Machine Learning Heuristics on Gingivobuccal Cancer Gene Datasets Reveals Key Candidate Attributes for Prognosis
Abstract
1. Introduction
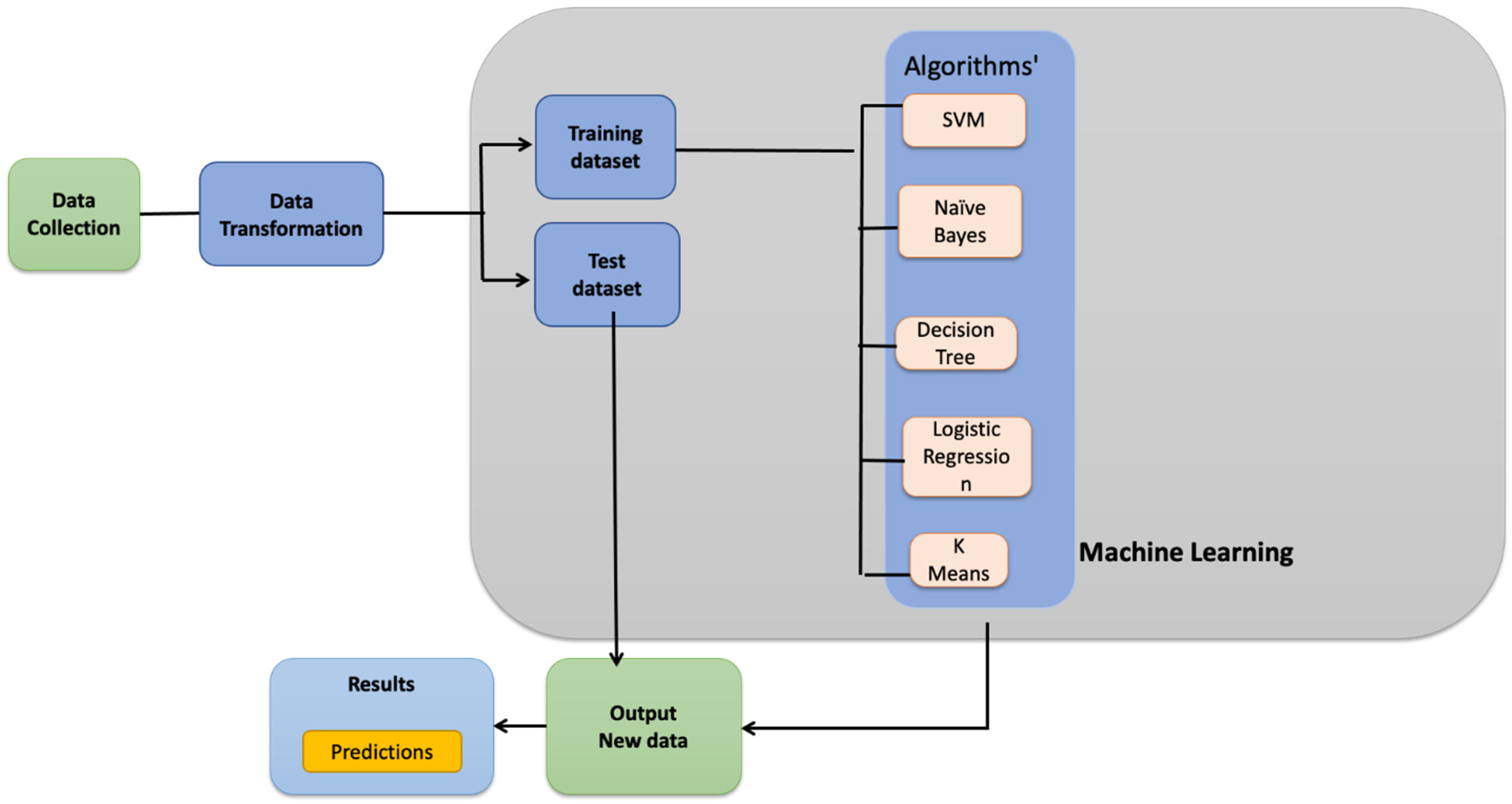
2. Materials and Methods
2.1. Datasets and Transformation
2.2. Experiments
2.3. Classifier Design and Training
2.4. Performance Evaluation
3. Results and Discussion
3.1. PIK3CA among the Select Genes with Highest Accuracy
3.2. Scatter Plot for K-Means
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mandlik, D.S.; Nair, S.S.; Patel, K.D.; Gupta, K.; Patel, P.; Patel, P.; Sharma, N.; Joshipura, A.; Patel, M. Squamous cell carcinoma of gingivobuccal complex: Literature, evidences and practice. J. Head Neck Physicians Surg. 2018, 6, 18–28. [Google Scholar] [CrossRef]
- Rivera, C. Essentials of oral cancer. Int. J. Clin. Exp. Pathol. 2015, 8, 11884–11894. [Google Scholar] [PubMed]
- Ali, J.; Sabiha, B.; Jan, H.U.; Haider, S.A.; Khan, A.A.; Ali, S.S. Genetic etiology of oral cancer. Oral Oncol. 2017, 70, 23–28. [Google Scholar] [CrossRef] [PubMed]
- Clayman, L. Malignant Odontogenic Tumors. In Holland-Frei Cancer Medicine, 6th ed.; Kufe, D.W., Pollock, R.E., Weichselbaum, R.R., Bast, R.C., Jr., Gansler, T.S., Holland, J.F., Frei, E., III, Eds.; BC Decker: Hamilton, ON, USA, 2003. Available online: https://www.ncbi.nlm.nih.gov/books/NBK13124/ (accessed on 30 October 2022).
