Abstract
Background: Chemoresistance is a significant barrier to combating head and neck cancer, and decoding this resistance can widen the therapeutic application of such chemotherapeutic drugs. This systematic review and meta-analysis explores the influence of microRNA (miRNA) expressions on chemoresistance in head and neck cancers (HNC). The objective is to evaluate the theragnostic effects of microRNA expressions on chemoresistance in HNC patients and investigate the utility of miRNAs as biomarkers and avenues for new therapeutic targets. Methods: We performed a comprehensive bibliographic search that included the SCOPUS, PubMed, and Science Direct bibliographic databases. These searches conformed to a predefined set of search strategies. Following the PRISMA guidelines, inclusion and exclusion criteria were framed upon completing the literature search. The data items extracted were tabulated and collated in MS Excel. This spreadsheet was used to determine the effect size estimation for the theragnostic effects of miRNA expressions on chemoresistance in HNC, the hazard ratio (HR), and 95% confidence intervals (95% CI). The comprehensive meta-analysis was performed using the random effects model. Heterogeneity among the data collected was assessed using the Q test, Tau2, I2, and Z measures. Publication bias of the included studies was checked using the Egger’s bias indicator test, Orwin and classic fail-safe N test, Begg and Mazumdar rank collection test, and Duval and Tweedie’s trim and fill methods. Results: After collating the data from 23 studies, dysregulation of 34 miRNAs was observed in 2189 people. These data were gathered from 23 studies. Out of the 34 miRNAs considered, 22 were up-regulated, while 12 were down-regulated. The TaqMan transcription kits were the most used miRNA profiling platform, and miR-200c was seen to have a mixed dysregulation. We measured the overall pooled effect estimate of HR to be 1.516 for the various analyzed miRNA at a 95% confidence interval of 1.303–1.765, with a significant p-value. The null hypothesis test’s Z value was 5.377, and the p-value was correspondingly noted to be less than 0.0001. This outcome indicates that the risk of death is determined to be higher in up-regulated groups than in down-regulated groups. Among the 34 miRNAs that were investigated, seven miRNAs were associated with an improved prognosis, especially with the overexpression of these seven miRNAs (miR15b-5p, miR-548b, miR-519d, miR-1278, miR-145, miR-200c, Hsa- miR139-3p). Discussion: The findings reveal that intricate relationships between miRNAs’ expression and chemotherapeutic resistance in HNC are more likely to exist and can be potential therapeutic targets. This review suggests the involvement of specific miRNAs as predictors of chemoresistance and sensitivity in HNC. The examination of the current study results illustrates the significance of miRNA expression as a theragnostic biomarker in medical oncology.
1. Introduction
1.1. Background
Head and neck cancer (HNC) is the sixth most common type of cancer [1]. The epithelial linings of the upper aero-digestive tract, including the oral cavity, oropharynx, hypopharynx, and larynx, are generally affected. HNC affects around 650,000 patients worldwide and accounts for more than 330,000 deaths annually [2]. Human papillomavirus (HPV)-induced oropharyngeal cancers are on the rise and are predominantly seen in young cohorts who are non-smokers and non-alcoholics [3,4,5,6]. The most common causes seem to be alcohol, smoking, and the high risk HPV variants [7,8]. This association is particularly the case for Type 16 (also known as HPV-16) and also occurs with Epstein-Barr viruses [9], which arise from the crypt epithelium of the palatine and lingual tonsils [10]. The standard form of treatment for this form of cancer includes radiotherapy, chemotherapy, or concurrent chemo/radiotherapy [9,11]. Chemotherapeutic drugs such as docetaxel, paclitaxel, and cisplatin treat HNCs [12].
1.2. Epidemiology
Worldwide, head and neck carcinoma contributes to more than 650,000 cases and 330,000 deaths every year and ranks as the sixth most common cancer globally [2,13]. In the United States, head and neck carcinomas represent about three percent of malignancies, with roughly 53,000 Americans developing HNC yearly and 10,800 deaths due to this disease. In Europe, there were approximately 250,000 cases (an expected four percent of the disease frequency) and 63,500 deaths in 2012. Males have an increased propensity to this disease with a male–female ratio ranging from 2:1 to 4:1.
The frequency rate in males surpasses 20 per 100,000 in France, Hong Kong, the Indian subcontinent, Central and Eastern Europe, Spain, Italy, Brazil, and African Americans in the United States. Oral cancers are more prevalent in India, and oropharyngeal cancers are more common in the western population [14,15]. The mortality of both laryngeal and oropharyngeal carcinoma is higher in African American men, reflecting the lower prevalence of human papillomavirus (HPV) positivity [16]. The chronic exposure of the upper aero-digestive tract to cancer-causing components such as tobacco use, alcohol consumption, and HPV can bring about dysplastic or premalignant sores/lesions in the oropharyngeal mucosa, which ultimately results in head and neck carcinoma [7,8,17,18,19,20]. The relative frequency of these risk factors adds to the variations in the observed distribution of HNC in various world zones [7].
Studies have shown that the dysregulation of miRNA plays a critical role in cancer progression and contributes to chemotherapeutic resistance or chemoresistance. Preclinical and clinical observational studies have revealed that miRNA expression profiling could improve the classification of high-risk patients with cancer who may develop chemoresistance [21]. The miRNA does so by targeting specific genes/pathways and inhibiting or accelerating those genes’ expression. For example, miR 200-b, miR 155, and miR 146-a, miR 422-a affect the multidrug resistance gene-1 (MDR-1) [22] and causes resistance to CDDP-CRTX and MMC-CRTX. Another study by Bonnin et al. [23] showed that multiple genes and pathways were affected, such as FOXG1, CD73/NT5E oncogene, adenosine receptor-dependent signaling overexpressing CD73.
1.3. Rationale
The data on the correlations between HNC chemoresistance/sensitivity and miRNA expression have currently not yielded clinically relevant solutions in the form of theragnostic biomarkers, regardless of the ongoing research in this field. Most of the publications on miRNA-specific chemoresistance of HNC are quite appropriate to the effects of particular miRNA [24,25,26,27,28,29]. The published studies were categorized into samples collected in a hospital for a specific region. After a detailed evaluation of the published literature globally, this systematic review was proffered. This systematic review and meta-analysis provides qualitative and quantitative data on miRNA and methodically assesses the pattern of specific chemoresistance in HNC. This clinical research team previously highlighted a systematic review and meta-analysis approach that permits us to collect the data across all published studies and possibly focus on the associated miRNAs, which have clinical relevance in decisions regarding chemotherapy in patients [21,30,31].
This systematic review and meta-analysis aims to assist researchers and clinicians by quantifying miRNA alterations associated with the chemotherapeutic response in HNC. Forthcoming studies can then identify their utility as predictors of chemotherapy response or theragnosis. This study collates the data on miRNAs and how their regulation can influence the sensitivity of chemotherapeutic drugs.
