Transcriptomic Profiling Unravels Novel Deregulated Gene Signatures Associated with Acute Myocardial Infarction: A Bioinformatics Approach
Abstract
1. Introduction
2. Materials and Methods
2.1. Microarray Datasets
2.2. Screening of Differentially Expressed Genes (DEGs)
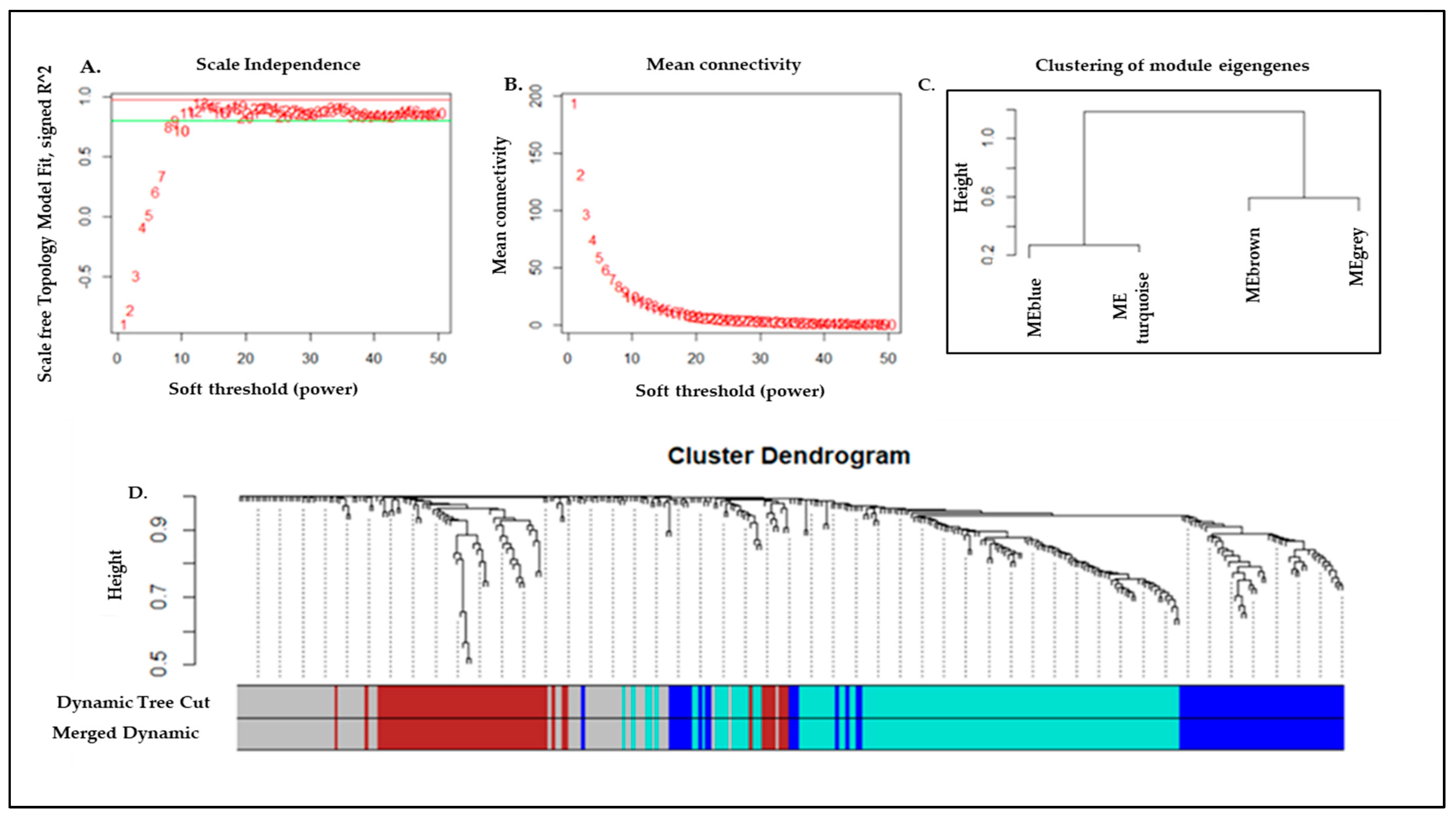
2.3. Weighted Gene Co-Expression Network Analysis (WGCNA) and Module Identification
2.4. Pathway Enrichment Analysis of Turquoise Module
2.5. Enrichment of TFs and microRNAs in Turquoise Module
3. Results
3.1. Identification of Densely Interconnected Genes
3.2. Key Pathway Identification Using Functional Analysis for Turquoise Module
3.3. Network of Genes and miRNAs of Turquoise Module
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cardiovascular Diseases. Available online: https://www.who.int/westernpacific/health-topics/cardiovascular-diseases (accessed on 6 March 2022).
