Novel Intronic Mutation in VMA21 Causing Severe Phenotype of X-Linked Myopathy with Excessive Autophagy—Case Report
Abstract
1. Introduction
2. Case Report
2.1. Clinical and Paraclinical Summary
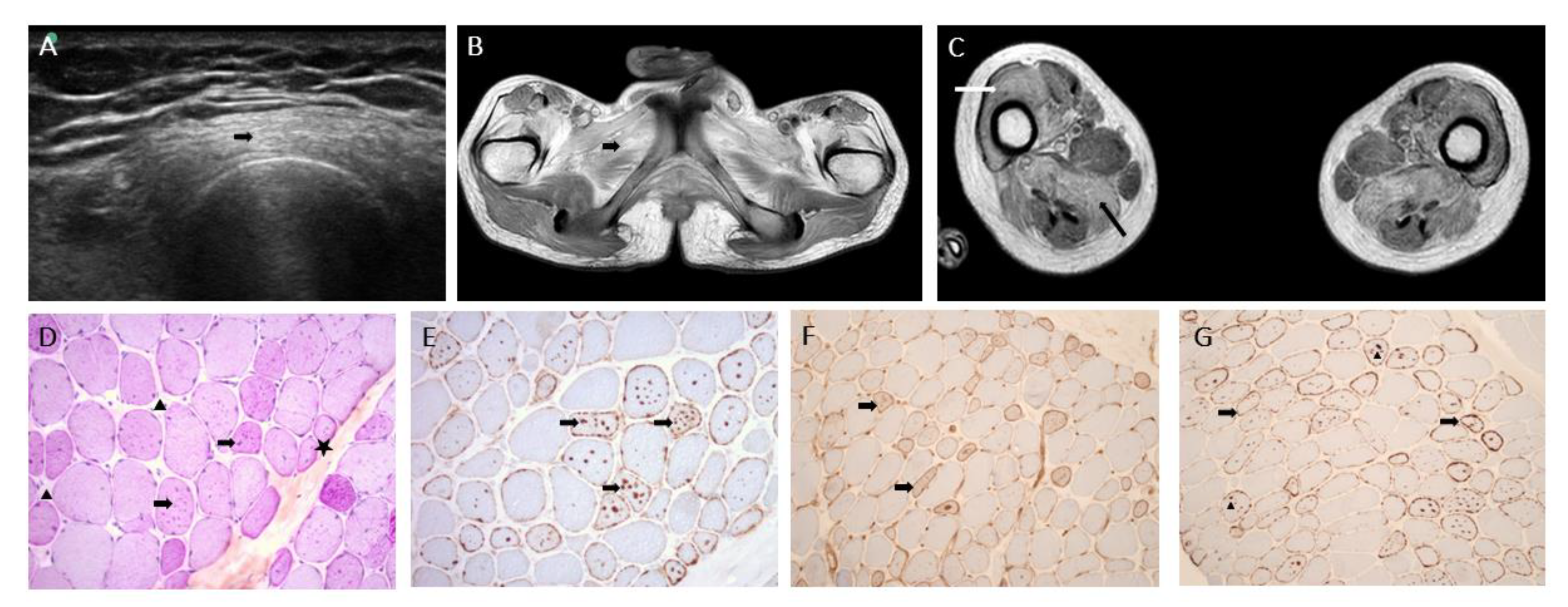
2.2. Muscle Biopsy
2.3. Genetic Analyses
2.4. mRNA and Protein Quantification from the Patient’s Fibroblasts
3. Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Khandia, R.; Dadar, M.; Munjal, A.; Dhama, K.; Karthik, K.; Tiwari, R.; Yatoo, M.I.; Iqbal, H.M.N.; Singh, K.P.; Joshi, S.K.; et al. A Comprehensive Review of Autophagy and Its Various Roles in Infectious, Non-Infectious, and Lifestyle Diseases: Current Knowledge and Prospects for Disease Prevention, Novel Drug Design, and Therapy. Cells 2019, 8, 674. [Google Scholar] [CrossRef] [PubMed]
- Dowling, J.J.; Moore, S.A.; Kalimo, H.; Minassian, B.A. X-Linked Myopathy with Excessive Autophagy: A Failure of Self-Eating. Acta Neuropathol. 2015, 129, 383–390. [Google Scholar] [CrossRef] [PubMed]
- Kalimo, H.; Savontaus, M.L.; Lang, H.; Paljärvi, L.; Sonninen, V.; Dean, P.B.; Katevuo, K.; Salminen, A. X-Linked Myopathy with Excessive Autophagy: A New Hereditary Muscle Disease. Ann. Neurol. 1988, 23, 258–265. [Google Scholar] [CrossRef] [PubMed]
- Ramachandran, N.; Munteanu, I.; Wang, P.; Ruggieri, A.; Rilstone, J.J.; Israelian, N.; Naranian, T.; Paroutis, P.; Guo, R.; Ren, Z.-P.; et al. VMA21 Deficiency Prevents Vacuolar ATPase Assembly and Causes Autophagic Vacuolar Myopathy. Acta Neuropathol. 2013, 125, 439–457. [Google Scholar] [CrossRef] [PubMed]
- Cotta, A.; Carvalho, E.; da-Cunha-Junior, A.L.; Navarro, M.M.; Menezes, M.M.; Paim, J.F.; Valicek, J.; Lima, M.I.; Velloso-Filho, R.; Freire-Lyra, M.H.; et al. Clinical, Imaging, Morphologic, and Molecular Features of X-Linked VMA21-Related Myopathy in Two Unrelated Brazilian Families. J. Neurol. Sci. 2020, 415, 116977. [Google Scholar] [CrossRef] [PubMed]
- Munteanu, I.; Ramachandran, N.; Ruggieri, A.; Awaya, T.; Nishino, I.; Minassian, B.A. Congenital Autophagic Vacuolar Myopathy Is Allelic to X-Linked Myopathy with Excessive Autophagy. Neurology 2015, 84, 1714–1716. [Google Scholar] [CrossRef] [PubMed]
- Kurashige, T.; Takahashi, T.; Yamazaki, Y.; Nagano, Y.; Kondo, K.; Nakamura, T.; Yamawaki, T.; Tsuburaya, R.; Hayashi, Y.K.; Nonaka, I.; et al. Elevated Urinary Β2 Microglobulin in the First Identified Japanese Family Afflicted by X-Linked Myopathy with Excessive Autophagy. Neuromuscul. Disord. 2013, 23, 911–916. [Google Scholar] [CrossRef] [PubMed]
- Crockett, C.D.; Ruggieri, A.; Gujrati, M.; Zallek, C.M.; Ramachandran, N.; Minassian, B.A.; Moore, S.A. Late Adult-Onset of X-Linked Myopathy with Excessive Autophagy. Muscle Nerve 2014, 50, 138–144. [Google Scholar] [CrossRef] [PubMed]
- Ruggieri, A.; Ramachandran, N.; Wang, P.; Haan, E.; Kneebone, C.; Manavis, J.; Morandi, L.; Moroni, I.; Blumbergs, P.; Mora, M.; et al. Non-Coding VMA21 Deletions Cause X-Linked Myopathy with Excessive Autophagy. Neuromuscul. Disord. 2015, 25, 207–211. [Google Scholar] [CrossRef] [PubMed]