Objectives
The primary objective is to qualitatively analyze the theragnostic effects of miRNA expressions in HNC patients across the world. The secondary objective of this proposed study is to evaluate the up- and down-regulation of miRNAs and evaluate the pooled estimated effect size on the prognosis of HNC patients and resistance in cell lines that may cause recurrence.
2. Search Strategy and Methods
The study was conducted by the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) statement [32] and was completed following a previously established protocol (PROSPERO registration number: CRD42018104657). The study protocol was already published elsewhere [33].
2.1. Review Questions
What effect does miRNA regulation have on chemotherapy?
What is the general prognosis of patients having miRNA-specific chemoresistance?
What are the miRNAs most responsible for chemoresistance in HNC patients?
What are the survival rates associated with each miRNA linked to chemoresistance, and how are they affected?
2.2. Study Design
Search Strategy
The PubMed and Science Direct databases were searched for publications published between 2008 and 2021. The Medical Subjective Heading (MeSH) search phrases were used in the search (Table 1). There were no limits on study participants regarding age, gender, ethnicity, country of origin, and morbidities (for patients and the general population). Four authors of this study (RJ, MRM, PS, and MR) independently assessed the titles and abstracts to see if the publications satisfied the inclusion criteria. In accordance with the protocol:

Table 1.
Key terms utilized in the search strategy.
The selected full-text papers were checked for studies that did not include abstracts.
The reference lists of the collected studies were manually searched to improve the robustness of the search results.
The cross-references from the selected studies were searched for additional articles.
When the relevant information was not available in the publication, we contacted the corresponding authors.
Any discrepancies were resolved through discussion and consensus with a third reviewer.
2.3. Selection Criteria
2.3.1. Inclusion Criteria
Studies analyzing the theragnostic effects of miRNA expressions in both HNC patients and cell lines were considered.
Studies analyzing miRNAs and chemoresistance/are performed in liquid biopsies (of plasma and saliva samples) were included.
Studies that discussed HNC patients’ clinicopathological characteristics and the hazard ratio (HR) or Kaplan–Meier curve were included.
Articles that discussed the survival outcomes of almost all stages of HNC patients were included in the meta-analysis.
Studies reporting miRNA profiling platform and miRNA expressions analysis using in vitro assays were included.
Genes and pathways involved in chemoresistance or sensitivity in HNC patients were also considered.
Studies appropriate to PRISMA guidelines for systematic review and meta-analysis were included.
2.3.2. Exclusion Criteria
Studies published in languages other than English were excluded.
Any information or results from letters to the editors, case studies, conference abstracts, case reports, and review articles of HNC were removed.
Studies performed only in patients or in vitro were excluded and were not considered for the systematic review.
Studies lacking proper discussion about miRNA profiling and pathways related to that were excluded.
Studies with no accessibility to survival outcomes, HR, or Kaplan–Meier (KM) curves were not considered for the meta-analysis.
Studies whose full texts were not accessible were excluded.
Duplicates were removed, and the study was excluded if it fell within the exclusion criteria.
2.4. Data Extraction and Management
All studies that satisfied the selection criteria were assessed, and all clinical and histological parameters for patients were extracted. Author names, year of publication, study location, study period, gender, sample size, source of a clinical sample, miRNAs profiling platform, follow-up period, miRNAs studied, histological type, lymph node metastasis/distant metastasis, clinical stages, and survival data were all sorted under the following headings: author names, year of publication, study location, study period, gender, sample size, source of a clinical sample, miRNAs profiling platform. The data from the studies that qualified for final inclusion were tabulated using a Microsoft Excel spreadsheet.
2.5. Assessment of Quality
The study quality of the literature extracted for the systematic review and meta-analyses were assessed using the Meta-Analysis of Observational Studies in Epidemiology (MOOSE) checklist. Studies that satisfied 0–33% of the 14 items on the checklist were considered to be of “poor” quality, with “satisfactory” study quality indicating an adherence of 33–66% of the study to the checklist, while “good” quality studies were in the range of 67–100% adherence. All studies included fell within either satisfactory or good study quality. The items specified in the MOOSE checklist are delineated in Table 2.

Table 2.
The Meta-Analysis of Observational Studies in Epidemiology (MOOSE) checklist.
2.6. Publication Bias
Publications bias indicators of the included studies were assessed using Orwin and classic fail-safe N test [34], Egger’s bias indicator test, Begg and Mazumdar Rank collection test, Duval and Tweedie’s trim fill calculation [35,36], and inverted funnel plot.
2.7. Comprehensive Meta-Analysis
Comprehensive meta-analysis was performed to estimate the pooled estimated effect size HR and 95% CI from the included studies using comprehensive meta-analysis (CMA) software version 3.0. Random effects models were used for meta-analysis. Cochran’s Q test, Tau square, Z value, and I2 statistic [37,38] were performed to assess the heterogeneity and hypothesis testing of the included studies. The random effects model was performed when the p-value > 0.05, and heterogeneity was observed. A forest plot was drawn to summarize the pooled HR estimate of the chemoresistance-specific miRNAs.
3. Results
3.1. Study Selection:
The selected and eligible studies for this systematic review and comprehensive meta-analysis through search results are shown in the flow chart in Figure 1. Studies were searched using critical terms, as seen in the protocol paper. Upward of 4610 records appeared upon merely searching MeSH keywords. After searching the duplicate records and records marked as ineligible, we had 459 papers. After shortlisting by scanning the titles and abstracts for relevant papers, we had 113 articles. After shortlisting the papers that did not match the selection criteria, we had 34 studies, out of which 23 were used for the meta-analysis as they had the required data. All these studies underwent quality assessment using the MOOSE checklist and were of acceptable quality for inclusion in a meta-analysis study.
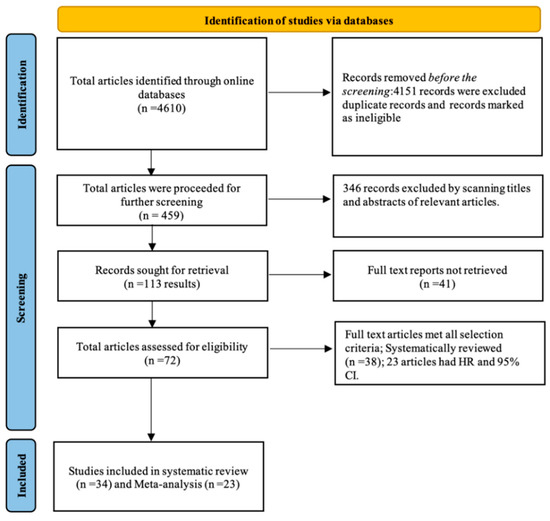
Figure 1.
Flow chart of the literature search.