- Mechanic, O.J.; Gavin, M.; Grossman, S.A. Acute Myocardial Infarction. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2022. [Google Scholar]
- Bajaj, A.; Sethi, A.; Rathor, P.; Suppogu, N.; Sethi, A. Acute Complications of Myocardial Infarction in the Current Era: Diagnosis and Management. J. Investig. Med. 2015, 63, 844–855. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Su, X.; Li, G.; Chen, J.; Tang, B.; Yang, Y. The Incidence of Acute Myocardial Infarction in Relation to Overweight and Obesity: A Meta-Analysis. Arch. Med. Sci. 2014, 10, 855–862. [Google Scholar] [CrossRef] [PubMed]
- Tavakol, M.; Ashraf, S.; Brener, S.J. Risks and Complications of Coronary Angiography: A Comprehensive Review. Glob. J. Health Sci. 2012, 4, 65–93. [Google Scholar] [CrossRef] [PubMed]
- Gorenoi, V.; Schönermark, M.P.; Hagen, A. CT Coronary Angiography vs. Invasive Coronary Angiography in CHD. GMS Health Technol. Assess. 2012, 8, Doc02. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.-Y.; Lu, P.-C.; Chen, P.-C. Utilization Trends in Traditional Chinese Medicine for Acute Myocardial Infarction. J. Ethnopharmacol. 2019, 241, 112010. [Google Scholar] [CrossRef]
- Oldgren, J.; Wernroth, L.; Stenestrand, U. RIKS-HIA registry, Sweden. Fibrinolytic Therapy and Bleeding Complications: Risk Predictors from RIKS-HIA. Heart 2010, 96, 1451–1457. [Google Scholar] [CrossRef]
- Aversano, T.; Aversano, L.T.; Passamani, E.; Knatterud, G.L.; Terrin, M.L.; Williams, D.O.; Forman, S.A. Atlantic Cardiovascular Patient Outcomes Research Team (C-PORT). Thrombolytic Therapy vs. Primary Percutaneous Coronary Intervention for Myocardial Infarction in Patients Presenting to Hospitals without On-Site Cardiac Surgery: A Randomized Controlled Trial. JAMA 2002, 287, 1943–1951. [Google Scholar] [CrossRef]
- Kiliszek, M.; Burzynska, B.; Michalak, M.; Gora, M.; Winkler, A.; Maciejak, A.; Leszczynska, A.; Gajda, E.; Kochanowski, J.; Opolski, G. Altered Gene Expression Pattern in Peripheral Blood Mononuclear Cells in Patients with Acute Myocardial Infarction. PLoS ONE 2012, 7, e50054. [Google Scholar] [CrossRef]
- Vanhaverbeke, M.; Vausort, M.; Veltman, D.; Zhang, L.; Wu, M.; Laenen, G.; Gillijns, H.; Moreau, Y.; Bartunek, J.; Van De Werf, F.; et al. Peripheral Blood RNA Levels of QSOX1 and PLBD1 Are New Independent Predictors of Left Ventricular Dysfunction After Acute Myocardial Infarction. Circ. Genom. Precis. Med. 2019, 12, e002656. [Google Scholar] [CrossRef]
