Dbf4 Zn-Finger Motif Is Specifically Required for Stimulation of Ctf19-Activated Origins in Saccharomyces cerevisiae
Abstract
1. Introduction
2. Materials and Methods
2.1. Plasmid and Yeast Strain Construction
2.2. Other Methods
2.3. Computation and Statistics
3. Results and Discussion
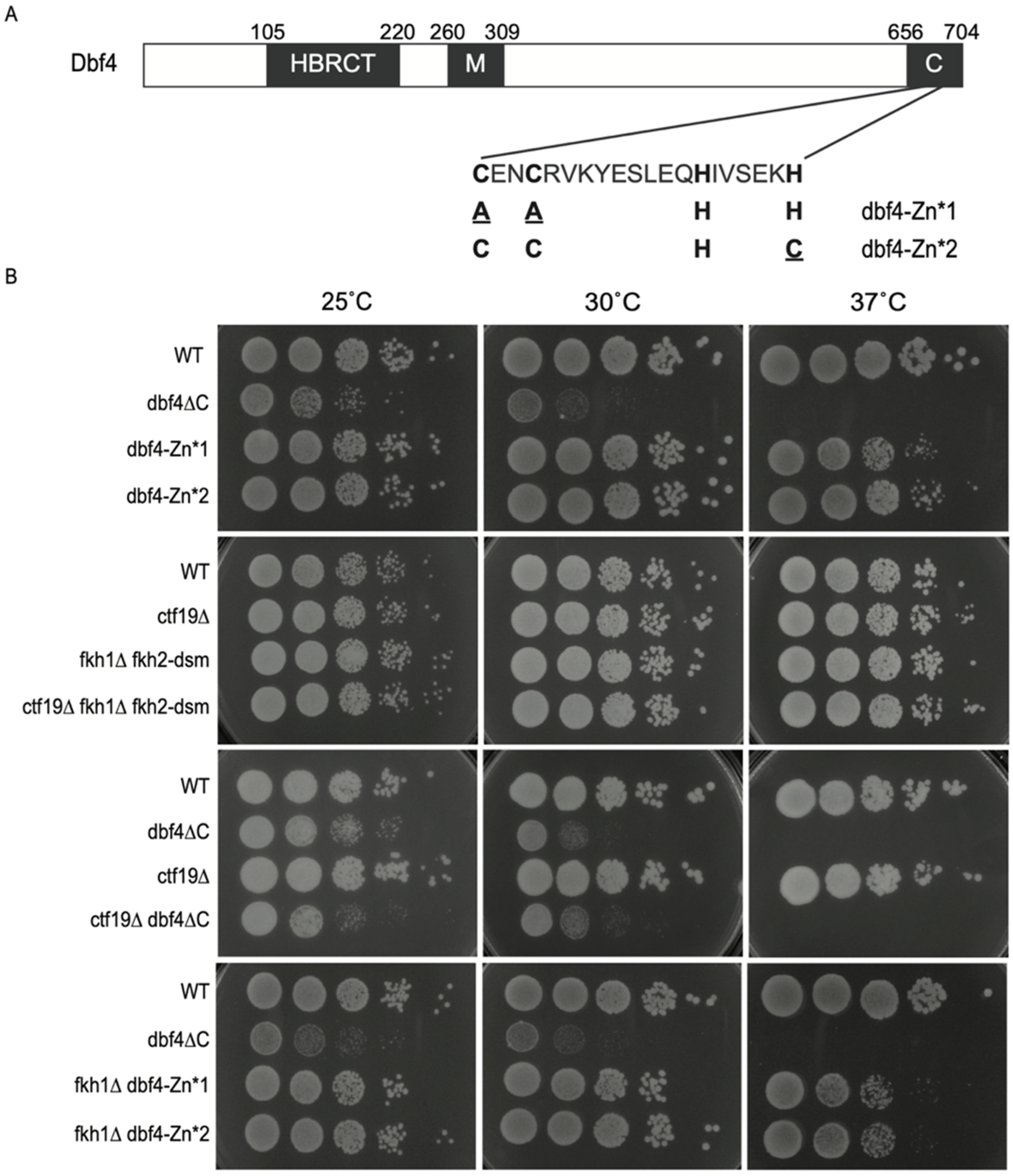
3.1. Dbf4∆C Is Defective in Essential Dbf4 Function(s) beyond Origin Targeting by Fkh1 and Ctf19
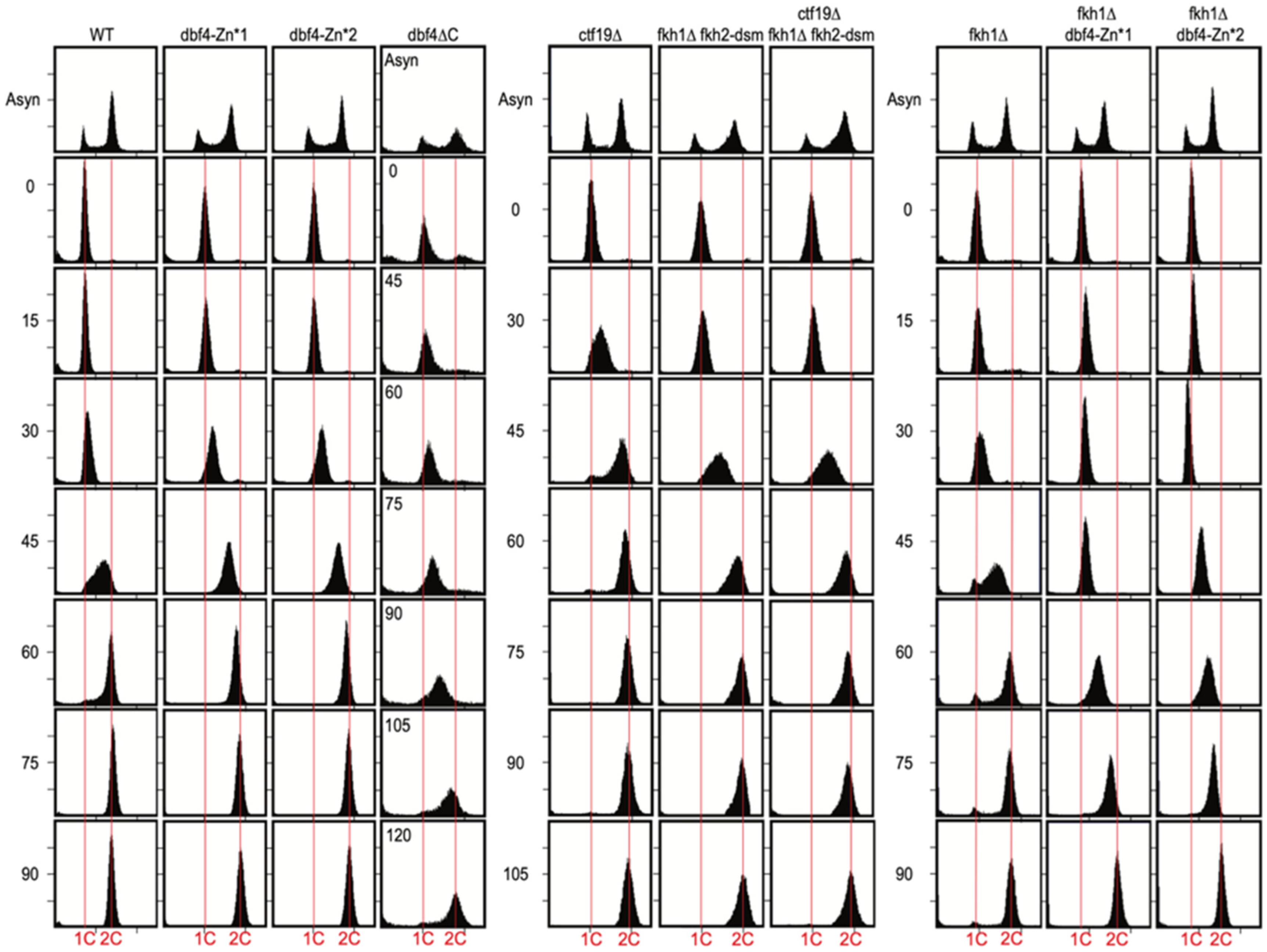
3.2. Dbf4∆C Is Defective in Overall Rate of Genome Replication While dbf4-Zn* and ctf19∆ Are Not
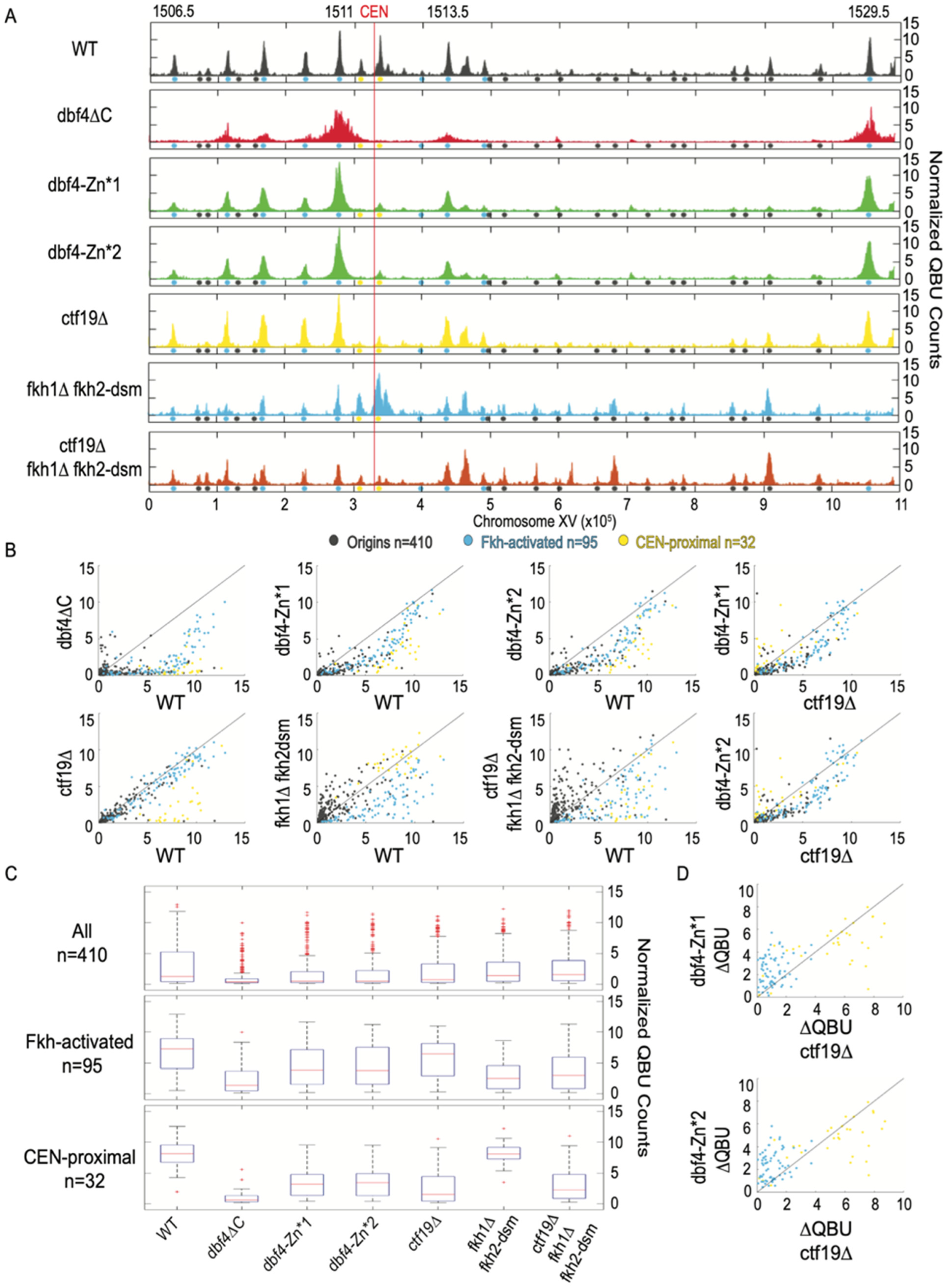
3.3. Dbf4-Zn* Mutations Specifically Eliminate Early Activation of CEN-Proximal Origins
3.4. CEN-Proximal Origins Are Differentially Sensitive to Loss of Ctf19 or Dbf4-Zn*
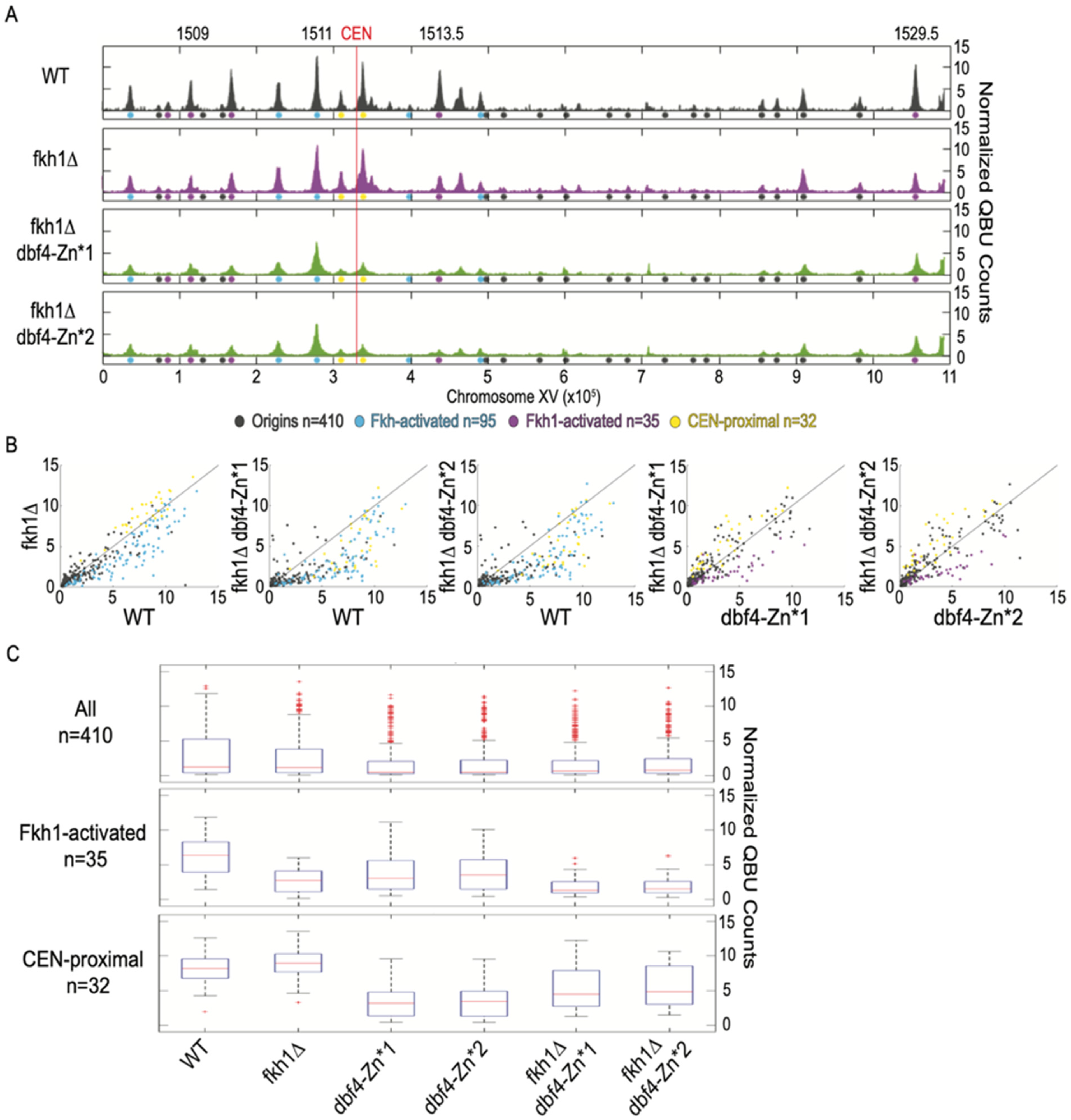
3.5. Dbf4-Zn* Retains Fkh1-Dependent Targeting to Fkh1-Activated Origins
3.6. Dbf4-Zn* Is Defective in Its Recruitment to CENs
4. Perspective
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gillespie, P.J.; Blow, J.J. DDK: The Outsourced Kinase of Chromosome Maintenance. Biology (Basel) 2022, 11, 877. [Google Scholar] [CrossRef]
- Natsume, T.; Muller, C.A.; Katou, Y.; Retkute, R.; Gierlinski, M.; Araki, H.; Blow, J.J.; Shirahige, K.; Nieduszynski, C.A.; Tanaka, T.U. Kinetochores coordinate pericentromeric cohesion and early DNA replication by Cdc7-Dbf4 kinase recruitment. Mol. Cell 2013, 50, 661–674. [Google Scholar] [CrossRef] [PubMed]