3.2. Study Characteristics
The results for the various parameters analyzed in this systematic review are shown in Table 3. As seen in the table, most of the studies were from China, Germany, and Japan. Thirty-four miRNAs were analyzed from a patient population of 2189 people. Tissue and serum samples were the most used in the studies analyzed in this review. In the studies analyzed, cisplatin was the most commonly used chemotherapeutic drug for which chemoresistance was observed. Out of the total 34 miRNAs, 22 miRNAs were up-regulated, while 12 miRNAs were down-regulated. The TaqMan transcription kit was the most commonly used of the miRNA profiling platforms. miRNA 200c was seen to have a mixed dysregulation; it was seen to be up-regulated in a study by Hamano R [39] and was seen to be down-regulated in a study by Song J [40].

Table 3.
Description of the 23 included studies.
3.2.1. In Vitro Assays
This section illustrates the commonly used in vitro assays collected from the studies represented by the following Figure 2 and Figure 3 below. In the 24 studies utilized in this review, 19 cell lines were used, OECM-1. Of these, the SAS cell lines were the most commonly used (Figure 3). Of the data collected from all the studies, the highest number of cell lines used in a single study was eight. Among the data collected, we also analyzed certain in vitro studies used in the collected studies. The most commonly used assays included qRT-PCR, cell proliferation assay, MTT assay for cell viability and cytotoxicity, luciferase reporter assays, Western blotting, apoptosis assay, clonogenic assay, scramble assay, immunoprecipitation as well as immunohistochemistry assays, RFLP assay, electrophoretic mobility shift assay, chemosensitivity, chromatin immunoprecipitation (ChIP) assay among others. Figure 2 summarizes how frequently the most common assays were used in the studies considered.
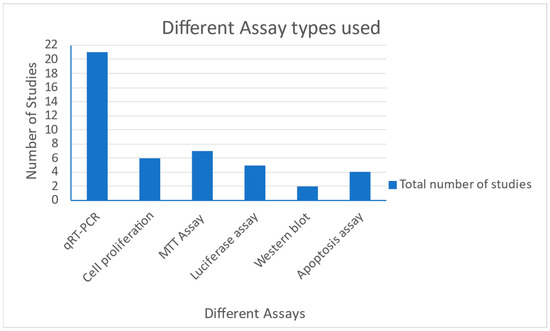
Figure 2.
Chart showing the various assays performed in the collected studies.
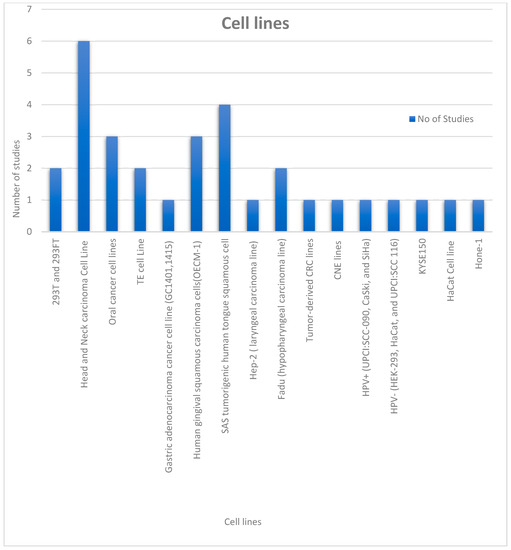
Figure 3.
Chart showing the various cell lines utilized in the collected studies.
3.2.2. Relation between miRNA Expression and Chemoresistance
The following data were obtained from the collected data analysis of the results. Out of the 34 miRNAs investigated in the study, the seven miRNAs (miR15b-5p, miR-548b, miR-519d, miR-1278, miR-145, miR-200c, Hsa- miR139-3p) were linked to better survival, while the rest of the 27 miRNAs were associated with poor survival. The following nine miRNAs were known to affect chemoresistance in HNC miR-200, miR-34a, miR-196a, miR-27a, miR-27b, miR-200c, miR-494, miR-1290, and miR-205. These mentioned miRNAs are up-regulated, except miR-34a, which is down-regulated. The following three miRNAs, miR-519d, miR-1278, and miR-29c, are known to inhibit chemoresistance and are noted to be down-regulated. The most commonly used chemotherapy drug among the nine drugs is cisplatin. Overall, 13 miRNAs were associated with regulating chemoresistance to chemotherapy drugs, as well as certain miRNAs such as miR-1290, which are known to affect the commonly used chemoradiotherapy (CRT).
3.2.3. Chemotherapy and HNC Patients
There were a total of nine drugs used as chemotherapy in the pooled studies: cisplatin (866 patients), 5-fluorouracil (646 patients), doxorubicin (260 patients), paclitaxel (151 patients), cetuximab (152 patients), oxaliplatin (33 patients), leucovorin (33 patients), silibinin (45 patients), and docetaxel (97 patients).
3.2.4. Drug Regulatory Pathways for miRNA-Mediated Chemosensitivity and Chemoresistance
Figure 4 below represents the various pathways affected in HNC and comprehensively illustrates the deregulation of miRNA. From the articles included in the study, 11 pathways and their associated genes were investigated and elaborated on in individual studies. Four pathways were described as associated with cell survival. In contrast, two pathways were related to apoptosis, four pathways were analyzed to be linked with cell differentiation and proliferation, while one was involved in angiogenesis.
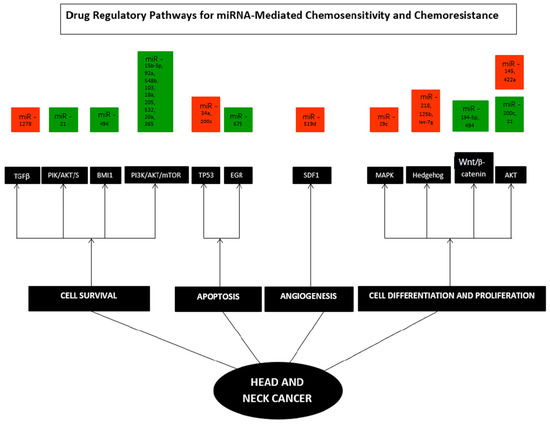
Figure 4.
Hallmarks of specific miRNAs in Head and Neck Cancer. TGFβ, PIK/AKT/S6, BMI1, PI3K/AKT/mTOR, TP53, EGR, SDF1, MAPK, Hedgehog, Wnt/β- catenin, and AKT pathways. Each hallmark depicts several examples of miRNAs that control specific cellular processes in HNC; some microRNAs influence multiple hallmarks, implying that they govern various pathways. Orange—down-regulated miRNA; Green—up-regulated miRNA.