- Niu, X.; Zhang, J.; Zhang, L.; Hou, Y.; Pu, S.; Chu, A.; Bai, M.; Zhang, Z. Weighted Gene Co-Expression Network Analysis Identifies Critical Genes in the Development of Heart Failure After Acute Myocardial Infarction. Front. Genet. 2019, 10, 1214. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R Package for Weighted Correlation Network Analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef]
- Chen, E.Y.; Tan, C.M.; Kou, Y.; Duan, Q.; Wang, Z.; Meirelles, G.V.; Clark, N.R.; Ma’ayan, A. Enrichr: Interactive and Collaborative HTML5 Gene List Enrichment Analysis Tool. BMC Bioinform. 2013, 14, 128. [Google Scholar] [CrossRef]
- McIntosh, V.J.; Lasley, R.D. Adenosine Receptor-Mediated Cardioprotection: Are All 4 Subtypes Required or Redundant? J. Cardiovasc. Pharmacol. Ther. 2012, 17, 21–33. [Google Scholar] [CrossRef]
- Liu, G.S.; Richards, S.C.; Olsson, R.A.; Mullane, K.; Walsh, R.S.; Downey, J.M. Evidence That the Adenosine A3 Receptor May Mediate the Protection Afforded by Preconditioning in the Isolated Rabbit Heart. Cardiovasc. Res. 1994, 28, 1057–1061. [Google Scholar] [CrossRef]
- Li, L.; Weng, Z.; Yao, C.; Song, Y.; Ma, T. Aquaporin-1 Deficiency Protects against Myocardial Infarction by Reducing both Edema and Apoptosis in Mice. Sci. Rep. 2015, 5, 13807. [Google Scholar] [CrossRef]
- Jiao, M.; Li, J.; Zhang, Q.; Xu, X.; Li, R.; Dong, P.; Meng, C.; Li, Y.; Wang, L.; Qi, W.; et al. Identification of Four Potential Biomarkers Associated with Coronary Artery Disease in Non-Diabetic Patients by Gene Co-Expression Network Analysis. Front. Genet. 2020, 11, 542. [Google Scholar] [CrossRef]
- Perini, E.D.; Schaefer, R.; Stöter, M.; Kalaidzidis, Y.; Zerial, M. Mammalian CORVET Is Required for Fusion and Conversion of Distinct Early Endosome Subpopulations. Traffic 2014, 15, 1366–1389. [Google Scholar] [CrossRef]
- Jonker, C.T.H.; Galmes, R.; Veenendaal, T.; Ten Brink, C.; van der Welle, R.E.N.; Liv, N.; de Rooij, J.; Peden, A.A.; van der Sluijs, P.; Margadant, C.; et al. Vps3 and Vps8 Control Integrin Trafficking from Early to Recycling Endosomes and Regulate Integrin-Dependent Functions. Nat. Commun. 2018, 9, 792. [Google Scholar] [CrossRef]