- Jaganathan, K.; Kyriazopoulou Panagiotopoulou, S.; McRae, J.F.; Darbandi, S.F.; Knowles, D.; Li, Y.I.; Kosmicki, J.A.; Arbelaez, J.; Cui, W.; Schwartz, G.B.; et al. Predicting Splicing from Primary Sequence with Deep Learning. Cell 2019, 176, 535–548.e24. [Google Scholar] [CrossRef] [PubMed]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and Guidelines for the Interpretation of Sequence Variants: A Joint Consensus Recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–424. [Google Scholar] [CrossRef] [PubMed]
- Rubin, D.I. Normal and Abnormal Spontaneous Activity. Handb. Clin. Neurol. 2019, 160, 257–279. [Google Scholar] [CrossRef] [PubMed]
- Munteanu, I.; Ackerley, C.A.; Mnatzakanian, G.N.; Kissel, J.T.; Minassian, B.A. Electrophysiology Extends the Phenotypic Spectrum of X-Linked Myopathy with Excessive Autophagy. Neurology 2005, 64, 927–928. [Google Scholar] [CrossRef] [PubMed]
- Jääskeläinen, S.K.; Juel, V.C.; Udd, B.; Villanova, M.; Liguori, R.; Minassian, B.A.; Falck, B.; Niemi, P.; Kalimo, H. Electrophysiological Findings in X-Linked Myopathy with Excessive Autophagy. Ann. Neurol. 2002, 51, 648–652. [Google Scholar] [CrossRef] [PubMed]
- Mercier, S.; Magot, A.; Caillon, F.; Isidor, B.; David, A.; Ferrer, X.; Vital, A.; Coquet, M.; Penttilä, S.; Udd, B.; et al. Muscle Magnetic Resonance Imaging Abnormalities in X-Linked Myopathy with Excessive Autophagy. Muscle Nerve 2015, 52, 673–680. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pegat, A.; Streichenberger, N.; Lacoste, N.; Hermier, M.; Menassa, R.; Coudert, L.; Theuriet, J.; Froissart, R.; Terrone, S.; Bouhour, F.; et al. Novel Intronic Mutation in VMA21 Causing Severe Phenotype of X-Linked Myopathy with Excessive Autophagy—Case Report. Genes 2022, 13, 2245. https://doi.org/10.3390/genes13122245
Pegat A, Streichenberger N, Lacoste N, Hermier M, Menassa R, Coudert L, Theuriet J, Froissart R, Terrone S, Bouhour F, et al. Novel Intronic Mutation in VMA21 Causing Severe Phenotype of X-Linked Myopathy with Excessive Autophagy—Case Report. Genes. 2022; 13(12):2245. https://doi.org/10.3390/genes13122245
Chicago/Turabian StylePegat, Antoine, Nathalie Streichenberger, Nicolas Lacoste, Marc Hermier, Rita Menassa, Laurent Coudert, Julian Theuriet, Roseline Froissart, Sophie Terrone, Francoise Bouhour, and et al. 2022. "Novel Intronic Mutation in VMA21 Causing Severe Phenotype of X-Linked Myopathy with Excessive Autophagy—Case Report" Genes 13, no. 12: 2245. https://doi.org/10.3390/genes13122245
APA StylePegat, A., Streichenberger, N., Lacoste, N., Hermier, M., Menassa, R., Coudert, L., Theuriet, J., Froissart, R., Terrone, S., Bouhour, F., Michel-Calemard, L., Schaeffer, L., & Jacquier, A. (2022). Novel Intronic Mutation in VMA21 Causing Severe Phenotype of X-Linked Myopathy with Excessive Autophagy—Case Report. Genes, 13(12), 2245. https://doi.org/10.3390/genes13122245