3.2.5. Association between miRNAs and Drug Regulatory Pathways of Chemoresistance
From the studies analyzed, it was noted that many miRNAs were up-regulated when chemoresistance was observed compared to the number of down-regulated miRNA. Chemoresistance to cetuximab was marked by up-regulation of nine miRNA, namely, miR-15b-5p, miR-92a, miR-548b, miR-103, miR-18a, miR-205, miR-532, miR-20a, and miR-365. Of these, miR 15b-5p affected p16, EGFR, and CD44; miR-15b-5p/TRIM-29/PTEN/AKT/mTOR signaling pathways; and others affected the PI3K/Akt/mTOR pathways. miR-218, Let-7g, and miR-125b were down-regulated during M4N treatment, and the Hedgehog and Wnt signaling cascades, SP1, MYC, and TP53 genes were seen to be affected. Up-regulation of miR-200b in CDDP-CRTX chemoresistance and up-regulation of miR-155 along with miR-146 in MMC-CRTX chemoresistance were all seen to affect the multidrug resistance gene-1. Chemoresistance towards silibinin, doxorubicin or cisplatin, or fluorouracil was marked by up-regulation of miRNA-494, which affected ZEB2 and β-catenin signaling, ADAM10, FOXM1, CD44, and ALDH1 pathways, and also Bmi1/ADAM10 pathways (Table 4).

Table 4.
The genetic pathways involved in chemoresistance.
miRNA 200c was seen to have mixed dysregulation, up-regulation of miR-200c was seen in the chemoresistance towards cisplatin, and the Akt pathway, MDR-1 gene, and PPP2R1B gene pathway were seen to be affected, while in another study, down-regulation of miR-200c was noted to affect the TP53 pathway.
3.3. Comprehensive Meta-Analysis
We analyzed the dysregulation of 34 miRNAs seen in 2189 HNC patients. These data were gathered from 23 studies. This analysis revealed that out of 34 miRNAs, 22 were up-regulated, while 12 were down-regulated. miRNA 200c was observed to be up-regulated in the Akt pathway of patients undergoing cisplatin treatment. In contrast, the same miRNA was seen to be down-regulated in the TP53 pathway in a different study. The overall pooled effect estimate for the various miRNAs was 1.516 with a 95% confidence interval of 1.303–1.765. The HR and the 95% CI for the studies included in this paper utilizing the fixed effect model are 1.302 (1.222–1.386). Moreover, the adjusted point estimate and the 95% CI values with the fixed model as reference are 1.263 (1.192–1.349). The HR and 95% CI values utilizing the random effects model are 1.516 (1.303–1.765). The point estimate and the 95% CI values with the random effects model are 1.336 (1.151–1.552). Table 5 depicts the publication bias indicators hypothesis testing and heterogeneity testing analysis of miRNA-specific chemoresistance in HNC.

Table 5.
Publication bias indicators and hypothesis testing and heterogeneity testing analysis of miRNA-specific chemoresistance in HNC.
3.4. Influence of miRNA Expression on the Survival of HNC Patients
In the results mentioned above, the Z value from the null hypothesis test was 5.377, and the p-value was correspondingly noted to be less than 0.0001. This result indicates that the risk of death is determined to be higher in up-regulated groups than in down-regulated groups. Among the 34 miRNAs that were investigated, 7 miRNA (miR15b-5p, miR-548b, miR-519d, miR-1278, miR-145, miR-200c, Hsa- miR139-3p) were associated with an improved prognosis (better survival). The other 27 miRNAs (miR-29c, miR-626, miR-5100, miR-21, miR-200b, miR-365, hsa-miR-194-5p, miR-200c(a), miR-200c(b), miR-532, miR-20a, miR-21, miR-155, miR-27a, miR-21, miR-494, miR-146a, miR-196a, miR 675(a), miR-149, miR-205, miR-18a, miR-103, miR-422a, miR-675(b), Let-7g(a), Let-7g(b), miR-1290(a), miR-1290(b), miR-92(a), miR 27b, miR-218) were associated with a poor prognosis (poor survival).
3.5. Extent of Variance of Estimated Effect Size across Included Studies
This study applied the Q-Statistics test, which assumes that all studies used in the analysis have the same impact size. With 39 degrees of freedom (df) and a noted p-value of less than 0.0001, the calculated Q value was 118.561. We cannot reject the null hypothesis as the true effect size was similar in all the included studies. In addition, the observed variation falls within the range assigned to the sampling error. The I2 statistic referred to the extent of observed variance that can effectively illustrate the differences in effect size instead of sampling error. I2 is 67.106% in this study. T2 (T = tau) (in log units) effectively denotes the variance of accurate effect sizes. In this study, the T2 value is 0.098. T stands for the standard deviation of actual effects (in log units). In this study, the T-value is 0.314.
3.6. Publication Bias and Sensitivity Analysis—Funnel Plot
Publication bias as an indicator is used because many studies that are complete are not published due to the outcome of the study, wherein the results may not be significant. A funnel plot was developed (Figure 5), which was asymmetric across survival outcomes. The asymmetry represents the presence of publication bias. The vertical axis represented the study size’s standard error and precision, and the horizontal axis represented the effect size. The dots represent individual studies, and one can appreciate that most of the studies are in the high significance region. This indicates the presence of publication bias.
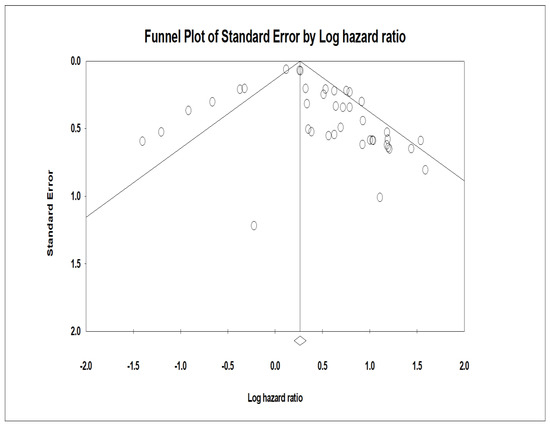
Figure 5.
Funnel plot of the studies included in this comprehensive meta-analysis of miRNA.. Funnel plot with inputted studies (studies included in this comprehensive meta-analysis of miRNA-specific chemoresistance in head and neck cancer). The black circles indicate imputed studies.
3.7. Orwin’s Fail-Safe N Test
In the studies in this meta-analysis review, the hazard ratio (HR) values were measured to be 1.30155. The mean hazard ratio cited in the results was 1.000 (generally can consist of any value other than a nil value). In this review, the HR observed to be 1.30155 is not placed between the mean HR of the missing studies, which is 1.000 [34].
3.8. Begg and Mazumdar Rank Correlation Test
This test is generally performed to correlate Kendall’s Rank with the standardized effects sizes and standard errors. The yield of a positive value in this test is indicative of a high degree of accuracy of the included studies in this meta-analysis. Kendall’s Tau (rank-order correlation) values were found to be 0.15897 (without continuity correction) and 0.15679 (with continuity correction). Subsequently, the p-tailed values for 1-tailed and 2-tailed were established as 0.07592 and 0.15184, respectively.