- Boya, P. Lysosomal Function and Dysfunction: Mechanism and Disease. Antioxid. Redox Signal. 2012, 17, 766–774. [Google Scholar] [CrossRef]
- Avissar, N.; Ornt, D.B.; Yagil, Y.; Horowitz, S.; Watkins, R.H.; Kerl, E.A.; Takahashi, K.; Palmer, I.S.; Cohen, H.J. Human Kidney Proximal Tubules Are the Main Source of Plasma Glutathione Peroxidase. Am. J. Physiol. 1994, 266, C367–C375. [Google Scholar] [CrossRef]
- Jin, R.C.; Mahoney, C.E.; Coleman Anderson, L.; Ottaviano, F.; Croce, K.; Leopold, J.A.; Zhang, Y.-Y.; Tang, S.-S.; Handy, D.E.; Loscalzo, J. Glutathione Peroxidase-3 Deficiency Promotes Platelet-Dependent Thrombosis in Vivo. Circulation 2011, 123, 1963–1973. [Google Scholar] [CrossRef] [PubMed]
- Clerk, A.; Meijles, D.N.; Hardyman, M.A.; Fuller, S.J.; Chothani, S.P.; Cull, J.J.; Cooper, S.T.E.; Alharbi, H.O.; Vanezis, K.; Felkin, L.E.; et al. Cardiomyocyte BRAF and Type 1 RAF Inhibitors Promote Cardiomyocyte and Cardiac Hypertrophy in Mice in Vivo. Biochem. J. 2022, 479, 401–424. [Google Scholar] [CrossRef] [PubMed]
- Muse, E.D.; Kramer, E.R.; Wang, H.; Barrett, P.; Parviz, F.; Novotny, M.A.; Lasken, R.S.; Jatkoe, T.A.; Oliveira, G.; Peng, H.; et al. A Whole Blood Molecular Signature for Acute Myocardial Infarction. Sci. Rep. 2017, 7, 12268. [Google Scholar] [CrossRef]
- Saenger, A.K.; Jaffe, A.S. Requiem for a Heavyweight: The Demise of Creatine Kinase-MB. Circulation 2008, 118, 2200–2206. [Google Scholar] [CrossRef]
- Devaux, Y.; Azuaje, F.; Vausort, M.; Yvorra, C.; Wagner, D.R. Integrated Protein Network and Microarray Analysis to Identify Potential Biomarkers after Myocardial Infarction. Funct. Integr. Genom. 2010, 10, 329–337. [Google Scholar] [CrossRef]
- Liu, Z.; Ma, C.; Gu, J.; Yu, M. Potential Biomarkers of Acute Myocardial Infarction Based on Weighted Gene Co-Expression Network Analysis. BioMedical Eng. OnLine 2019, 18, 9. [Google Scholar] [CrossRef]
- Guo, S.; Wu, J.; Zhou, W.; Liu, X.; Liu, Y.; Zhang, J.; Jia, S.; Li, J.; Wang, H. Identification and Analysis of Key Genes Associated with Acute Myocardial Infarction by Integrated Bioinformatics Methods. Medicine 2021, 100, e25553. [Google Scholar] [CrossRef]
- Shao, G. Integrated RNA Gene Expression Analysis Identified Potential Immune-Related Biomarkers and RNA Regulatory Pathways of Acute Myocardial Infarction. PLoS ONE 2022, 17, e0264362. [Google Scholar] [CrossRef]