- Elzay, R.P. Classification of primary intraosseous carcinoma. Oral Surg. Oral Med. Oral Pathol. 1982, 54, 299–303. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, A.T.; Luu, M.; Clair, J.M.-S.; Mita, A.C.; Scher, K.S.; Lu, D.J.; Shiao, S.L.; Ho, A.S.; Zumsteg, Z.S. Comparison of Survival after Transoral Robotic Surgery vs Nonrobotic Surgery in Patients with Early-Stage Oropharyngeal Squamous Cell Carcinoma. JAMA Oncol. 2020, 6, 1555. [Google Scholar] [CrossRef] [PubMed]
- Deshpande, A.M.; Wong, D.T. Molecular mechanisms of head and neck cancer. Expert Rev. Anticancer Ther. 2008, 8, 799–809. [Google Scholar] [CrossRef] [PubMed]
- Mohri, M.; Rostamizadeh, A.; Talwalkar, A. Foundations of Machine Learning; Adaptive Computation and Machine Learning Series; MIT Press: Cambridge, MA, USA, 2012. [Google Scholar]
- Ijaq, J.; Malik, G.; Kumar, A.; Das, P.S.; Meena, N.; Bethi, N.; Sundararajan, V.S.; Suravajhala, P. A model to predict the function of hypothetical proteins through a nine-point classification scoring schema. BMC Bioinform. 2019, 20, 14. [Google Scholar] [CrossRef] [PubMed]
- Malik, G.; Gulati, I.K. Little Motion, Big Results: Using Motion Magnification to Reveal Subtle Tremors in Infants. In Proceedings of the Workshop on Artificial Intelligence for Healthcare in 24th European Conference on Artificial Intelligence, Santiago de Compostela, Spain, 4 September 2020. [Google Scholar]
- Malik, G.; Linsley, D.; Serre, T.; Mingolla, E. The Challenge of Appearance-Free Object Tracking with Feedforward Neural Networks. In Proceedings of the CVPR Workshop on Dynamic Neural Networks Meet Computer Vision, Virtual, 20 June 2021. [Google Scholar]
- Alabi, R.O.; Almangush, A.; Elmusrati, M.; Mäkitie, A.A. Deep Machine Learning for Oral Cancer: From Precise Diagnosis to Precision Medicine. Front. Oral Health 2022, 2, 794248. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, N.; Abbasi, M.S.; Zuberi, F.; Qamar, W.; Halim, M.S.B.; Maqsood, A.; Alam, M.K. Artificial Intelligence Techniques: Analysis, Application, and Outcome in Dentistry—A Systematic Review. BioMed Res. Int. 2021, 2021, 9751564. [Google Scholar] [CrossRef] [PubMed]