In Figure 6, CMA software was used to calculate and analyze the HR values’ pooled hazard ratios for HNC prognostic data. The meta-analysis was conducted by analyzing 23 studies involving 34 miRNAs from a combined patient pool of 2189 HNC patients. The analysis yielded a Z value of 8.63 with a p-value of less than 0.001.
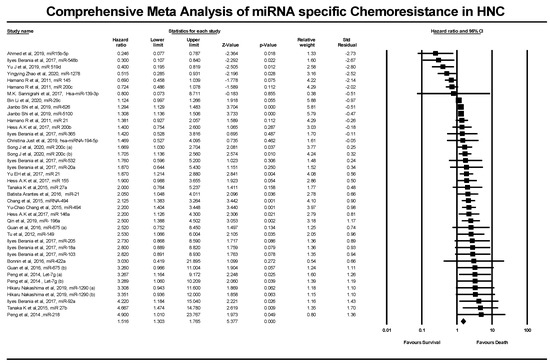
Figure 6.
Forest plot of the studies included in this comprehensive meta-analysis of miRNA-specific chemoresistance in head and neck cancer.
3.9. Egger’s Test of Intercept
This study yielded an intercept of 0.89103 at 95% CI (0.14381–1.63825), t-value = 2.41400, and 38 degrees of freedom. The p-value generated for the one-tailed test was 0.01305, and the p-value for the two-tailed test was 0.0207.
3.10. Duval and Tweedie’s Trim and Fill Test
This test is instrumental in the study as it helps diminish the effect of publication bias. This test is generally performed when the funnel plot observed is asymmetrical [35]. The studies that contribute to the asymmetry are trimmed from the right side of the funnel plot to pinpoint the unbiased effect. This is then filled back by re-inserting the trimmed studies on the right and the imputed studies on the left side of the mean effect. In this review, approximately ten studies that produced asymmetry in the plot were trimmed and filled. This funnel plot was created using CMA software (Englewood, NJ 07631 USA) and illustrates the trimmed and imputed studies.
4. Discussion
Head and neck cancers (HNC) are plagued by the inherent chemoresistance towards the most commonly used drugs in HNC, such as cisplatin and cetuximab. This drug resistance tends to lead to rapid deterioration of the long-term prognosis of the patient [25,57]. This study aims to evaluate the potential role of miRNAs, which are one type among several types of small non-coding RNAs known to play a specific role in cancer progression, including the development of chemoresistance [25,33]. The regulation of chemoresistance specific miRNA is significant in the genesis of cancer as well as in the prognosis of the affected patient. miRNAs are also known to play a significant role in apoptosis, DNA repair, and epithelial–mesenchymal regulation in the cell cycle. The previous meta-analysis on HNC illustrated the role of miRNAs in targeting patient survival [25]. Dai et al. performed an earlier descriptive review on HNC that investigated the role of miRNAs in targeting drug regulatory receptors; however, the authors of this study highlighted only three miRNAs related to chemoresistance in HNC [58]. Hence a systematic review that included a comprehensive meta-analysis of thirty-four miRNAs that impact chemoresistance to drugs in HNC was needed. This systematic review was performed using 459 articles obtained through MeSH PubMed key search terms, among which 34 publications were included for a systematic review, and 23 articles were included for a comprehensive meta-analysis based on selection criteria.
The pathological parameters were evaluated and analyzed to effectively correlate and understand the risk factors that may affect or aggravate the disease progression. The hazard ratio values and the 95% CI values were also collected and tabulated to create forest plots that illustrate the role of each miRNA influencing the patients’ prognosis. These miRNAs showed chemoresistance to malignant cells by silencing or inactivating pathways that promote chemoresistance directly or indirectly. For instance, in a study conducted by Martz et al. [59], activation of certain pathways such as Notch-1, phosphoinositide 3-kinase (PI3K), and mammalian target of rapamycin (mTOR), PI3K/AKT, and estrogen receptor (ER) signaling pathways tend to induce chemoresistance to various drugs used for treatment.
4.1. Strengths of the Study
Global literature-based meta-analysis: The studies collected for this systematic review and meta-analysis are abreast with the recent global literature. The impact of certain miRNA on treatment regimens for different HNC patients was looked at from studies collected worldwide. Best research practice in the HNC field: This study adheres to apposite research practice and statistical guidelines. The study’s findings were reported according to the PRISMA guidelines and were registered in PROSPERO.
Clinical recommendation for future studies: This review provides a template for future studies exploring the clinical utility of miRNA.
Methodologically sound analysis: Most of the studies included in this review were of acceptable quality, and the application of quality evaluation tools proved the study’s methodological quality.
Publication bias indicators: A detailed evaluation of publication bias indicators is a fundamental parameter of meta-analysis, which aids any biases in reporting original literature-based meta-analyses of previously published studies. In addition, as per the PRISMA guidelines, an additional investigation of publication bias indicators for small and missing studies was recommended.
First comprehensive meta-analysis study: The authors identify that this is one of the first systematic reviews and meta-analyses on chemoresistance-specific miRNAs in HNC patients.
4.2. Limitations of this Study
Despite the retrospective data collated globally, a significant proportion of the included studies arose primarily from China, Canada, Japan, and Germany, limiting the widespread applicability of the studies. In some studies, HR and the 95% confidence interval data were not directly provided and had to be extracted from the Kaplan–Meier Curves, leading to estimation errors. Each study used varying analysis procedures, such as different techniques and sample sources. This leads to inherent heterogeneity between the studies and could contribute to bias.
5. Conclusions
This comprehensive review and meta-analysis offer conclusive evidence on the role of the miRNAs that affect the survival of patients by affecting the chemoresistance and the disease progression in patients. The regulation of these miRNA is crucial in terms of prognosis and survival. Using forest plots and other statistical methods, we conclusively cement our findings that certain miRNA may negatively affect the patient’s survival leading to a poor prognosis. Future longitudinal research with patient-based meta-analysis is essential to demonstrate the specific miRNAs that may be intricately involved in chemoresistance in HNC.
Author Contributions
Conceptualisation: R.J.; methodology, R.J.; resources, R.J., K.P., M.K., G.K.M., S.S., S.B., R.R.M., V.P., A.K.J., A.K.B. and C.P.; writing—original draft preparation, all authors; writing—review and editing, all authors; visualisation, R.J. and S.K.; project administration, R.J. All authors have read and agreed to the published version of the manuscript.
Funding
Funded in part by the National Institutes of Health (NIH)/National Institute of Dental and Craniofacial Research (NIDCR) R01DE028105 grant to S.K.