- Peart, J.N.; Headrick, J.P. Adenosinergic Cardioprotection: Multiple Receptors, Multiple Pathways. Pharmacol. Ther. 2007, 114, 208–221. [Google Scholar] [CrossRef]
- Burgoyne, J.R.; Mongue-Din, H.; Eaton, P.; Shah, A.M. Redox Signaling in Cardiac Physiology and Pathology. Circ. Res. 2012, 111, 1091–1106. [Google Scholar] [CrossRef]
- Salonen, J.T.; Ylä-Herttuala, S.; Yamamoto, R.; Butler, S.; Korpela, H.; Salonen, R.; Nyyssönen, K.; Palinski, W.; Witztum, J.L. Autoantibody against Oxidised LDL and Progression of Carotid Atherosclerosis. Lancet 1992, 339, 883–887. [Google Scholar] [CrossRef]
- Duarte, T.; Gonçalves, S.; Sá, C.; Rodrigues, R.; Marinheiro, R.; Fonseca, M.; Seixo, F.; Caria, R. Prognostic Impact of Iron Metabolism Changes in Patients with Acute Coronary Syndrome. Arq. Bras. Cardiol. 2018, 111, 144–150. [Google Scholar] [CrossRef]
- Kautz, L.; Meynard, D.; Monnier, A.; Darnaud, V.; Bouvet, R.; Wang, R.-H.; Deng, C.; Vaulont, S.; Mosser, J.; Coppin, H.; et al. Iron Regulates Phosphorylation of Smad1/5/8 and Gene Expression of Bmp6, Smad7, Id1, and Atoh8 in the Mouse Liver. Blood 2008, 112, 1503–1509. [Google Scholar] [CrossRef]
- Baars, T.; Neumann, U.; Jinawy, M.; Hendricks, S.; Sowa, J.-P.; Kälsch, J.; Riemenschneider, M.; Gerken, G.; Erbel, R.; Heider, D.; et al. In Acute Myocardial Infarction Liver Parameters Are Associated with Stenosis Diameter. Medicine 2016, 95, e2807. [Google Scholar] [CrossRef]
- Banach, J.; Gilewski, W.; Słomka, A.; Buszko, K.; Błażejewski, J.; Karasek, D.; Rogowicz, D.; Żekanowska, E.; Sinkiewicz, W. Bone Morphogenetic Protein 6-a Possible New Player in Pathophysiology of Heart Failure. Clin. Exp. Pharmacol. Physiol. 2016, 43, 1247–1250. [Google Scholar] [CrossRef]
- Iwata, K.; Nishinaka, T.; Matsuno, K.; Yabe-Nishimura, C. Increased Gene Expression of Glutathione Peroxidase-3 in Diabetic Mouse Heart. Biol. Pharm. Bull. 2006, 29, 1042–1045. [Google Scholar] [CrossRef]
- Pastori, D.; Pignatelli, P.; Farcomeni, A.; Menichelli, D.; Nocella, C.; Carnevale, R.; Violi, F. Aging-Related Decline of Glutathione Peroxidase 3 and Risk of Cardiovascular Events in Patients with Atrial Fibrillation. J. Am. Heart Assoc. 2016, 5, e003682. [Google Scholar] [CrossRef]
