Characterization of the Complete Mitochondrial Genome of the Spotted Catfish Arius maculatus (Thunberg, 1792) and Its Phylogenetic Implications
Abstract
1. Introduction
2. Materials and Methods
2.1. Samples and Mitogenome Sequencing
2.2. Genome Assembly and Annotation
2.3. Genome Sequence Analysis
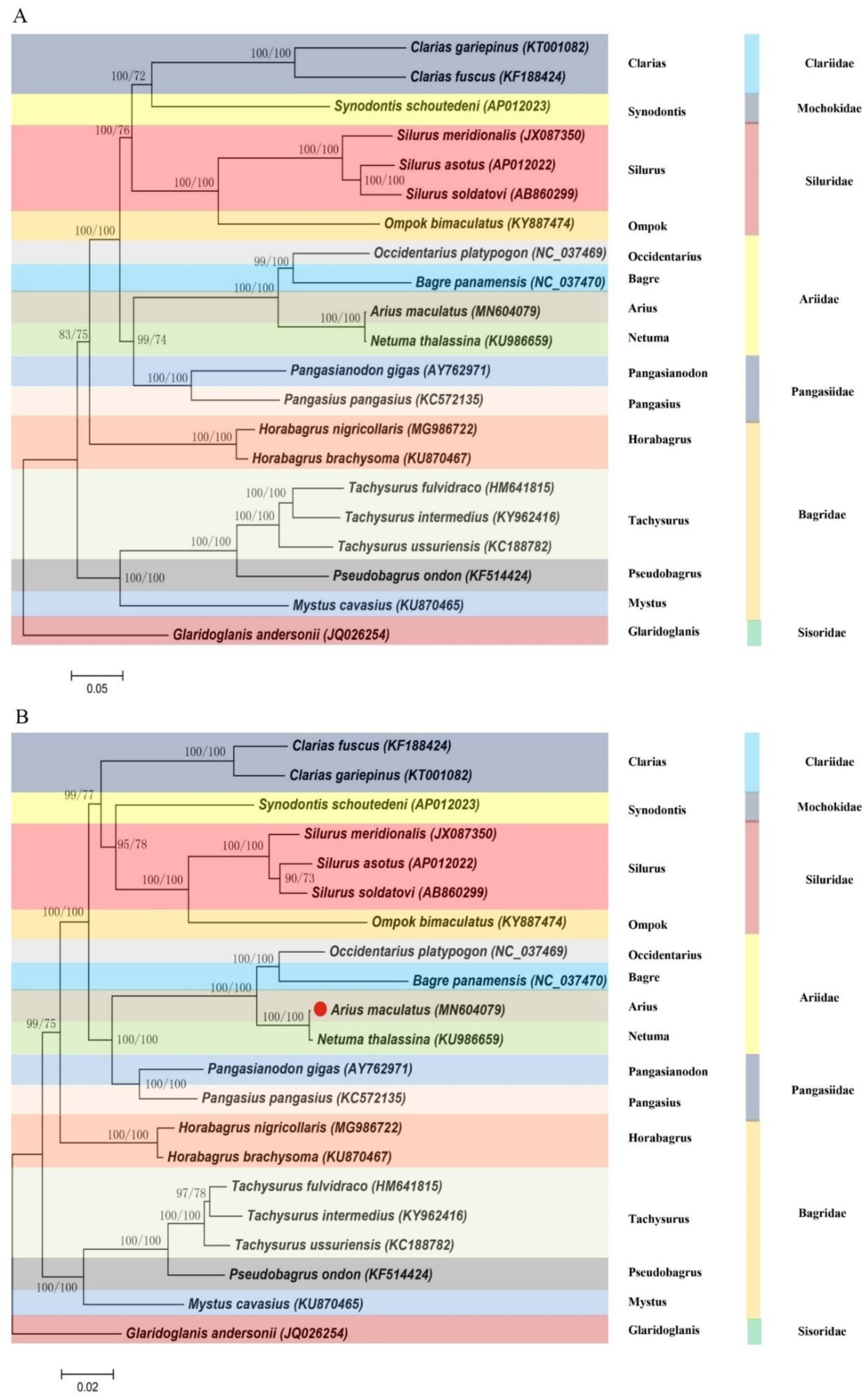
2.4. Phylogenetic Analysis
3. Results and Discussion
3.1. Genome Size and Organization
3.2. Protein-Coding Gene Features
3.3. Transfer RNAs and Ribosomal RNAs
3.4. The Control Region
3.5. Overlapping and Intergenic Spacer Regions
3.6. Synonymous and Nonsynonymous Substitutions
3.7. Phylogeny
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wolstenholme, D.R. Animal mitochondrial DNA: Structure and evolution. Int. Rev. Cytol. 1992, 141, 173–216. [Google Scholar] [CrossRef] [PubMed]
- Lei, R.; Shore, G.D.; Brenneman, R.A.; Engberg, S.E.; Sitzmann, B.D.; Bailey, C.A.; Kimmel, L.M.; Randriamampionona, R.; Ranaivoarisoa, J.F.; Louis, E.J. Complete sequence and gene organization of the mitochondrial genome for Hubbard′s sportive lemur (Lepilemur hubbardorum). Gene 2010, 464, 44–49. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.C.; Gong, F.H.; Liu, T.T.; Guo, H.Y.; Zhang, N.; Zhu, K.C.; Jiang, S.G. Shotgun assembly of the mitochondrial genome from Fenneropenaeus penicillatus with phylogenetic consideration. Mar. Genom. 2015, 24, 379–386. [Google Scholar] [CrossRef] [PubMed]
- Krzywinski, J.; Li, C.; Morris, M.; Conn, J.E.; Lima José, B.; Povoa, M.M.; Wilkerson, R.C. Analysis of the evolutionary forces shaping mitochondrial genomes of a Neotropical malaria vector complex. Mol. Phylogenet. Evol. 2011, 58, 469–477. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhu, K.C.; Liang, Y.Y.; Wu, N.; Guo, H.Y.; Zhang, N.; Jiang, S.G.; Zhang, D.C. Sequencing and characterization of the complete mitochondrial genome of Japanese Swellshark (Cephalloscyllium umbratile). Sci. Rep. 2017, 7, 15299. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Ma, C.; Li, X.; Xu, Z.; Feng, N.; Ma, L. The complete mitochondrial genome sequence and gene organization of the mud crab (Scylla paramamosain) with phylogenetic consideration. Gene 2013, 519, 120–127. [Google Scholar] [CrossRef]
- Cameron, S.L. Insect mitochondrial genomics: Implications for evolution and phylogeny. Annu. Rev. Entomol. 2014, 59, 95–117. [Google Scholar] [CrossRef]
