.; An, H.; Gazave, E.; Gore, M.A.; Pires, J.C.; Robertson, L.D.; Labate, J.A. Population structure and phylogenetic relationships in a diverse panel of Brassica rapa L. Front. Plant Sci. 2017, 8, 321. [Google Scholar] [CrossRef] [Green Version]
- Lohani, N.; Jain, D.; Singh, M.B.; Bhalla, P.L. Engineering multiple abiotic stress tolerance in canola, Brassica napus. Front. Plant Sci. 2020, 11, 3. [Google Scholar] [CrossRef]
- Haider, S.; Iqbal, J.; Naseer, S.; Yaseen, T.; Shaukat, M.; Bibi, H.; Ahmad, Y.; Daud, H.; Abbasi, N.L.; Mahmood, T. Molecular mechanisms of plant tolerance to heat stress: Current landscape and future perspectives. Plant Cell Rep. 2021, 40, 2247–2271. [Google Scholar] [CrossRef]
- Rahaman, M.; Mamidi, S.; Rahman, M. Genome-wide association study of heat stress-tolerance traits in spring-type Brassica napus L. under controlled conditions. Crop J. 2018, 6, 115–125. [Google Scholar] [CrossRef]
- Wang, H.; Chen, J.-G.; Chang, Y. Identification, expression, and interaction analysis of ovate family proteins in Populus trichocarpa reveals a role of PtOFP1 regulating drought stress response. Front. Plant Sci. 2021, 12, 1–15. [Google Scholar] [CrossRef] [PubMed]
| # | Trait | Abbreviation | Unit | Evaluation Method | Stage of Observation |
|---|---|---|---|---|---|
| 1 | Bud temperature depression (difference between bud and ambient temperature) | T1 | °C | Leaf and bud temperatures were measured with an infrared thermometer (Impac® Model IN 15 plus); a separate digital thermometer with a 1 s response time was used to measure ambient temperature. | Early flowering stage |
| 2 | Leaf temperature depression (difference between leaf and ambient temperature) | T2 | °C | Early flowering stage | |
| 3 | Temperature difference between bud and leaf | T3 | °C | Early flowering stage | |
| 4 | Leaf conductance | LC | mmol m−2 s−1 | Sum of the adaxial and abaxial stomatal conductance of the youngest fully expanded leaf measured by SC1 leaf porometer. | Early flowering stage |
| 5 | Chlorophyll content index | CI | SPAD unit | Ratio of leaf transmittance in red light at 650 nm and near-infrared light at 940 nm, as measured on intact leaves with a portable chlorophyll meter SPAD-502Plus | Early flowering stage |
| 6 | Above-ground biomass | BM | g | Weight of all plant materials above the ground at harvest | Maturity |
| 7 | Seed yield whole plant | YWP | g | Weight of seed harvested from each plant | Maturity |
| 8 | Harvest index | HI | % | Ratio of seed yield vs biomass | Maturity |
| 9 | Seed yield of five pods | Y5P | g | Seed yield from five bagged floral buds on the main stem | Maturity |
| Trait | Random Effects | Fixed Effects | Predicted Value | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Line | Line × Treatment | Error | Treatment | C | H | %C | ||||
| Yield-related traits at maturity | ||||||||||
| Y5P | 0.0002 ± 0.0007 | NS | 0.0065 ± 0.0009 | *** | 0.0031 ± 0.0002 | 112.14 | *** | 0.1148 ± 0.0076 | 0.0022 ± 0.0076 | −98.08 |
| YWP | 0.1836 ± 0.0479 | *** | 0.1913 ± 0.0421 | *** | 0.4106 ± 0.0260 | 30.307 | *** | 1.0026 ± 0.0625 | 0.6107 ± 0.0625 | −39.09 |
