Human Genetic Research in Wallacea and Sahul: Recent Findings and Future Prospects
Abstract
1. Introduction
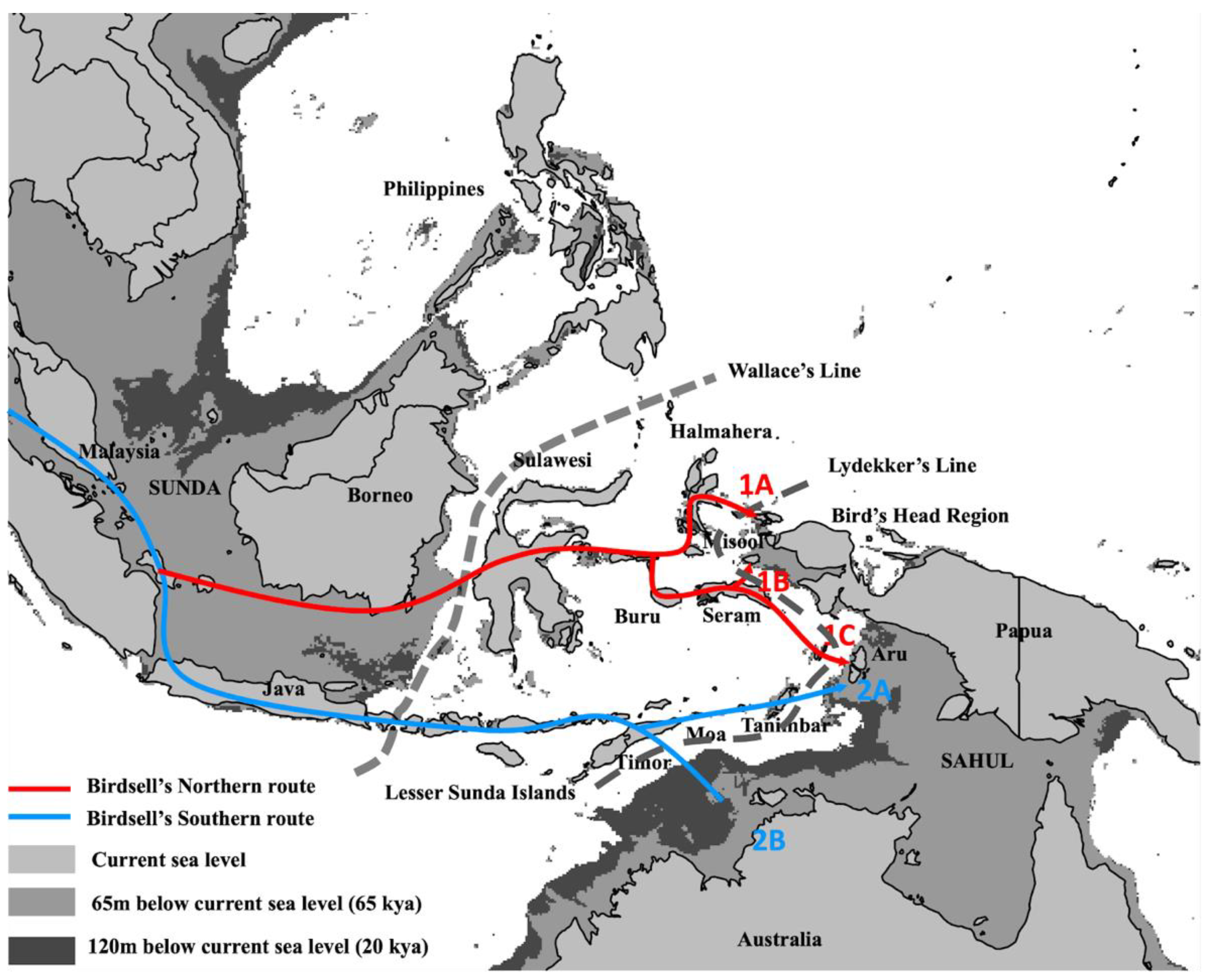
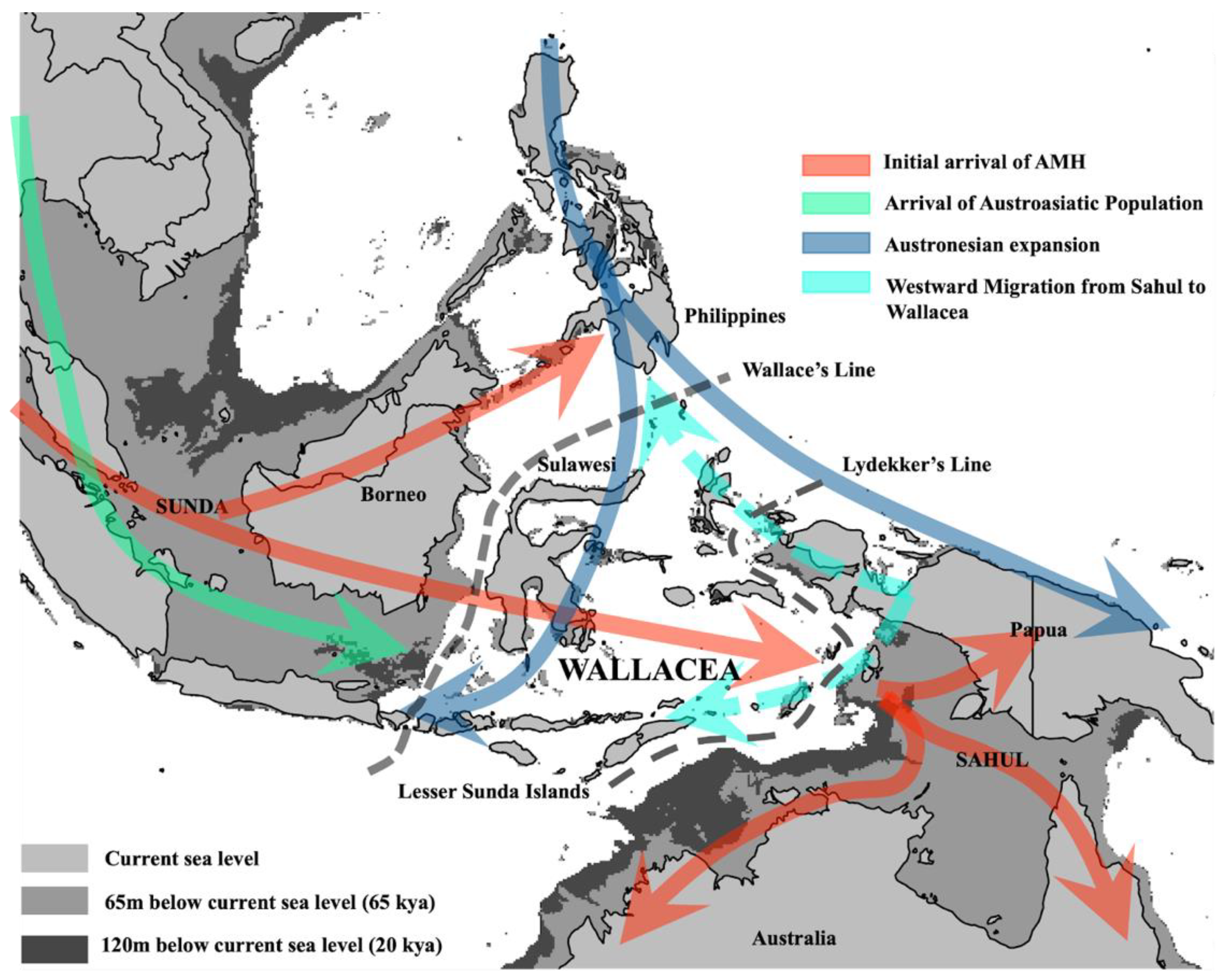
2. Genetic Evidence for AMH Arrival in Wallacea and Sahul
3. Extensive Migrations and Mixing in Holocene Island Southeast Asia (ISEA)
4. Wallacean Paleogenomic Studies
5. Archaic Hominin Introgression
6. Conclusions and Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zaim, Y.; Ciochon, R.L.; Polanski, J.M.; Grine, F.E.; Bettis, E.A., 3rd; Rizal, Y.; Franciscus, R.G.; Larick, R.R.; Heizler, M.; Aswan; et al. New 1.5 Million-Year-Old Homo Erectus Maxilla from Sangiran (Central Java, Indonesia). J. Hum. Evol. 2011, 61, 363–376. [Google Scholar] [CrossRef] [PubMed]
- Rizal, Y.; Westaway, K.E.; Zaim, Y.; van den Bergh, G.D.; Bettis, E.A., 3rd; Morwood, M.J.; Huffman, O.F.; Grün, R.; Joannes-Boyau, R.; Bailey, R.M.; et al. Last Appearance of Homo Erectus at Ngandong, Java, 117,000–108,000 Years Ago. Nature 2020, 577, 381–385. [Google Scholar] [CrossRef]
- Brown, P.; Sutikna, T.; Morwood, M.J.; Soejono, R.P.; Jatmiko; Saptomo, E.W.; Due, R.A. A New Small-Bodied Hominin from the Late Pleistocene of Flores, Indonesia. Nature 2004, 431, 1055–1061. [Google Scholar] [CrossRef] [PubMed]
- Morwood, M.J.; Brown, P.; Jatmiko; Sutikna, T.; Saptomo, E.W.; Westaway, K.E.; Due, R.A.; Roberts, R.G.; Maeda, T.; Wasisto, S.; et al. Further Evidence for Small-Bodied Hominins from the Late Pleistocene of Flores, Indonesia. Nature 2005, 437, 1012–1017. [Google Scholar] [CrossRef] [PubMed]
- Brumm, A.; van den Bergh, G.D.; Storey, M.; Kurniawan, I.; Alloway, B.V.; Setiawan, R.; Setiyabudi, E.; Grün, R.; Moore, M.W.; Yurnaldi, D.; et al. Age and Context of the Oldest Known Hominin Fossils from Flores. Nature 2016, 534, 249–253. [Google Scholar] [CrossRef]
- Détroit, F.; Mijares, A.S.; Corny, J.; Daver, G.; Zanolli, C.; Dizon, E.; Robles, E.; Grün, R.; Piper, P.J. A New Species of Homo from the Late Pleistocene of the Philippines. Nature 2019, 568, 181–186. [Google Scholar] [CrossRef]
- Westaway, K.E.; Louys, J.; Awe, R.D.; Morwood, M.J.; Price, G.J.; Zhao, J.-X.; Aubert, M.; Joannes-Boyau, R.; Smith, T.M.; Skinner, M.M.; et al. An Early Modern Human Presence in Sumatra 73,000–63,000 Years Ago. Nature 2017, 548, 322–325. [Google Scholar] [CrossRef]