Institutional Review Board Statement
This systemic review paper is based on a dataset comprised of anonymized and preexisting data compiled from numerous previously published articles. As a result, no official ethical approval for human research (HREC) is required for this comprehensive meta-analysis study.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Poddar, A.; Aranha, R.; Royam, M.M.; Gothandam, K.M.; Nachimuthu, R.; Jayaraj, R. Incidence, prevalence, and mortality associated with head and neck cancer in India: Protocol for a systematic review. Indian J. Cancer 2019, 56, 101. [Google Scholar]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394. [Google Scholar] [CrossRef] [PubMed]
- Jayaraj, R.; Kumarasamy, C.; Royam, M.M.; Sabarimurugan, S.; Baxi, S. Prognostic implications of pathologic lymph nodes in HPV-positive oropharyngeal cancers: Clinical validity and strategies for routine clinical practice. Oral Oncol. 2019, 92, 99–100. [Google Scholar] [CrossRef]
- Jayaraj, R.; Kumarasamy, C.; Madhav, M.R.; Shetty, S. Clinical validation of the Salivary HPV DNA assessment and its link to the locoregional disease burden in advanced HPV associated oropharyngeal cancer. Oral Oncol. 2019, 97, 149–150. [Google Scholar] [CrossRef] [PubMed]
- Jayaraj, R.; Kumarasamy, C. Comment on, “Survival for HPV-positive oropharyngeal squamous cell carcinoma with surgical versus non-surgical treatment approach: A systematic review and meta-analysis”. Oral Oncol. 2019, 90, 137–138. [Google Scholar] [CrossRef] [PubMed]
- Jayaraj, R.; Kumarasamy, C. Comment on “Increased risk of second cancers at sites associated with HPV after a prior HPV-associated malignancy, a systematic review and meta-analysis”. Br. J. Cancer 2019, 120, 954–955. [Google Scholar] [CrossRef]
- Poddar, A.; Aranha, R.R.; Muthukaliannan, G.K.; Nachimuthu, R.; Jayaraj, R. Head and neck cancer risk factors in India: Protocol for systematic review and meta-analysis. BMJ Open 2018, 8, e020014. [Google Scholar] [CrossRef]
- Singh, J.; Ramamoorthi, R.; Baxi, S.; Thomas, M. The risk factors of head and neck cancer and their general patterns in Australia: A descriptive review and update. J. Environ. Pathol. Toxicol. Oncol. 2014, 33, 45–57. [Google Scholar] [CrossRef]
- Ahmad, P.; Sana, J.; Slávik, M.; Gurín, D.; Radová, L.; Gablo, N.A.; Kazda, T.; Smilek, P.; Horáková, Z.; Gal, B.; et al. MicroRNA-15b-5p predicts locoregional relapse in head and neck carcinoma patients treated with intensity-modulated radiotherapy. Cancer Genom. Proteom. 2019, 16, 139–146. [Google Scholar] [CrossRef] [PubMed]
- Boscolo-Rizzo, P.; Zorzi, M.; Del Mistro, A.; Da Mosto, M.C.; Tirelli, G.; Buzzoni, C.; Rugge, M.; Polesel, J.; Guzzinati, S.; Group, A.W. The evolution of the epidemiological landscape of head and neck cancer in Italy: Is there evidence for an increase in the incidence of potentially HPV-related carcinomas? PLoS ONE 2018, 13, e0192621. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.-C.; Jan, C.-I.; Peng, C.-Y.; Lai, Y.-C.; Hu, F.-W.; Yu, C.-C. Activation of microRNA-494-targeting Bmi1 and ADAM10 by silibinin ablates cancer stemness and predicts favourable prognostic value in head and neck squamous cell carcinomas. Oncotarget 2015, 6, 24002. [Google Scholar] [CrossRef] [PubMed]
- Seiwert, T.Y.; Salama, J.K.; Vokes, E.E. The chemoradiation paradigm in head and neck cancer. Nat. Clin. Pract. Oncol. 2007, 4, 156–171. [Google Scholar] [CrossRef] [PubMed]
- Jayaraj, R.; Singh, J.; Baxi, S.; Ramamoorthi, R.; Thomas, M. Trends in incidence of head and neck cancer in the northern territory, Australia, between 2007 and 2010. Asian Pac. J. Cancer Prev. 2014, 15, 7753–7756. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bray, F.; Ren, J.S.; Masuyer, E.; Ferlay, J. Global estimates of cancer prevalence for 27 sites in the adult population in 2008. Int. J. Cancer 2013, 132, 1133–1145. [Google Scholar] [CrossRef]
- Lambert, R.; Sauvaget, C.; de Camargo Cancela, M.; Sankaranarayanan, R. Epidemiology of cancer from the oral cavity and oropharynx. Eur. J. Gastroenterol. Hepatol. 2011, 23, 633–641. [Google Scholar] [CrossRef]
- Hashim, D.; Genden, E.; Posner, M.; Hashibe, M.; Boffetta, P. Head and neck cancer prevention: From primary prevention to impact of clinicians on reducing burden. Ann. Oncol. 2019, 30, 744–756. [Google Scholar] [CrossRef]
- Jayaraj, R.; Shetty, S.S.; Kumaraswamy, C.; Gothandam, K.; Ram, M.R.; Shaw, P. Clinical interpretation of findings from a systematic review and a comprehensive meta-analysis on clinicopathological and prognostic characteristics of oral squamous cell carcinomas (OSCC) arising in patients with oral lichen planus (OLP). Oral Oncol. 2020, 108, 104974. [Google Scholar] [CrossRef]
- Jayaraj, R.; Shetty, S.; Kumaraswamy, C.; Raymond, G.; Govind, S.K.; Shaw, P. Clinical validity and conceptual interpretation of systematic review and meta-analysis on elective neck dissection (END) versus observation for early-stage oral squamous cell carcinoma (OSCC). Oral Oncol. 2020, 109, 104764. [Google Scholar] [CrossRef]
- Jayaraj, R.; Shetty, S.; Kumaraswamy, C.; Raymond, G.; Govind, S.K.; Chandramoorthy, H.C.; Shaw, P. Clinical validity of clinicopathological and prognostic significance of circulating tumor cells in head and neck squamous cell carcinoma. Oral Oncol. 2020, 109, 104727. [Google Scholar] [CrossRef]