| BM | 33.14 ± 4.621 | *** | 2.289 ± 1.099 | * | 17.34 ± 1.095 | 6.85 | ** | 10.77 ± 0.5579 | 11.70 ± 0.5570 | 8.64 |
| HI | 59.31 ± 17.43 | *** | 33.70 ± 17.77 | * | 268.27 ± 17.65 | 24.78 | *** | 14.11 ± 1.215 | 7.051 ± 1.191 | −50.03 |
| Physiological traits during heat stress | ||||||||||
| T1 | 0.0001 ± 0.0000 | NS | 0.2433 ± 0.0369 | *** | 0.1605 ± 0.0100 | 116.39 | *** | −1.496 ± 0.0475 | −2.184 ± 0.0472 | −45.99 |
| T2 | 0.0787 ± 0.0691 | NS | 0.5825 ± 0.08879 | *** | 0.3770 ± 0.0235 | 234.70 | *** | −2.501 ± 0.0770 | −4.082 ± 0.0768 | −63.21 |
| T3 | 0.1133 ± 0.0455 | ** | 0.2303 ± 0.0500 | *** | 0.4778 ± 0.0300 | 124.61 | *** | 1.007 ± 0.0621 | 1.868 ± 0.0616 | 85.50 |
| CI | 42.09 ± 6.684 | *** | 14.96 ± 2.791 | *** | 21.65 ± 1.351 | 9.22 | ** | 33.36 ± 0.6948 | 35.12 ± 0.6936 | 5.28 |
| LC | 5365.07 ± 5546.58 | NS | 52,488.21 ± 7164.96 | *** | 16,796.12 ± 1048.64 | 165.86 | *** | 515.91 ± 21.82 | 894.97 ± 21.79 | 73.47 |
| Trait | S1 (67 Lines) | S2 (26 Lines) | Tol1 (35 Lines) | Tol2 (6 Lines) | ||||
|---|---|---|---|---|---|---|---|---|
| %C | STI | %C | STI | %C | STI | %C | STI | |
| Yield-related traits at maturity | ||||||||
| Y5P | −98.871 | 0.016 | −99.370 | 0.014 | −99.536 | 0.015 | −82.303 | 0.481 |
| YWP | −53.673 | 0.342 | −42.482 | 1.410 | −8.375 | 0.968 | −22.807 | 1.703 |
| BM | 7.447 | 0.908 | 4.687 | 3.591 | 22.798 | 0.638 | 15.532 | 0.799 |
| HI | −60.615 | 0.274 | −58.859 | 0.334 | −37.822 | 1.756 | −43.814 | 1.045 |
| Physiological traits during heat stress | ||||||||
| T1 | −50.850 | 1.472 | −49.365 | 1.443 | −60.430 | 1.523 | −65.560 | 1.540 |
| T2 | −63.507 | 1.682 | −63.447 | 1.603 | −86.770 | 1.604 | −98.057 | 1.637 |
| T3 | 111.502 | 2.088 | 128.260 | 1.862 | 140.279 | 1.787 | 152.271 | 1.830 |
| CI | 6.581 | 1.036 | 1.135 | 1.334 | 10.004 | 0.995 | 16.235 | 1.174 |
| LC | 82.134 | 1.794 | 56.748 | 1.699 | 128.071 | 1.703 | 86.578 | 1.934 |
| Chromosome | Position | Marker | Trait Associated | LOD | Phenotypic Variance Explained (%) |
|---|---|---|---|---|---|
| STI and %C based on YWP | |||||
| A03 | 5622193 | UQnapus1553 | YWP_%C | 3.81 | 15.11 |
| A04 | 14473945 | UQnapus5039 | YWP_%C | 3.26 | 9.45 |
| A05 | 7032029 | UQnapus2055 | YWP_%C | 3.57 | 12.10 |
| A05 | 21539755 | UQnapus5640 | YWP_STI | 4.37 | 19.06 |
| A07 | 9559418 | UQnapus0132 | YWP_%C | 3.55 | 10.98 |
| A07 | 13661428 | UQnapus0012 | YWP_%C | 4.56 | 13.26 |
| Under heat stress condition | |||||
| A01 | 23298355 | UQnapus5153 | HI_H | 3.72 | 8.38 |
| A01 | 24614901 | UQnapus1279 | BM_H | 3.71 | 14.18 |
| A02 | 11002571 | UQnapus1420 | BM_H | 3.42 | 10.61 |
| A02 | 25794385 | UQnapus1090 | HI_H | 5.06 | 15.72 |
| A03 | 5898488 | UQnapus1557 | BM_H | 4.30 | 12.85 |
| A03 | 9882877 | UQnapus2837 | HI_H | 3.26 | 12.29 |
| A03 | 18737164 | UQnapus1661 | Y5P_H | 6.07 | 19.61 |
| A03 | 19296951 | UQnapus1669 | BM_H | 3.24 | 10.96 |
| A03 | 20865276 | UQnapus1680 | BM_H | 3.12 | 6.52 |
| A03 | 29591540 | UQnapus5276 | BM_H | 3.05 | 9.98 |
| A04 | 11451264 | UQnapus1923 | Y5P_H | 3.07 | 11.44 |