- Marwick, B.; Clarkson, C.; O’Connor, S.; Collins, S. Early Modern Human Lithic Technology from Jerimalai, East Timor. J. Hum. Evol. 2016, 101, 45–64. [Google Scholar] [CrossRef]
- O’Connell, J.F.; Allen, J.; Williams, M.A.J.; Williams, A.N.; Turney, C.S.M.; Spooner, N.A.; Kamminga, J.; Brown, G.; Cooper, A. When Did Homo Sapiens First Reach Southeast Asia and Sahul? Proc. Natl. Acad. Sci. USA 2018, 115, 8482–8490. [Google Scholar] [CrossRef]
- Clarkson, C.; Jacobs, Z.; Marwick, B.; Fullagar, R.; Wallis, L.; Smith, M.; Roberts, R.G.; Hayes, E.; Lowe, K.; Carah, X.; et al. Human Occupation of Northern Australia by 65,000 Years Ago. Nature 2017, 547, 306–310. [Google Scholar] [CrossRef]
- Hawkins, S.; O’Connor, S.; Maloney, T.R.; Litster, M.; Kealy, S.; Fenner, J.N.; Aplin, K.; Boulanger, C.; Brockwell, S.; Willan, R.; et al. Oldest Human Occupation of Wallacea at Laili Cave, Timor-Leste, Shows Broad-Spectrum Foraging Responses to Late Pleistocene Environments. Quat. Sci. Rev. 2017, 171, 58–72. [Google Scholar] [CrossRef]
- Norman, K.; Inglis, J.; Clarkson, C.; Faith, J.T.; Shulmeister, J.; Harris, D. An Early Colonisation Pathway into Northwest Australia 70–60,000 Years Ago. Quat. Sci. Rev. 2018, 180, 229–239. [Google Scholar] [CrossRef]
- O’Connor, S.; Ono, R.; Clarkson, C. Pelagic Fishing at 42,000 Years before the Present and the Maritime Skills of Modern Humans. Science 2011, 334, 1117–1121. [Google Scholar] [CrossRef] [PubMed]
- Brumm, A.; Oktaviana, A.A.; Burhan, B.; Hakim, B.; Lebe, R.; Zhao, J.-X.; Sulistyarto, P.H.; Ririmasse, M.; Adhityatama, S.; Sumantri, I.; et al. Oldest Cave Art Found in Sulawesi. Sci. Adv. 2021, 7, eabd4648. [Google Scholar] [CrossRef]
- Norman, K.; Shipton, C.; O’Connor, S.; Malanali, W.; Collins, P.; Wood, R.; Saktura, W.M.; Roberts, R.G.; Jacobs, Z. Human Occupation of the Kimberley Coast of Northwest Australia 50,000 Years Ago. Quat. Sci. Rev. 2022, 288, 107577. [Google Scholar] [CrossRef]
- Kealy, S.; Louys, J.; O’Connor, S. Reconstructing Palaeogeography and Inter-Island Visibility in the Wallacean Archipelago during the Likely Period of Sahul Colonization, 65-45 000 Years Ago: Reconstructing Palaeogeography and Intervisibility in Wallacea. Archaeol. Prospect. 2017, 24, 259–272. [Google Scholar] [CrossRef]