- Sheu, Y.J.; Stillman, B. The Dbf4-Cdc7 kinase promotes S phase by alleviating an inhibitory activity in Mcm4. Nature 2010, 463, 113–117. [Google Scholar] [CrossRef]
- Mantiero, D.; Mackenzie, A.; Donaldson, A.; Zegerman, P. Limiting replication initiation factors execute the temporal programme of origin firing in budding yeast. EMBO J. 2011, 30, 4805–4814. [Google Scholar] [CrossRef]
- Patel, P.K.; Kommajosyula, N.; Rosebrock, A.; Bensimon, A.; Leatherwood, J.; Bechhoefer, J.; Rhind, N. The Hsk1(Cdc7) replication kinase regulates origin efficiency. Mol. Biol. Cell 2008, 19, 5550–5558. [Google Scholar] [CrossRef]
- Tanaka, S.; Nakato, R.; Katou, Y.; Shirahige, K.; Araki, H. Origin association of Sld3, Sld7, and Cdc45 proteins is a key step for determination of origin-firing timing. Curr. Biol. 2011, 21, 2055–2063. [Google Scholar] [CrossRef] [PubMed]
- Aparicio, O.M. Location, location, location: Its all in the timing for replication origins. Genes Dev. 2013, 27, 117–128. [Google Scholar] [CrossRef] [PubMed]
- Knott, S.R.; Peace, J.M.; Ostrow, A.Z.; Gan, Y.; Rex, A.E.; Viggiani, C.J.; Tavare, S.; Aparicio, O.M. Forkhead transcription factors establish origin timing and long-range clustering in S. cerevisiae. Cell 2012, 148, 99–111. [Google Scholar] [CrossRef]