| A04 | 18294405 | UQnapus2000 | HI_H | 3.72 | 13.64 |
| A05 | 4109774 | UQnapus2563 | HI_H | 4.58 | 15.90 |
| A05 | 10231305 | UQnapus5012 | BM_H | 3.24 | 10.34 |
| A05 | 17371711 | UQnapus0273 | Y5P_H | 3.18 | 10.80 |
| A06 | 3956415 | UQnapus5652 | BM_H | 3.33 | 10.46 |
| A07 | 6234486 | UQnapus2420 | HI_H | 4.00 | 14.11 |
| A07 | 11614849 | UQnapus2473 | Y5P_H | 4.69 | 15.67 |
| A07 | 17122140 | UQnapus0069 | Y5P_H | 5.16 | 9.23 |
| A08 | 2617392 | UQnapus2637 | Y5P_H | 3.80 | 13.02 |
| A09 | 8745612 | UQnapus2826 | YWP_H | 3.03 | 8.04 |
| A09 | 36005032 | UQnapus2957 | Y5P_H | 3.14 | 10.93 |
| A10 | 6601391 | UQnapus4615 | BM_H | 5.04 | 15.61 |
| A10 | 8771845 | UQnapus3070 | BM_H | 3.13 | 8.86 |
| Under control condition | |||||
| A01 | 6681985 | UQnapus5365 | BM_C | 3.52 | 14.03 |
| A01 | 14706490 | UQnapus1205 | Y5P_C | 3.94 | 12.20 |
| A01 | 16284432 | UQnapus3105 | BM_C | 3.08 | 11.89 |
| A01 | 20392779 | UQnapus1241 | HI_C | 3.81 | 12.05 |
| A02 | 1250723 | UQnapus5063 | BM_C | 3.85 | 7.05 |
| A02 | 8703496 | UQnapus1394 | YWP_C | 3.12 | 8.64 |
| A02 | 8884662 | UQnapus5442 | YWP_C | 3.08 | 10.53 |
| A02 | 8949907 | UQnapus1398 | HI_C | 3.57 | 11.64 |
| A03 | 5898488 | UQnapus1557 | BM_C | 3.67 | 13.07 |
| A03 | 16861693 | UQnapus1638 | HI_C | 7.46 | 22.77 |
| A04 | 11451264 | UQnapus1923 | HI_C | 3.68 | 12.20 |
| A05 | 4109774 | UQnapus2563 | HI_C | 6.31 | 20.35 |
| A05 | 17680875 | UQnapus2144 | YWP_C | 3.38 | 9.73 |
| A06 | 1530076 | UQnapus2220 | BM_C | 3.23 | 13.05 |
| A06 | 2902967 | UQnapus5649 | YWP_C | 3.79 | 11.01 |
| A06 | 3186312 | UQnapus5341 | HI_C | 3.84 | 13.01 |
| A06 | 15891262 | UQnapus5014 | HI_C | 3.53 | 12.31 |
| A07 | 9119865 | UQnapus2455 | BM_C | 3.43 | 13.20 |
| A07 | 11614849 | UQnapus2473 | YWP_C | 3.71 | 10.81 |
| A07 | 17157679 | UQnapus0091 | YWP_C | 3.22 | 10.48 |
| A08 | 8043536 | UQnapus2097 | Y5P_C | 3.83 | 19.78 |
| A08 | 20418844 | UQnapus2762 | YWP_C | 3.01 | 8.85 |
| A09 | 798560 | UQnapus2777 | HI_C | 7.46 | 22.82 |
| A09 | 798560 | UQnapus2777 | YWP_C | 3.19 | 9.04 |
| A09 | 1372115 | UQnapus2783 | YWP_C | 3.20 | 10.48 |
| A09 | 6624646 | UQnapus2806 | YWP_C | 3.29 | 10.48 |
| A09 | 7086176 | UQnapus2807 | HI_C | 3.13 | 10.25 |
| A09 | 7452134 | UQnapus2809 | YWP_C | 3.34 | 9.13 |
| A09 | 27135256 | UQnapus2916 | HI_C | 4.30 | 15.47 |
| A09 | 35718978 | UQnapus3633 | HI_C | 4.16 | 23.43 |
| A10 | 4433065 | UQnapus1779 | YWP_C | 3.84 | 11.94 |
| A10 | 6507394 | UQnapus1763 | HI_C | 3.03 | 10.47 |
| A10 | 6601391 | UQnapus4615 | BM_C | 4.65 | 16.14 |
| # | B. rapa | B. napus | At Ortholog | Gene | Annotation/Function (GO/At Ortholog Function) |
|---|---|---|---|---|---|
| 1 | Bra015593 | BnaA07g01650D | AT3G48800.1 | Sterile α motif (SAM) domain-containing protein | Sterile α motif (SAM) domain-containing protein |
| 2 | Bra038828 | BnaA07g01660D | AT2G18890.1 | Protein kinase superfamily protein | Protein kinase superfamily protein; FUNCTIONS IN: protein serine/threonine kinase activity, protein kinase activity, ATP binding; INVOLVED IN: protein amino acid phosphorylation |