- Kealy, S.; Louys, J.; O’Connor, S. Least-Cost Pathway Models Indicate Northern Human Dispersal from Sunda to Sahul. J. Hum. Evol. 2018, 125, 59–70. [Google Scholar] [CrossRef]
- Bird, M.I.; Beaman, R.J.; Condie, S.A.; Cooper, A.; Ulm, S.; Veth, P. Palaeogeography and Voyage Modeling Indicates Early Human Colonization of Australia Was Likely from Timor-Roti. Quat. Sci. Rev. 2018, 191, 431–439. [Google Scholar] [CrossRef]
- Birdsell, J.B. The Recalibration of a Paradigm for the First Peopling of Greater Australia. Sunda and Sahul: Prehistoric Studies in Southeast Asia, Melanesia, and Australia; Academic Press: London, UK, 1977; pp. 113–167. [Google Scholar]
- Ingman, M.; Gyllensten, U. Mitochondrial Genome Variation and Evolutionary History of Australian and New Guinean Aborigines. Genome Res. 2003, 13, 1600–1606. [Google Scholar] [CrossRef]
- van Holst Pellekaan, S.M.; Ingman, M.; Roberts-Thomson, J.; Harding, R.M. Mitochondrial Genomics Identifies Major Haplogroups in Aboriginal Australians. Am. J. Phys. Anthropol. 2006, 131, 282–294. [Google Scholar] [CrossRef]
- Nagle, N.; van Oven, M.; Wilcox, S.; van Holst Pellekaan, S.; Tyler-Smith, C.; Xue, Y.; Ballantyne, K.N.; Wilcox, L.; Papac, L.; The Genographic Consortium; et al. Aboriginal Australian Mitochondrial Genome Variation—An Increased Understanding of Population Antiquity and Diversity. Sci. Rep. 2017, 7, 43041. [Google Scholar] [CrossRef] [PubMed]
- Tobler, R.; Rohrlach, A.; Soubrier, J.; Bover, P.; Llamas, B.; Tuke, J.; Bean, N.; Abdullah-Highfold, A.; Agius, S.; O’Donoghue, A.; et al. Aboriginal Mitogenomes Reveal 50,000 Years of Regionalism in Australia. Nature 2017, 544, 180–184. [Google Scholar] [CrossRef] [PubMed]
- Duggan, A.T.; Evans, B.; Friedlaender, F.R.; Friedlaender, J.S.; Koki, G.; Andrew Merriwether, D.; Kayser, M.; Stoneking, M. Maternal History of Oceania from Complete mtDNA Genomes: Contrasting Ancient Diversity with Recent Homogenization Due to the Austronesian Expansion. Am. J. Hum. Genet. 2014, 94, 721–733. [Google Scholar] [CrossRef] [PubMed]
- Pedro, N.; Brucato, N.; Fernandes, V.; André, M.; Saag, L.; Pomat, W.; Besse, C.; Boland, A.; Deleuze, J.-F.; Clarkson, C.; et al. Papuan Mitochondrial Genomes and the Settlement of Sahul. J. Hum. Genet. 2020, 65, 875–887. [Google Scholar] [CrossRef]