- Fang, D.; Lengronne, A.; Shi, D.; Forey, R.; Skrzypczak, M.; Ginalski, K.; Yan, C.; Wang, X.; Cao, Q.; Pasero, P.; et al. Dbf4 recruitment by forkhead transcription factors defines an upstream rate-limiting step in determining origin firing timing. Genes Dev. 2017, 31, 2405–2415. [Google Scholar] [CrossRef]
- Zhang, H.; Petrie, M.V.; He, Y.; Peace, J.M.; Chiolo, I.E.; Aparicio, O.M. Dynamic relocalization of replication origins by Fkh1 requires execution of DDK function and Cdc45 loading at origins. Elife 2019, 8, e45512. [Google Scholar] [CrossRef]
- Looke, M.; Kristjuhan, K.; Varv, S.; Kristjuhan, A. Chromatin-dependent and -independent regulation of DNA replication origin activation in budding yeast. EMBO Rep. 2012, 14, 191–198. [Google Scholar] [CrossRef]
- Ostrow, A.Z.; Nellimoottil, T.; Knott, S.R.; Fox, C.A.; Tavare, S.; Aparicio, O.M. Fkh1 and Fkh2 bind multiple chromosomal elements in the S. cerevisiae genome with distinct specificities and cell cycle dynamics. PLoS ONE 2014, 9, e87647. [Google Scholar] [CrossRef] [PubMed]
- Reinapae, A.; Jalakas, K.; Avvakumov, N.; Looke, M.; Kristjuhan, K.; Kristjuhan, A. Recruitment of Fkh1 to replication origins requires precisely positioned Fkh1/2 binding sites and concurrent assembly of the pre-replicative complex. PLoS Genet. 2017, 13, e1006588. [Google Scholar] [CrossRef]
- Jones, D.R.; Prasad, A.A.; Chan, P.K.; Duncker, B.P. The Dbf4 motif C zinc finger promotes DNA replication and mediates resistance to genotoxic stress. Cell Cycle 2010, 9, 2018–2026. [Google Scholar] [CrossRef]