- Einarson, T.R.; Acs, A.; Ludwig, C.; Panton, U.H. Prevalence of Cardiovascular Disease in Type 2 Diabetes: A Systematic Literature Review of Scientific Evidence from across the World in 2007–2017. Cardiovasc. Diabetol. 2018, 17, 83. [Google Scholar] [CrossRef]
- Nistri, S.; Sassoli, C.; Bani, D. Notch Signaling in Ischemic Damage and Fibrosis: Evidence and Clues from the Heart. Front. Pharmacol. 2017, 8, 187. [Google Scholar] [CrossRef]
- Gude, N.A.; Emmanuel, G.; Wu, W.; Cottage, C.T.; Fischer, K.; Quijada, P.; Muraski, J.A.; Alvarez, R.; Rubio, M.; Schaefer, E.; et al. Activation of Notch-Mediated Protective Signaling in the Myocardium. Circ. Res. 2008, 102, 1025–1035. [Google Scholar] [CrossRef]
- Li, Y.; Hiroi, Y.; Ngoy, S.; Okamoto, R.; Noma, K.; Wang, C.-Y.; Wang, H.-W.; Zhou, Q.; Radtke, F.; Liao, R.; et al. Notch1 in Bone Marrow-Derived Cells Mediates Cardiac Repair after Myocardial Infarction. Circulation 2011, 123, 866–876. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Wan, L.; Xu, Q.; Zhao, Y.; Liu, J. Notch Signaling Activation Contributes to Cardioprotection Provided by Ischemic Preconditioning and Postconditioning. J. Transl. Med. 2013, 11, 251. [Google Scholar] [CrossRef] [PubMed]
- Sassoli, C.; Pini, A.; Mazzanti, B.; Quercioli, F.; Nistri, S.; Saccardi, R.; Zecchi-Orlandini, S.; Bani, D.; Formigli, L. Mesenchymal Stromal Cells Affect Cardiomyocyte Growth through Juxtacrine Notch-1/Jagged-1 Signaling and Paracrine Mechanisms: Clues for Cardiac Regeneration. J. Mol. Cell. Cardiol. 2011, 51, 399–408. [Google Scholar] [CrossRef] [PubMed]
- Qin, C.; Buxton, K.D.; Pepe, S.; Cao, A.H.; Venardos, K.; Love, J.E.; Kaye, D.M.; Yang, Y.H.; Morand, E.F.; Ritchie, R.H. Reperfusion-Induced Myocardial Dysfunction Is Prevented by Endogenous Annexin-A1 and Its N-Terminal-Derived Peptide Ac-ANX-A12-26. Br. J. Pharmacol. 2013, 168, 238–252. [Google Scholar] [CrossRef] [PubMed]
- Sabatine, M.S.; Liu, E.; Morrow, D.A.; Heller, E.; McCarroll, R.; Wiegand, R.; Berriz, G.F.; Roth, F.P.; Gerszten, R.E. Metabolomic Identification of Novel Biomarkers of Myocardial Ischemia. Circulation 2005, 112, 3868–3875. [Google Scholar] [CrossRef]
- Emmrich, S.; Henke, K.; Hegermann, J.; Ochs, M.; Reinhardt, D.; Klusmann, J.-H. MiRNAs can Increase the Efficiency of Ex Vivo Platelet Generation. Ann. Hematol. 2012, 91, 1673–1684. [Google Scholar] [CrossRef]