- Hudjashov, G.; Kivisild, T.; Underhill, P.A.; Endicott, P.; Sanchez, J.J.; Lin, A.A.; Shen, P.; Oefner, P.; Renfrew, C.; Villems, R.; et al. Revealing the Prehistoric Settlement of Australia by Y Chromosome and mtDNA Analysis. Proc. Natl. Acad. Sci. USA 2007, 104, 8726–8730. [Google Scholar] [CrossRef]
- Bergström, A.; Nagle, N.; Chen, Y.; McCarthy, S.; Pollard, M.O.; Ayub, Q.; Wilcox, S.; Wilcox, L.; van Oorschot, R.A.H.; McAllister, P.; et al. Deep Roots for Aboriginal Australian Y Chromosomes. Curr. Biol. 2016, 26, 809–813. [Google Scholar] [CrossRef]
- Bergström, A.; Stringer, C.; Hajdinjak, M.; Scerri, E.M.L.; Skoglund, P. Origins of Modern Human Ancestry. Nature 2021, 590, 229–237. [Google Scholar] [CrossRef]
- Rasmussen, M.; Guo, X.; Wang, Y.; Lohmueller, K.E.; Rasmussen, S.; Albrechtsen, A.; Skotte, L.; Lindgreen, S.; Metspalu, M.; Jombart, T.; et al. An Aboriginal Australian Genome Reveals Separate Human Dispersals into Asia. Science 2011, 334, 94–98. [Google Scholar] [CrossRef]
- Malaspinas, A.-S.; Westaway, M.C.; Muller, C.; Sousa, V.C.; Lao, O.; Alves, I.; Bergström, A.; Athanasiadis, G.; Cheng, J.Y.; Crawford, J.E.; et al. A Genomic History of Aboriginal Australia. Nature 2016, 538, 207–214. [Google Scholar] [CrossRef]
- Mallick, S.; Li, H.; Lipson, M.; Mathieson, I.; Gymrek, M.; Racimo, F.; Zhao, M.; Chennagiri, N.; Nordenfelt, S.; Tandon, A.; et al. The Simons Genome Diversity Project: 300 Genomes from 142 Diverse Populations. Nature 2016, 538, 201–206. [Google Scholar] [CrossRef]
- Reich, D.; Patterson, N.; Kircher, M.; Delfin, F.; Nandineni, M.R.; Pugach, I.; Ko, A.M.-S.; Ko, Y.-C.; Jinam, T.A.; Phipps, M.E.; et al. Denisova Admixture and the First Modern Human Dispersals into Southeast Asia and Oceania. Am. J. Hum. Genet. 2011, 89, 516–528. [Google Scholar] [CrossRef] [PubMed]
- Reich, D.; Green, R.E.; Kircher, M.; Krause, J.; Patterson, N.; Durand, E.Y.; Viola, B.; Briggs, A.W.; Stenzel, U.; Johnson, P.L.F.; et al. Genetic History of an Archaic Hominin Group from Denisova Cave in Siberia. Nature 2010, 468, 1053–1060. [Google Scholar] [CrossRef]
- Meyer, M.; Kircher, M.; Gansauge, M.-T.; Li, H.; Racimo, F.; Mallick, S.; Schraiber, J.G.; Jay, F.; Prüfer, K.; de Filippo, C.; et al. A High-Coverage Genome Sequence from an Archaic Denisovan Individual. Science 2012, 338, 222–226. [Google Scholar] [CrossRef]