- Sikorski, R.S.; Hieter, P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 1989, 122, 19–27. [Google Scholar] [CrossRef]
- Goldstein, A.L.; Pan, X.; McCusker, J.H. Heterologous URA3MX cassettes for gene replacement in Saccharomyces cerevisiae. Yeast 1999, 15, 507–511. [Google Scholar] [CrossRef]
- Ito, H.; Fukuda, Y.; Murata, K.; Kimura, A. Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 1983, 153, 163–168. [Google Scholar] [CrossRef]
- Longtine, M.S.; McKenzie, A., 3rd; Demarini, D.J.; Shah, N.G.; Wach, A.; Brachat, A.; Philippsen, P.; Pringle, J.R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 1998, 14, 953–961. [Google Scholar] [CrossRef]
- Viggiani, C.J.; Aparicio, O.M. New vectors for simplified construction of BrdU-Incorporating strains of Saccharomyces cerevisiae. Yeast 2006, 23, 1045–1051. [Google Scholar] [CrossRef]
- He, Y.; Petrie, M.P.; Zhang, H.; Peace, J.M.; Aparicio, O.M. Rpd3 regulates single-copy origins independently of the rDNA array by opposing Fkh1-mediated origin stimulation. Proc. Natl. Acad. Sci. USA 2022, 119, e2212134119. [Google Scholar] [CrossRef]
- Haye-Bertolozzi, J.E.; Aparicio, O.M. Quantitative Bromodeoxyuridine Immunoprecipitation Analyzed by High-Throughput Sequencing (qBrdU-seq or QBU). Methods Mol. Biol.-Genome Instab. Methods Protoc. 2018, 1672, 209–225. [Google Scholar]