- Jayaraj, R.; Shetty, S.; Kumaraswamy, C.; Raghul, S.; Gothandam, K.M.; Raymond, G.; Ravishankar, R.M.; Shaw, P. Clinical comments on prognostic and clinicopathological significance of PD-L1 overexpression in oral squamous cell carcinoma (OSCC). Oral Oncol. 2020, 104886. [Google Scholar] [CrossRef]
- Royam, M.M.; Kumarasamy, C.; Baxi, S.; Gupta, A.; Ramesh, N.; Muthukaliannan, G.K.; Jayaraj, R. Current evidence on miRNAs as potential theranostic markers for detecting chemoresistance in colorectal cancer: A systematic review and meta-analysis of preclinical and clinical studies. Mol. Diagn. Ther. 2019, 23, 65–82. [Google Scholar] [CrossRef]
- Hess, A.-K.; Müer, A.; Mairinger, F.D.; Weichert, W.; Stenzinger, A.; Hummel, M.; Budach, V.; Tinhofer, I. MiR-200b and miR-155 as predictive biomarkers for the efficacy of chemoradiation in locally advanced head and neck squamous cell carcinoma. Eur. J. Cancer 2017, 77, 3–12. [Google Scholar] [CrossRef] [PubMed]
- Bonnin, N.; Armandy, E.; Carras, J.; Ferrandon, S.; Battiston-Montagne, P.; Aubry, M.; Guihard, S.; Meyronet, D.; Foy, J.-P.; Saintigny, P.; et al. MiR-422a promotes loco-regional recurrence by targeting NT5E/CD73 in head and neck squamous cell carcinoma. Oncotarget 2016, 7, 44023. [Google Scholar] [CrossRef] [PubMed]
- Jayaraj, R.; Kumaraswamy, C.; Raymond, G.; Ram, M.R.; Govind, S.K.; Chandramoorthy, H.C.; Shaw, P. Diagnostic implications of miRNAs in Liquid Biopsy for Oral Squamous Cell Carcinoma (OSCC): Clinical validity and interpretation. Oral Oncol. 2020, 109, 104634. [Google Scholar] [CrossRef]
- Kumarasamy, C.; Madhav, M.R.; Sabarimurugan, S.; Krishnan, S.; Baxi, S.; Gupta, A.; Gothandam, K.; Jayaraj, R. Prognostic value of miRNAs in head and neck cancers: A comprehensive systematic and meta-analysis. Cells 2019, 8, 772. [Google Scholar] [CrossRef] [PubMed]
- Jayaraj, R.; Kumarasamy, C.; Madhav, M.R.; Pandey, V.; Sabarimurugan, S.; Ramesh, N.; Gothandam, K.; Baxi, S. Comment on “systematic review and meta-analysis of diagnostic accuracy of miRNAs in patients with pancreatic cancer”. Dis. Markers 2018, 2018, 6904569. [Google Scholar] [CrossRef]
- Sabarimurugan, S.; Madurantakam Royam, M.; Das, A.; Das, S.; KM, G.; Jayaraj, R. Systematic review and meta-analysis of the prognostic significance of miRNAs in melanoma patients. Mol. Diagn. Ther. 2018, 22, 653–669. [Google Scholar] [CrossRef]
- Jayaraj, R.; Kumarasamy, C.; Sabarimurugan, S.; Baxi, S. Commentary: Blood-derived microRNAs for pancreatic cancer diagnosis: A narrative review and meta-analysis. Front. Physiol. 2019, 9, 1896. [Google Scholar] [CrossRef]
- Jayaraj, R.; Kumarasamy, C.; Sabarimurugan, S.; Baxi, S. Diagnostic and prognostic value of microRNAs for cancers-strategies and approaches to improve the clinical utility. J Cancer 2019, 10, 1252–1253. [Google Scholar] [CrossRef]
- Jayaraj, R.; Raymond, G.; Krishnan, S.; Tzou, K.S.; Baxi, S.; Ram, M.R.; Govind, S.K.; Chandramoorthy, H.C.; Abu-Khzam, F.N.; Shaw, P. Clinical theragnostic potential of diverse miRNA expressions in prostate cancer: A systematic review and meta-analysis. Cancers 2020, 12, 1199. [Google Scholar] [CrossRef]
- Madurantakam Royam, M.; Ramesh, R.; Shanker, R.; Sabarimurugan, S.; Kumarasamy, C.; Ramesh, N.; Gothandam, K.M.; Baxi, S.; Gupta, A.; Krishnan, S.; et al. Mirna predictors of pancreatic cancer chemotherapeutic response: A systematic review and meta-analysis. Cancers 2019, 11, 900. [Google Scholar] [CrossRef]
- Item Page, C. PRISMA-P (Preferred Reporting Items for Systematic review and Meta-Analysis Protocols) 2015 checklist: Recommended items to address in a systematic review protocol. Methods 2015, 5, 6. [Google Scholar]
- Shaw, P.; Raymond, G.; Senthilnathan, R.; Kumarasamy, C.; Baxi, S.; Suresh, D.; Shetty, S.; Ram M, R.; Chandramoorthy, H.C.; Sivanandy, P.; et al. Clinical Theragnostic Relationship between Chemotherapeutic Resistance, and Sensitivity and miRNA Expressions in Head and Neck Cancers: A Systematic Review and Meta-Analysis Protocol. Genes 2021, 12, 2029. [Google Scholar] [CrossRef]
- Orwin, R.G. A fail-safe N for effect size in meta-analysis. J. Educ. Stat. 1983, 8, 157–159. [Google Scholar] [CrossRef]
- Duval, S.; Tweedie, R. A nonparametric “trim and fill” method of accounting for publication bias in meta-analysis. J. Am. Stat. Assoc. 2000, 95, 89–98. [Google Scholar]
- Egger, M.; Smith, G.D.; Schneider, M.; Minder, C. Bias in meta-analysis detected by a simple, graphical test. Bmj 1997, 315, 629–634. [Google Scholar] [CrossRef]
- Cochran, W.G. The combination of estimates from different experiments. Biometrics 1954, 10, 101–129. [Google Scholar] [CrossRef]
- Higgins, J.P.; Thompson, S.G.; Deeks, J.J.; Altman, D.G. Measuring inconsistency in meta-analyses. Bmj 2003, 327, 557–560. [Google Scholar] [CrossRef] [PubMed]
- Hamano, R.; Miyata, H.; Yamasaki, M.; Kurokawa, Y.; Hara, J.; ho Moon, J.; Nakajima, K.; Takiguchi, S.; Fujiwara, Y.; Mori, M.; et al. Overexpression of miR-200c induces chemoresistance in esophageal cancers mediated through activation of the Akt signaling pathway. Clin. Cancer Res. 2011, 17, 3029–3038. [Google Scholar] [CrossRef]
- Song, J.; Zhang, N.; Cao, L.; Xiao, D.; Ye, X.; Luo, E.; Zhang, Z. Down-regulation of miR-200c associates with poor prognosis of oral squamous cell carcinoma. Int. J. Clin. Oncol. 2020, 25, 1072–1078. [Google Scholar] [CrossRef]