- McEvoy, B.P.; Lind, J.M.; Wang, E.T.; Moyzis, R.K.; Visscher, P.M.; van Holst Pellekaan, S.M.; Wilton, A.N. Whole-Genome Genetic Diversity in a Sample of Australians with Deep Aboriginal Ancestry. Am. J. Hum. Genet. 2010, 87, 297–305. [Google Scholar] [CrossRef] [PubMed]
- Pugach, I.; Delfin, F.; Gunnarsdóttir, E.; Kayser, M.; Stoneking, M. Genome-Wide Data Substantiate Holocene Gene Flow from India to Australia. Proc. Natl. Acad. Sci. USA 2013, 110, 1803–1808. [Google Scholar] [CrossRef] [PubMed]
- Wollstein, A.; Lao, O.; Becker, C.; Brauer, S.; Trent, R.J.; Nürnberg, P.; Stoneking, M.; Kayser, M. Demographic History of Oceania Inferred from Genome-Wide Data. Curr. Biol. 2010, 20, 1983–1992. [Google Scholar] [CrossRef] [PubMed]
- Lipson, M.; Loh, P.-R.; Patterson, N.; Moorjani, P.; Ko, Y.-C.; Stoneking, M.; Berger, B.; Reich, D. Reconstructing Austronesian Population History in Island Southeast Asia. Nat. Commun. 2014, 5, 4689. [Google Scholar] [CrossRef]
- Purnomo, G.A.; Mitchell, K.J.; O’Connor, S.; Kealy, S.; Taufik, L.; Schiller, S.; Rohrlach, A.; Cooper, A.; Llamas, B.; Sudoyo, H.; et al. Mitogenomes Reveal Two Major Influxes of Papuan Ancestry across Wallacea Following the Last Glacial Maximum and Austronesian Contact. Genes 2021, 12, 965. [Google Scholar] [CrossRef]
- Karmin, M.; Flores, R.; Saag, L.; Hudjashov, G.; Brucato, N.; Crenna-Darusallam, C.; Larena, M.; Endicott, P.L.; Jakobsson, M.; Lansing, J.S.; et al. Episodes of Diversification and Isolation in Island Southeast Asian and Near Oceanian Male Lineages. Mol. Biol. Evol. 2022, 39, msac045. [Google Scholar] [CrossRef]
- Brucato, N.; André, M.; Tsang, R.; Saag, L.; Kariwiga, J.; Sesuki, K.; Beni, T.; Pomat, W.; Muke, J.; Meyer, V.; et al. Papua New Guinean Genomes Reveal the Complex Settlement of North Sahul. Mol. Biol. Evol. 2021, 38, 5107–5121. [Google Scholar] [CrossRef]
- Nagle, N.; Ballantyne, K.N.; van Oven, M.; Tyler-Smith, C.; Xue, Y.; Wilcox, S.; Wilcox, L.; Turkalov, R.; van Oorschot, R.A.H.; van Holst Pellekaan, S.; et al. Mitochondrial DNA Diversity of Present-Day Aboriginal Australians and Implications for Human Evolution in Oceania. J. Hum. Genet. 2017, 62, 343–353. [Google Scholar] [CrossRef] [PubMed]