| 3 | BnaA07g01670D | AT2G15740.1 AT5G42640.1 | C2H2 and C2H2-like zinc fingers superfamily protein | FUNCTIONS IN: sequence-specific DNA binding transcription factor activity, zinc ion binding, nucleic acid binding; INVOLVED IN: regulation of transcription; LOCATED IN: intracellular | |
| 4 | Bra039606 | BnaA07g01680D | AT2G18490.1 AT2G15740.1 AT5G42640.1 | ||
| 5 | Bra039605 | BnaA07g01690D | AT5G09920.1 | RNA polymerase II, Rpb4, core protein | Non-catalytic subunit specific to DNA-dependent RNA polymerase II; the ortholog of budding yeast RPB4 |
| 6 | Bra039604 | BnaA07g01700D | AT2G18500.1 | Ovate family protein 7 (OFP7) | INVOLVED IN: biological process unknown; LOCATED IN: plasma membrane; EXPRESSED IN: shoot apex, embryo, hypocotyl, flower, seed |
| 7 | Bra039603 | BnaA07g01710D | AT2G18510.1 | Embryo defective 2444 (emb2444), RNA-binding (RRM/RBD/RNP motifs) family protein | FUNCTIONS IN: RNA binding, nucleotide binding, nucleic acid binding; INVOLVED IN: embryo development ending in seed dormancy; LOCATED IN: nucleolus |
| 8 | Bra028100 | BnaA07g01720D | AT4G36710.1 | GRAS family transcription factor | CONTAINS InterPro DOMAIN/s: Transcription factor GRAS (InterPro:IPR005202) |
| 9 | Bra028098 | BnaA07g01730D | AT2G18570.1 AT2G18560.1 | UDP-Glycosyltransferase superfamily protein | FUNCTIONS IN: transferase activity, transferring glycosyl groups; INVOLVED IN: metabolic process; LOCATED IN: cellular component unknown |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, S.; Hayward, A.; Dey, S.S.; Choudhary, M.; Witt Hmon, K.P.; Inturrisi, F.C.; Dolatabadian, A.; Neik, T.X.; Yang, H.; Siddique, K.H.M.; et al. Quantitative Trait Loci for Heat Stress Tolerance in Brassica rapa L. Are Distributed across the Genome and Occur in Diverse Genetic Groups, Flowering Phenologies and Morphotypes. Genes 2022, 13, 296. https://doi.org/10.3390/genes13020296
Chen S, Hayward A, Dey SS, Choudhary M, Witt Hmon KP, Inturrisi FC, Dolatabadian A, Neik TX, Yang H, Siddique KHM, et al. Quantitative Trait Loci for Heat Stress Tolerance in Brassica rapa L. Are Distributed across the Genome and Occur in Diverse Genetic Groups, Flowering Phenologies and Morphotypes. Genes. 2022; 13(2):296. https://doi.org/10.3390/genes13020296
Chicago/Turabian StyleChen, Sheng, Alice Hayward, Shyam S. Dey, Mukesh Choudhary, Khaing P. Witt Hmon, Fabian C. Inturrisi, Aria Dolatabadian, Ting Xiang Neik, Hua Yang, Kadambot H. M. Siddique, and et al. 2022. "Quantitative Trait Loci for Heat Stress Tolerance in Brassica rapa L. Are Distributed across the Genome and Occur in Diverse Genetic Groups, Flowering Phenologies and Morphotypes" Genes 13, no. 2: 296. https://doi.org/10.3390/genes13020296
APA StyleChen, S., Hayward, A., Dey, S. S., Choudhary, M., Witt Hmon, K. P., Inturrisi, F. C., Dolatabadian, A., Neik, T. X., Yang, H., Siddique, K. H. M., Batley, J., & Cowling, W. A. (2022). Quantitative Trait Loci for Heat Stress Tolerance in Brassica rapa L. Are Distributed across the Genome and Occur in Diverse Genetic Groups, Flowering Phenologies and Morphotypes. Genes, 13(2), 296. https://doi.org/10.3390/genes13020296