- Ostrow, A.Z.; Kalhor, R.; Gan, Y.; Villwock, S.K.; Linke, C.; Barberis, M.; Chen, L.; Aparicio, O.M. Conserved forkhead dimerization motif controls DNA replication timing and spatial organization of chromosomes in S. cerevisiae. Proc. Natl. Acad. Sci. USA 2017, 114, E2411–E2419. [Google Scholar] [CrossRef] [PubMed]
- Ostrow, A.Z.; Viggiani, C.J.; Aparicio, J.G.; Aparicio, O.M. ChIP-Seq to Analyze the Binding of Replication Proteins to Chromatin. Methods Mol. Biol. 2015, 1300, 155–168. [Google Scholar]
- Nieduszynski, C.A.; Hiraga, S.; Ak, P.; Benham, C.J.; Donaldson, A.D. OriDB: A DNA replication origin database. Nucleic Acids Res. 2007, 35, D40–D46. [Google Scholar] [CrossRef]
- Zhong, Y.; Nellimoottil, T.; Peace, J.M.; Knott, S.R.; Villwock, S.K.; Yee, J.M.; Jancuska, J.M.; Rege, S.; Tecklenburg, M.; Sclafani, R.A.; et al. The level of origin firing inversely affects the rate of replication fork progression. J. Cell Biol. 2013, 201, 373–383. [Google Scholar] [CrossRef] [PubMed]

| Name | Sequence | Source |
|---|---|---|
| ∆N-dbf4_F | ATAGGGCGAATTGGAGCTCCACCGCGGTGGCGGCCGCTCTACAAACTTAGACGAACACC | This Study |
| ∆N-dbf4_R | AAAGCTGGGTACCGGGCCCCCCCTCGAGGTCGACGGTATCTCAATACCAGCTTTCTAGC | " |
| dbf4∆C_F | GACAGCACAGACAGCACAGCCGGTGAAGAAAGAAACGGTAtgaggcgcgccacttctaaa | " |
| dbf4∆C_R | GATTTTATCACTAAAAGCTACTGCACTTTACGTCGTGTCCcggcgttagtatcgaatcga | " |
| CTF19∆_F | GTGTGATCTTGTTGATACTAGGTCGGCAAAGAACGCAAATCGATCCCCGGGTTAATTAA | " |