- Ogawa, T.; Saiki, Y.; Shiga, K.; Chen, N.; Fukushige, S.; Sunamura, M.; Nagase, H.; Hashimoto, S.; Matsuura, K.; Saijo, S.; et al. mi R-34a is downregulated in cis-diamminedichloroplatinum treated sinonasal squamous cell carcinoma patients with poor prognosis. Cancer Sci. 2012, 103, 1737–1743. [Google Scholar] [CrossRef]
- Yu, E.-H.; Tu, H.-F.; Wu, C.-H.; Yang, C.-C.; Chang, K.-W. MicroRNA-21 promotes perineural invasion and impacts survival in patients with oral carcinoma. J. Chin. Med. Assoc. 2017, 80, 383–388. [Google Scholar] [CrossRef]
- Qin, X.; Guo, H.; Wang, X.; Zhu, X.; Yan, M.; Wang, X.; Xu, Q.; Shi, J.; Lu, E.; Chen, W.; et al. Exosomal miR-196a derived from cancer-associated fibroblasts confers cisplatin resistance in head and neck cancer through targeting CDKN1B and ING5. Genome Biol. 2019, 20, 12. [Google Scholar] [CrossRef]
- Tanaka, K.; Miyata, H.; Sugimura, K.; Fukuda, S.; Kanemura, T.; Yamashita, K.; Miyazaki, Y.; Takahashi, T.; Kurokawa, Y.; Yamasaki, M.; et al. miR-27 is associated with chemoresistance in esophageal cancer through transformation of normal fibroblasts to cancer-associated fibroblasts. Carcinogenesis 2015, 36, 894–903. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Li, Y.; Wang, X.; Yang, Y. Dysregulation of MiR-519d affects oral squamous cell carcinoma invasion and metastasis by targeting MMP3. J. Cancer 2019, 10, 2720. [Google Scholar] [CrossRef]
- Just, C.; Knief, J.; Lazar-Karsten, P.; Petrova, E.; Hummel, R.; Röcken, C.; Wellner, U.; Thorns, C. MicroRNAs as potential biomarkers for chemoresistance in adenocarcinomas of the esophagogastric junction. J. Oncol. 2019, 2019, 4903152. [Google Scholar] [CrossRef] [PubMed]
- Arantes, L.M.R.B.; Laus, A.C.; Melendez, M.E.; de Carvalho, A.C.; Sorroche, B.P.; De Marchi, P.R.M.; Evangelista, A.F.; Scapulatempo-Neto, C.; de Souza Viana, L.; Carvalho, A.L. MiR-21 as prognostic biomarker in head and neck squamous cell carcinoma patients undergoing an organ preservation protocol. Oncotarget 2017, 8, 9911. [Google Scholar] [CrossRef]
- Guan, G.-F.; Zhang, D.-J.; Wen, L.-J.; Xin, D.; Liu, Y.; Yu, D.-J.; Su, K.; Zhu, L.; Guo, Y.-Y.; Wang, K. Overexpression of lncRNA H19/miR-675 promotes tumorigenesis in head and neck squamous cell carcinoma. Int. J. Med. Sci. 2016, 13, 914. [Google Scholar] [CrossRef] [PubMed]
- Tu, H.-F.; Liu, C.-J.; Chang, C.-L.; Wang, P.-W.; Kao, S.-Y.; Yang, C.-C.; Yu, E.-H.; Lin, S.-C.; Chang, K.-W. The association between genetic polymorphism and the processing efficiency of miR-149 affects the prognosis of patients with head and neck squamous cell carcinoma. PLoS ONE 2012, 7, e51606. [Google Scholar] [CrossRef]
- Shi, J.; Bao, X.; Liu, Z.; Zhang, Z.; Chen, W.; Xu, Q. Serum miR-626 and miR-5100 are promising prognosis predictors for oral squamous cell carcinoma. Theranostics 2019, 9, 920. [Google Scholar] [CrossRef]
- Peng, S.-C.; Liao, C.-T.; Peng, C.-H.; Cheng, A.-J.; Chen, S.-J.; Huang, C.-G.; Hsieh, W.-P.; Yen, T.-C. MicroRNAs MiR-218, MiR-125b, and Let-7g predict prognosis in patients with oral cavity squamous cell carcinoma. PLoS ONE 2014, 9, e102403. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, H.; Yoshida, R.; Hirosue, A.; Kawahara, K.; Sakata, J.; Arita, H.; Yamamoto, T.; Toya, R.; Murakami, R.; Hiraki, A.; et al. Circulating miRNA-1290 as a potential biomarker for response to chemoradiotherapy and prognosis of patients with advanced oral squamous cell carcinoma: A single-center retrospective study. Tumor Biol. 2019, 41, 1010428319826853. [Google Scholar] [CrossRef] [PubMed]
- Berania, I.; Cardin, G.B.; Clément, I.; Guertin, L.; Ayad, T.; Bissada, E.; Nguyen-Tan, P.F.; Filion, E.; Guilmette, J.; Gologan, O.; et al. Four PTEN-targeting co-expressed mi RNA s and ACTN 4-targeting mi R-548b are independent prognostic biomarkers in human squamous cell carcinoma of the oral tongue. Int. J. Cancer 2017, 141, 2318–2328. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Wang, P.; Wu, Q. miR-1278 sensitizes nasopharyngeal carcinoma cells to cisplatin and suppresses autophagy via targeting ATG2B. Mol. Cell. Probes 2020, 53, 101597. [Google Scholar] [CrossRef] [PubMed]
- Sannigrahi, M.K.; Sharma, R.; Singh, V.; Panda, N.K.; Rattan, V.; Khullar, M. Role of host miRNA hsa-mir-139-3p in hpv-16–induced carcinomas. Clin. Cancer Res. 2017, 23, 3884–3895. [Google Scholar] [CrossRef]
- Li, B.; Hong, P.; Zheng, C.-C.; Dai, W.; Chen, W.-Y.; Yang, Q.-S.; Han, L.; Tsao, S.W.; Chan, K.T.; Lee, N.P.Y.; et al. Identification of miR-29c and its target FBXO31 as a key regulatory mechanism in esophageal cancer chemoresistance: Functional validation and clinical significance. Theranostics 2019, 9, 1599. [Google Scholar] [CrossRef] [PubMed]
- Kumarasamy, C.; Devi, A.; Jayaraj, R. Prognostic value of microRNAs in head and neck cancers: A systematic review and meta-analysis protocol. Syst. Rev. 2018, 7, 150. [Google Scholar] [CrossRef]
- Dai, F.; Dai, L.; Zheng, X.; Guo, Y.; Zhang, Y.; Niu, M.; Lu, Y.; Li, H.; Hou, R.; Zhang, Y.; et al. Non-coding RNAs in drug resistance of head and neck cancers: A review. Biomed. Pharmacother. 2020, 127, 110231. [Google Scholar] [CrossRef]
- Magee, P.; Shi, L.; Garofalo, M. Role of microRNAs in chemoresistance. Ann. Transl. Med. 2015, 3, 332. [Google Scholar] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).