- Larena, M.; Sanchez-Quinto, F.; Sjödin, P.; McKenna, J.; Ebeo, C.; Reyes, R.; Casel, O.; Huang, J.-Y.; Hagada, K.P.; Guilay, D.; et al. Multiple Migrations to the Philippines during the Last 50,000 Years. Proc. Natl. Acad. Sci. USA 2021, 118, e2026132118. [Google Scholar] [CrossRef]
- Patterson, N.; Moorjani, P.; Luo, Y.; Mallick, S.; Rohland, N.; Zhan, Y.; Genschoreck, T.; Webster, T.; Reich, D. Ancient Admixture in Human History. Genetics 2012, 192, 1065–1093. [Google Scholar] [CrossRef] [PubMed]
- Hudjashov, G.; Karafet, T.M.; Lawson, D.J.; Downey, S.; Savina, O.; Sudoyo, H.; Lansing, J.S.; Hammer, M.F.; Cox, M.P. Complex Patterns of Admixture across the Indonesian Archipelago. Mol. Biol. Evol. 2017, 34, 2439–2452. [Google Scholar] [CrossRef]
- Higuchi, R.; Bowman, B.; Freiberger, M.; Ryder, O.A.; Wilson, A.C. DNA Sequences from the Quagga, an Extinct Member of the Horse Family. Nature 1984, 312, 282–284. [Google Scholar] [CrossRef] [PubMed]
- Olalde, I.; Posth, C. Latest Trends in Archaeogenetic Research of West Eurasians. Curr. Opin. Genet. Dev. 2020, 62, 36–43. [Google Scholar] [CrossRef]
- McColl, H.; Racimo, F.; Vinner, L.; Demeter, F.; Gakuhari, T.; Moreno-Mayar, J.V.; van Driem, G.; Gram Wilken, U.; Seguin-Orlando, A.; de la Fuente Castro, C.; et al. The Prehistoric Peopling of Southeast Asia. Science 2018, 361, 88–92. [Google Scholar] [CrossRef]
- Heupink, T.H.; Subramanian, S.; Wright, J.L.; Endicott, P.; Westaway, M.C.; Huynen, L.; Parson, W.; Millar, C.D.; Willerslev, E.; Lambert, D.M. Ancient mtDNA Sequences from the First Australians Revisited. Proc. Natl. Acad. Sci. USA 2016, 113, 6892–6897. [Google Scholar] [CrossRef]
- Posth, C.; Nägele, K.; Colleran, H.; Valentin, F.; Bedford, S.; Kami, K.W.; Shing, R.; Buckley, H.; Kinaston, R.; Walworth, M.; et al. Language Continuity despite Population Replacement in Remote Oceania. Nat. Ecol. Evol. 2018, 2, 731–740. [Google Scholar] [CrossRef]
- Lipson, M.; Skoglund, P.; Spriggs, M.; Valentin, F.; Bedford, S.; Shing, R.; Buckley, H.; Phillip, I.; Ward, G.K.; Mallick, S.; et al. Population Turnover in Remote Oceania Shortly after Initial Settlement. Curr. Biol. 2018, 28, 1157–1165.e7. [Google Scholar] [CrossRef]