- Jakob, P.; Kacprowski, T.; Briand-Schumacher, S.; Heg, D.; Klingenberg, R.; Stähli, B.E.; Jaguszewski, M.; Rodondi, N.; Nanchen, D.; Räber, L.; et al. Profiling and Validation of Circulating MicroRNAs for Cardiovascular Events in Patients Presenting with ST-Segment Elevation Myocardial Infarction. Eur. Heart J. 2017, 38, 511–515. [Google Scholar] [CrossRef]
- Bye, A.; Røsjø, H.; Nauman, J.; Silva, G.J.J.; Follestad, T.; Omland, T.; Wisløff, U. Circulating MicroRNAs Predict Future Fatal Myocardial Infarction in Healthy Individuals—The HUNT Study. J. Mol. Cell. Cardiol. 2016, 97, 162–168. [Google Scholar] [CrossRef]
- Welten, S.M.J.; Bastiaansen, A.J.N.M.; de Jong, R.C.M.; de Vries, M.R.; Peters, E.A.B.; Boonstra, M.C.; Sheikh, S.P.; La Monica, N.; Kandimalla, E.R.; Quax, P.H.A.; et al. Inhibition of 14q32 MicroRNAs MiR-329, MiR-487b, MiR-494, and MiR-495 Increases Neovascularization and Blood Flow Recovery after Ischemia. Circ. Res. 2014, 115, 696–708. [Google Scholar] [CrossRef]
- Johnson, J.L. Elucidating the Contributory Role of MicroRNA to Cardiovascular Diseases (a Review). Vascul. Pharmacol. 2019, 114, 31–48. [Google Scholar] [CrossRef]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: A Comprehensive Gene Set Enrichment Analysis Web Server 2016 Update. Nucleic Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef]
- Wincewicz, A.; Sulkowski, S. Stat Proteins as Intracellular Regulators of Resistance to Myocardial Injury in the Context of Cardiac Remodeling and Targeting for Therapy. Adv. Clin. Exp. Med. 2017, 26, 703–708. [Google Scholar] [CrossRef]
- McCormick, J.; Suleman, N.; Scarabelli, T.; Knight, R.; Latchman, D.; Stephanou, A. STAT1 Deficiency in the Heart Protects against Myocardial Infarction by Enhancing Autophagy. J. Cell. Mol. Med. 2012, 16, 386–393. [Google Scholar] [CrossRef]
- Song, N.; Li, X.-M.; Luo, J.-Y.; Zhai, H.; Zhao, Q.; Zhou, X.-R.; Liu, F.; Zhang, X.-H.; Gao, X.-M.; Li, X.-M.; et al. Construction and Analysis for Differentially Expressed Long Non-Coding RNAs and MRNAs in Acute Myocardial Infarction. Sci. Rep. 2020, 10, 6989. [Google Scholar] [CrossRef]
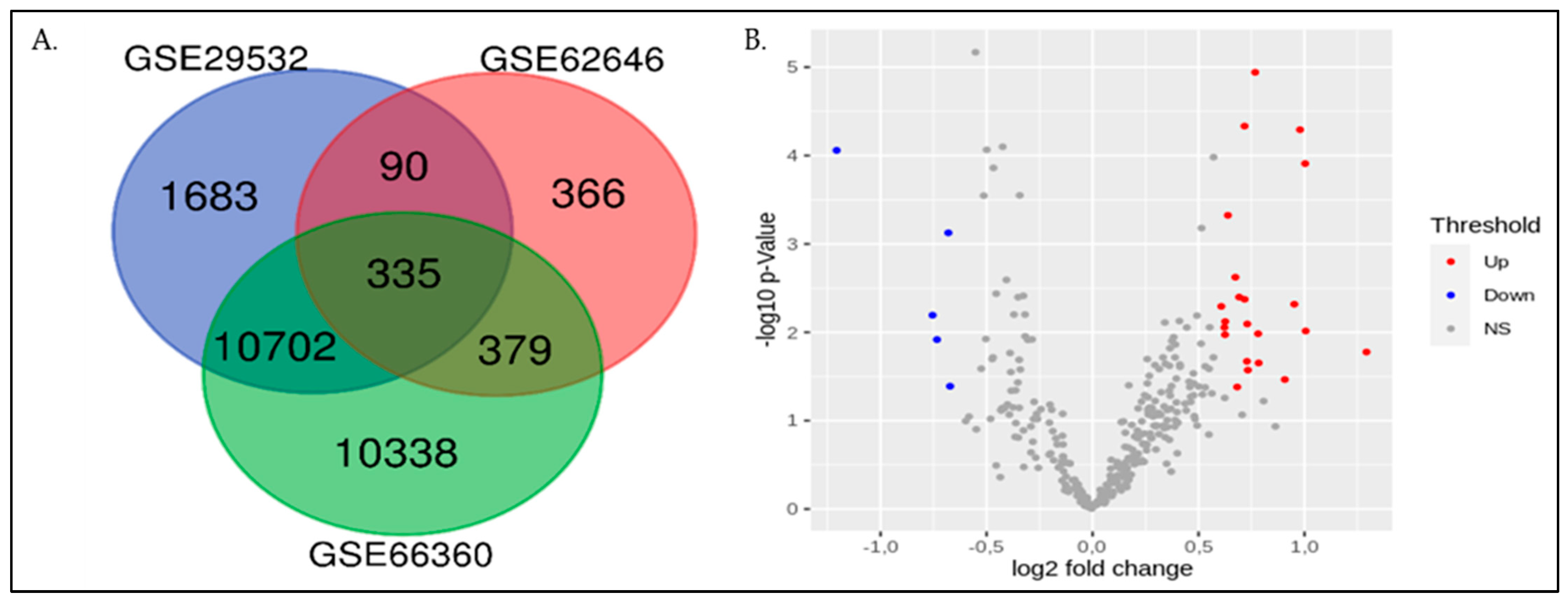


| GEO Dataset ID | GEO Platform Accession Number | Subjects | Types of Sample | Microarray Platform | |