| CTF19∆_R | GTTTAAGCAAGCCGTCCAGTTGGCAATGCAAATGGAACAGAATTCGAGCTCGTTTAAAC | " |
| dbf4-Zn-aaHH | CGGTAAAAAATTCCGGATACgcTGAAAATgcTCGTGTAAAATACG | " |
| dbf4-Zn-CCHc | CATAGTTTCTGAGAAGtgTTTGTCTTTCGCTGAAAACG | " |
| TB_FLAG-swap | GATGTCATGATCTTTATAATCACCGTCATGGTCTTTGTATCCATTTTCTTCTTTCTTTTCTAAA | " |
| swap-FLAG-Dbf4 | GATTATAAAGATCATGACATCGATTACAAGGATGACGATGACAAGGGTGACGGTGCTGGT TTAAG | " |
| Name | Genotype | Source |
|---|---|---|
| SSy161 | MATa ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100 bar1∆::hisG | Viggiani et al. 2006 |
| SSy162 | MATα | " |
| CVy63 | leu2::BrdU-Inc(LEU2) | " |
| CVy70 | MATα ura3::BrdU-Inc(URA3) | " |
| EAy1 | 3xFLAG-DBF4 ADE2::FLOPv2x2 | This Study |
| EAy2 | 3xFLAG-dbf4-Zn*1 (C661A C664A) ADE2::FLOPv2x2 | " |
| HYy143 | MATα fkh1Δ::KANMX fkh2-dsm | " |
| HYy176 | dbf4∆C::KANMX leu2::BrdU-Inc(LEU2) | " |
| HYy207 | ctf19Δ::TRP1(Kl) fkh1Δ::KANMX fkh2-dsm ura3::BrdU-Inc(URA3) | " |
| HYy210 | ctf19Δ::TRP1(Kl) ura3::BrdU-Inc(URA3) | " |
| HYy211 | dbf4∆C::HIS3MX ctf19Δ::TRP1 ura3::BrdU-Inc(URA3) | " |
| HYy215 | dbf4∆C::HIS3MX ura3::BrdU-Inc(URA3) | " |
| HYy217 | fkh1Δ::KANMX ura3::BrdU-Inc(URA3) | " |
| HYy218 | fkh1Δ::KANMX fkh2-dsm ura3::BrdU-Inc(URA3) | " |
| JOSHy1 | dbf4∆C::HIS3MX leu2::BrdU-Inc(LEU2) | " |
| JOSHy2 | dbf4∆C::HIS3MX ura3::BrdU-Inc(URA3) | " |
| MPy35 | ADE2::FLOPv2x2 | " |