- Wright, J.L.; Wasef, S.; Heupink, T.H.; Westaway, M.C.; Rasmussen, S.; Pardoe, C.; Fourmile, G.G.; Young, M.; Johnson, T.; Slade, J.; et al. Ancient Nuclear Genomes Enable Repatriation of Indigenous Human Remains. Sci. Adv. 2018, 4, eaau5064. [Google Scholar] [CrossRef] [PubMed]
- Carlhoff, S.; Duli, A.; Nägele, K.; Nur, M.; Skov, L.; Sumantri, I.; Oktaviana, A.A.; Hakim, B.; Burhan, B.; Syahdar, F.A.; et al. Genome of a Middle Holocene Hunter-Gatherer from Wallacea. Nature 2021, 596, 543–547. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, S.; Nägele, K.; Carlhoff, S.; Pugach, I.; Koesbardiati, T.; Hübner, A.; Meyer, M.; Oktaviana, A.A.; Takenaka, M.; Katagiri, C.; et al. Ancient Genomes from the Last Three Millennia Support Multiple Human Dispersals into Wallacea. Nat. Ecol. Evol. 2022, 6, 1024–1034. [Google Scholar] [CrossRef] [PubMed]
- Green, R.E.; Krause, J.; Briggs, A.W.; Maricic, T.; Stenzel, U.; Kircher, M.; Patterson, N.; Li, H.; Zhai, W.; Fritz, M.H.-Y.; et al. A Draft Sequence of the Neandertal Genome. Science 2010, 328, 710–722. [Google Scholar] [CrossRef]
- Prüfer, K.; Racimo, F.; Patterson, N.; Jay, F.; Sankararaman, S.; Sawyer, S.; Heinze, A.; Renaud, G.; Sudmant, P.H.; de Filippo, C.; et al. The Complete Genome Sequence of a Neanderthal from the Altai Mountains. Nature 2014, 505, 43–49. [Google Scholar] [CrossRef]
- Browning, S.R.; Browning, B.L.; Zhou, Y.; Tucci, S.; Akey, J.M. Analysis of Human Sequence Data Reveals Two Pulses of Archaic Denisovan Admixture. Cell 2018, 173, 53–61.e9. [Google Scholar] [CrossRef]
- Mondal, M.; Bertranpetit, J.; Lao, O. Approximate Bayesian Computation with Deep Learning Supports a Third Archaic Introgression in Asia and Oceania. Nat. Commun. 2019, 10, 246. [Google Scholar] [CrossRef]
- Vernot, B.; Tucci, S.; Kelso, J.; Schraiber, J.G.; Wolf, A.B.; Gittelman, R.M.; Dannemann, M.; Grote, S.; McCoy, R.C.; Norton, H.; et al. Excavating Neandertal and Denisovan DNA from the Genomes of Melanesian Individuals. Science 2016, 352, 235–239. [Google Scholar] [CrossRef]
- Teixeira, J.C.; Cooper, A. Using Hominin Introgression to Trace Modern Human Dispersals. Proc. Natl. Acad. Sci. USA 2019, 116, 15327–15332. [Google Scholar] [CrossRef]
- Larena, M.; McKenna, J.; Sanchez-Quinto, F.; Bernhardsson, C.; Ebeo, C.; Reyes, R.; Casel, O.; Huang, J.-Y.; Hagada, K.P.; Guilay, D.; et al. Philippine Ayta Possess the Highest Level of Denisovan Ancestry in the World. Curr. Biol. 2021, 31, 4219–4230.e10. [Google Scholar] [CrossRef]