|---|---|---|---|---|---|
| Patients | Controls | ||||
| GSE66360 | GPL570 | 49 | 50 | Whole blood | Affymetrix Human Genome U133 Plus 2.0 Array |
| GSE29532 | GPL5175 | 8 | 6 | Whole blood cells | Affymetrix Human Exon 1.0 ST Array |
| GSE62646 | GPL6244 | 28 | 14 | Peripheral Blood Mononuclear Cells | Affymetrix Human Gene 1.0 ST Array |
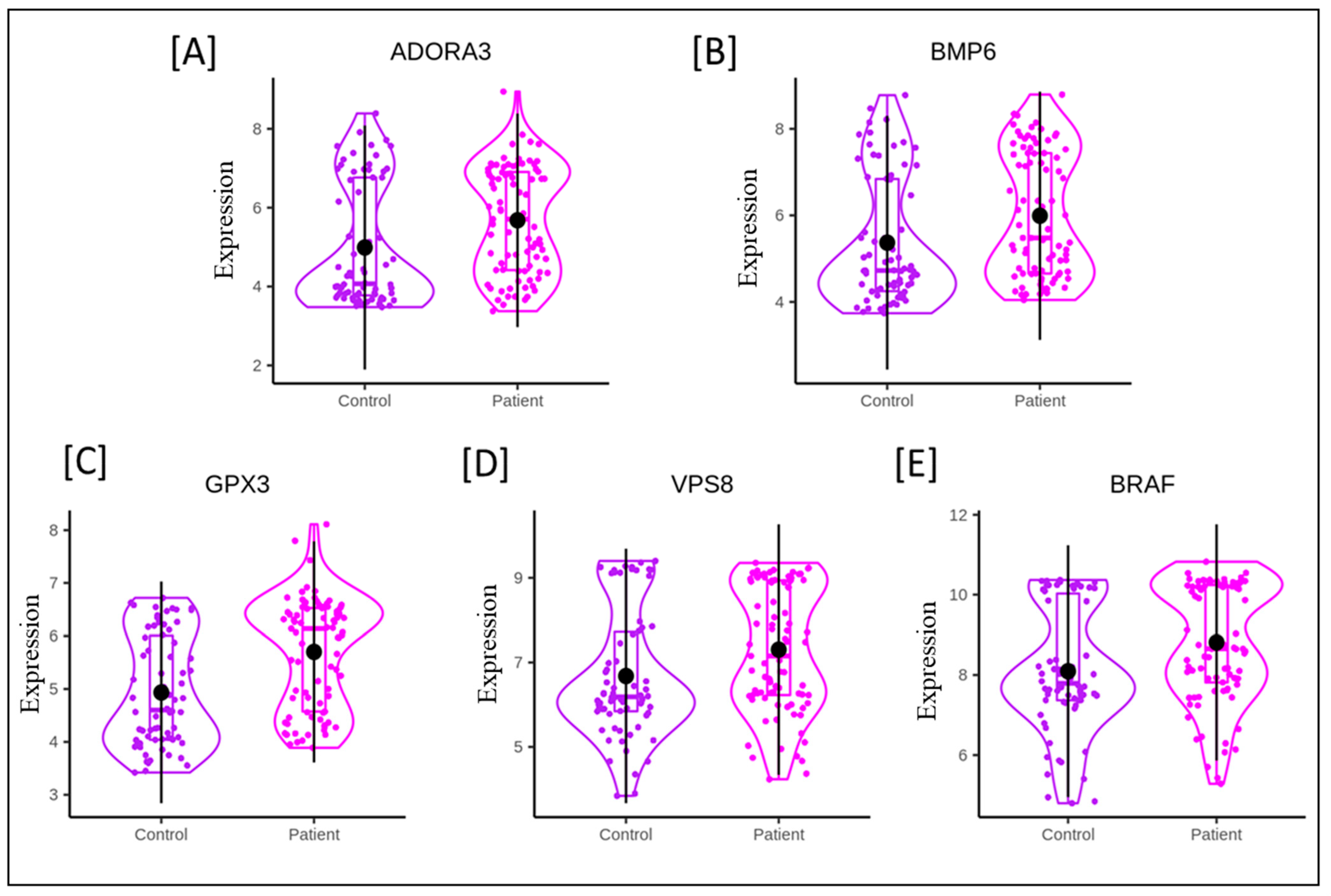
| S.No. | Name of Gene | Expression Level | References |
|---|---|---|---|
| 1. | ADORA3 (Adenosine A3 receptor ADOR) | Upregulated | [15,16] |
| 2. | AQP1 (aquaporin 1) | Upregulated | [17] |
| 3. | BMP6 (Bone Morphogenetic Protein 5) | Upregulated | [18] |
| 4. | VPS8 (Vacuolar Protein Sorting-Associated Protein 8 Homolog) | Upregulated | [19,20,21] |
| 5. | (GPx3) Glutathione Peroxidase 3 | Upregulated | [22,23] |
| 6. | BRAF (B-Raf Proto-Oncogene, a serine/threonine kinase) | Upregulated | [24] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kumar, S.; Shih, C.-M.; Tsai, L.-W.; Dubey, R.; Gupta, D.; Chakraborty, T.; Sharma, N.; Singh, A.V.; Swarup, V.; Singh, H.N. Transcriptomic Profiling Unravels Novel Deregulated Gene Signatures Associated with Acute Myocardial Infarction: A Bioinformatics Approach. Genes 2022, 13, 2321. https://doi.org/10.3390/genes13122321
Kumar S, Shih C-M, Tsai L-W, Dubey R, Gupta D, Chakraborty T, Sharma N, Singh AV, Swarup V, Singh HN. Transcriptomic Profiling Unravels Novel Deregulated Gene Signatures Associated with Acute Myocardial Infarction: A Bioinformatics Approach. Genes. 2022; 13(12):2321. https://doi.org/10.3390/genes13122321
Chicago/Turabian StyleKumar, Sanjay, Chun-Ming Shih, Lung-Wen Tsai, Rajni Dubey, Deepika Gupta, Tanmoy Chakraborty, Naveen Sharma, Abhishek Vikram Singh, Vishnu Swarup, and Himanshu Narayan Singh. 2022. "Transcriptomic Profiling Unravels Novel Deregulated Gene Signatures Associated with Acute Myocardial Infarction: A Bioinformatics Approach" Genes 13, no. 12: 2321. https://doi.org/10.3390/genes13122321
APA StyleKumar, S., Shih, C.-M., Tsai, L.-W., Dubey, R., Gupta, D., Chakraborty, T., Sharma, N., Singh, A. V., Swarup, V., & Singh, H. N. (2022). Transcriptomic Profiling Unravels Novel Deregulated Gene Signatures Associated with Acute Myocardial Infarction: A Bioinformatics Approach. Genes, 13(12), 2321. https://doi.org/10.3390/genes13122321