| MPy74 | dbf4-Zn*1 (C661A C664A) leu2::BrdU-Inc(LEU2) | " |
| MPy76 | dbf4-Zn*2 (H680C) leu2::BrdU-Inc(LEU2) | " |
| MPy86 | fkh1Δ::URA3MX dbf4-Zn*1 (C661A C664A) leu2::BrdU-Inc(LEU2) | " |
| MPy90 | fkh1Δ::URA3MX dbf4-Zn*2 (H680C) leu2::BrdU-Inc(LEU2) | " |
| MPy125 | 3xFLAG-DBF4 ADE2::FLOPv2x2 | " |
| OAy1123 | MATα fkh1Δ::URA3MX fkh2Δ::HIS3MX | " |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Petrie, M.V.; Zhang, H.; Arnold, E.M.; Gan, Y.; Aparicio, O.M. Dbf4 Zn-Finger Motif Is Specifically Required for Stimulation of Ctf19-Activated Origins in Saccharomyces cerevisiae. Genes 2022, 13, 2202. https://doi.org/10.3390/genes13122202
Petrie MV, Zhang H, Arnold EM, Gan Y, Aparicio OM. Dbf4 Zn-Finger Motif Is Specifically Required for Stimulation of Ctf19-Activated Origins in Saccharomyces cerevisiae. Genes. 2022; 13(12):2202. https://doi.org/10.3390/genes13122202
Chicago/Turabian StylePetrie, Meghan V., Haiyang Zhang, Emily M. Arnold, Yan Gan, and Oscar M. Aparicio. 2022. "Dbf4 Zn-Finger Motif Is Specifically Required for Stimulation of Ctf19-Activated Origins in Saccharomyces cerevisiae" Genes 13, no. 12: 2202. https://doi.org/10.3390/genes13122202
APA StylePetrie, M. V., Zhang, H., Arnold, E. M., Gan, Y., & Aparicio, O. M. (2022). Dbf4 Zn-Finger Motif Is Specifically Required for Stimulation of Ctf19-Activated Origins in Saccharomyces cerevisiae. Genes, 13(12), 2202. https://doi.org/10.3390/genes13122202