- Teixeira, J.C.; Jacobs, G.S.; Stringer, C.; Tuke, J.; Hudjashov, G.; Purnomo, G.A.; Sudoyo, H.; Cox, M.P.; Tobler, R.; Turney, C.S.M.; et al. Widespread Denisovan Ancestry in Island Southeast Asia but No Evidence of Substantial Super-Archaic Hominin Admixture. Nat. Ecol. Evol. 2021, 5, 616–624. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, G.S.; Hudjashov, G.; Saag, L.; Kusuma, P.; Darusallam, C.C.; Lawson, D.J.; Mondal, M.; Pagani, L.; Ricaut, F.-X.; Stoneking, M.; et al. Multiple Deeply Divergent Denisovan Ancestries in Papuans. Cell 2019, 177, 1010–1021.e32. [Google Scholar] [CrossRef] [PubMed]
- Jinam, T.A.; Phipps, M.E.; Aghakhanian, F.; Majumder, P.P.; Datar, F.; Stoneking, M.; Sawai, H.; Nishida, N.; Tokunaga, K.; Kawamura, S.; et al. Discerning the Origins of the Negritos, First Sundaland People: Deep Divergence and Archaic Admixture. Genome Biol. Evol. 2017, 9, 2013–2022. [Google Scholar] [CrossRef] [PubMed]
- van den Bergh, G.D.; Li, B.; Brumm, A.; Grün, R.; Yurnaldi, D.; Moore, M.W.; Kurniawan, I.; Setiawan, R.; Aziz, F.; Roberts, R.G.; et al. Earliest Hominin Occupation of Sulawesi, Indonesia. Nature 2016, 529, 208–211. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Taufik, L.; Teixeira, J.C.; Llamas, B.; Sudoyo, H.; Tobler, R.; Purnomo, G.A. Human Genetic Research in Wallacea and Sahul: Recent Findings and Future Prospects. Genes 2022, 13, 2373. https://doi.org/10.3390/genes13122373
Taufik L, Teixeira JC, Llamas B, Sudoyo H, Tobler R, Purnomo GA. Human Genetic Research in Wallacea and Sahul: Recent Findings and Future Prospects. Genes. 2022; 13(12):2373. https://doi.org/10.3390/genes13122373
Chicago/Turabian StyleTaufik, Leonard, João C. Teixeira, Bastien Llamas, Herawati Sudoyo, Raymond Tobler, and Gludhug A. Purnomo. 2022. "Human Genetic Research in Wallacea and Sahul: Recent Findings and Future Prospects" Genes 13, no. 12: 2373. https://doi.org/10.3390/genes13122373
APA StyleTaufik, L., Teixeira, J. C., Llamas, B., Sudoyo, H., Tobler, R., & Purnomo, G. A. (2022). Human Genetic Research in Wallacea and Sahul: Recent Findings and Future Prospects. Genes, 13(12), 2373. https://doi.org/10.3390/genes13122373