- Mahendran, N.; Durai Raj Vincent, P.M.; Srinivasan, K.; Chang, C.-Y. Machine Learning Based Computational Gene Selection Models: A Survey, Performance Evaluation, Open Issues, and Future Research Directions. Front. Genet. 2020, 11, 603808. [Google Scholar] [CrossRef] [PubMed]
- Tseng, Y.; Wang, H.; Lin, T.; Lu, J.; Hsieh, C.; Liao, C. Development of a Machine Learning Model for Survival Risk Stratification of Patients With Advanced Oral Cancer. JAMA Netw. Open 2020, 3, e2011768. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.W.; Lee, S.; Kwon, S.; Nam, W.; Cha, I.-H.; Kim, H.J. Deep learning-based survival prediction of oral cancer patients. Sci. Rep. 2019, 9, 6994. [Google Scholar] [CrossRef] [PubMed]
- Eibe, F.; Hall, M.A.; Witten, I.H. The WEKA Workbench. Online Appendix for “Data Mining: Practical Machine Learning Tools and Techniques”, 4th ed.; Morgan Kaufmann: San Francisco, CA, USA, 2016. [Google Scholar]
- Witten, I.H.; Frank, E. Data Mining: Practical Machine Learning Tools and Techniques with Java Implementations; Morgan Kaufmann: San Francisco, CA, USA, 2000. [Google Scholar]
- Sengupta, N.; Sarode, S.C.; Sarode, G.S.; Ghone, U. Scarcity of publicly available oral cancer image datasets for machine learning research. Oral Oncol. 2022, 126, 105737. [Google Scholar] [CrossRef] [PubMed]
- Ngiam, J.; Khosla, A.; Kim, M.; Nam, J.; Lee, H.; Ng, A.Y. Multimodal deep learning. In Proceedings of the 28th International Conference on Machine Learning, Bellevue, DC, USA, 28 June–2 July 2011; pp. 689–696. [Google Scholar]
- De Guia, J.M.; Devaraj, M.; Leung, C.K. DeepGx: Deep learning using gene expression for cancer classification. In Proceedings of the 2019 IEEE/ACM International Conference on Advances in Social Networks Analysis and Mining, Vancouver, BC, Canada, 27–30 August 2019; pp. 913–920. [Google Scholar]
- Iddamalgoda, L.; Das, P.S.; Aponso, A.; Sundararajan, V.S.; Suravajhala, P.; Valadi, J.K. Data Mining and Pattern Recognition Models for Identifying Inherited Diseases: Challenges and Implications. Front. Genet. 2016, 7, 136. [Google Scholar] [CrossRef] [PubMed][Green Version]
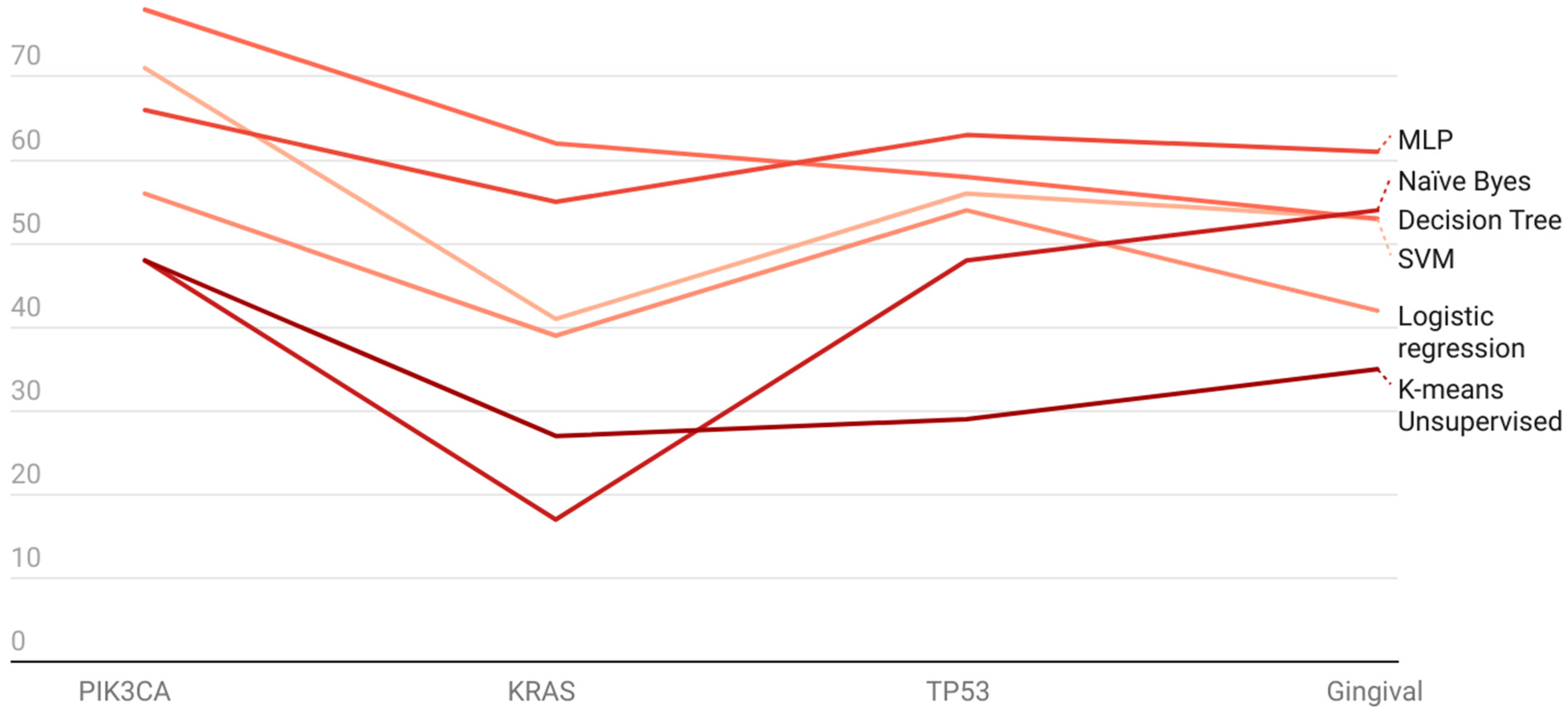

| ML Algorithms [Accuracies %] | |||||||
|---|---|---|---|---|---|---|---|
| SVM | MLP | Logistic Regression | Naïve Byes | Decision Tree | K-Means Unsupervised | ||
| Genes | PIK3CA | 71 | 66 | 56 | 48 | 78 | 48 |
| KRAS | 41 | 55 | 39 | 17 | 62 | 27 | |
| TP53 | 56 | 63 | 54 | 48 | 58 | 29 | |
| Gingival | 53 | 61 | 42 | 54 | 53 | 35 | |
| Genes | SVM | MLP | Logistic Regression | Naïve Bayes | Decision Tree | K-Mean |
|---|---|---|---|---|---|---|
| PIK3CA | 88% | 83% | 76% | 100% | 87% | 21% |
| KRAS | 82% | 70% | 74% | 19% | 92% | 8% |
| TP53 | 51% | 57% | 63% | 89% | 57% | 53% |
| Gingival | 47% | 48% | 53% | 95% | 45% | 55% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Singh, T.; Malik, G.; Someshwar, S.; Le, H.T.T.; Polavarapu, R.; Chavali, L.N.; Melethadathil, N.; Sundararajan, V.S.; Valadi, J.; Kavi Kishor, P.B.; et al. Machine Learning Heuristics on Gingivobuccal Cancer Gene Datasets Reveals Key Candidate Attributes for Prognosis. Genes 2022, 13, 2379. https://doi.org/10.3390/genes13122379
Singh T, Malik G, Someshwar S, Le HTT, Polavarapu R, Chavali LN, Melethadathil N, Sundararajan VS, Valadi J, Kavi Kishor PB, et al. Machine Learning Heuristics on Gingivobuccal Cancer Gene Datasets Reveals Key Candidate Attributes for Prognosis. Genes. 2022; 13(12):2379. https://doi.org/10.3390/genes13122379
Chicago/Turabian StyleSingh, Tanvi, Girik Malik, Saloni Someshwar, Hien Thi Thu Le, Rathnagiri Polavarapu, Laxmi N. Chavali, Nidheesh Melethadathil, Vijayaraghava Seshadri Sundararajan, Jayaraman Valadi, P. B. Kavi Kishor, and et al. 2022. "Machine Learning Heuristics on Gingivobuccal Cancer Gene Datasets Reveals Key Candidate Attributes for Prognosis" Genes 13, no. 12: 2379. https://doi.org/10.3390/genes13122379
APA StyleSingh, T., Malik, G., Someshwar, S., Le, H. T. T., Polavarapu, R., Chavali, L. N., Melethadathil, N., Sundararajan, V. S., Valadi, J., Kavi Kishor, P. B., & Suravajhala, P. (2022). Machine Learning Heuristics on Gingivobuccal Cancer Gene Datasets Reveals Key Candidate Attributes for Prognosis. Genes, 13(12), 2379. https://doi.org/10.3390/genes13122379