- Chen, H.; Lin, L.L.; Chen, X.; Ai, W.M.; Chen, S.B. Complete mitochondrial genome and the phylogenetic position of the Blotchy swell shark Cephaloscyllium umbratile. Mitochondrial DNA A 2016, 27, 3045–3047. [Google Scholar] [CrossRef]
- Gissi, C.; Iannelli, F.; Pesole, G. Evolution of the mitochondrial genome of metazoa as exemplified by comparison of congeneric species. Heredity 2008, 101, 301–320. [Google Scholar] [CrossRef]
- Oliveira, M.T.; Barau, J.G.; Martins-Junqueira, A.C.; Feijao, P.C.; da Rosa, A.C.; Abreu, C.F.; Azeredo-Espin, A.M.; Lessinger, A.C. Structure and evolution of the mitochondrial genomes of Haematobia irritans and Stomoxis calcitrans: The Muscidae (Diptera: Calyptratae) perspective. Mol. Phylogenet. Evol. 2008, 48, 850–857. [Google Scholar] [CrossRef]
- Behura, S.K.; Lobo, N.F.; Haas, B.; Debruyn, B.; Lovin, D.D.; Shumway, M.F.; Puiu, D.; Romero-Severson, J.; Nene, V.; Severson, D.W. Complete sequences of mitochondria genomes of Aedes aegypti and Culex quinquefasciatus and comparative analysis of mitochondrial DNA fragments inserted in the nuclear genomes. Insect. Biochem. Mol. Biol. 2011, 41, 770–777. [Google Scholar] [CrossRef]
- Cameron, S.L. How to sequence and annotate insect mitochondrial genomes for systematic and comparative genomics research. Syst. Entomol. 2014, 39, 400–411. [Google Scholar] [CrossRef]
- Stepien, C.A.; Kocher, T.D. Molecules and morphology in studies of fish evolution. In Molecular Systematics of Fishes; Elsevier: New York, NY, USA, 1997; pp. 1–11. [Google Scholar]
- Miya, M.; Nishida, M. Use of mitogenomic information in teleostean molecular phylogenetics: A tree-based exploration under the maximumparsimony optimality criterion. Mol. Phylogenet. Evol. 2000, 17, 437–455. [Google Scholar] [CrossRef] [PubMed]
- Chu, W.S.; Hou, Y.Y.; Ueng, Y.T.; Wang, J.P.; Chen, H.C. Estimates of age, growth and mortality of spotted catfish, Arius maculatus (Thunberg, 1792), off the Coast of Yunlin, Southwestern Taiwan. Afr. J. Biotechnol. 2011, 10, 15416–15421. [Google Scholar] [CrossRef]
- Maitra, S.; Harikrishnan, M.; Nidhin, B. Feeding strategy, dietary overlap and resource partitioning among four mesopredatory catfishes of a tropical estuary. J. Fish Biol. 2019, 96, 130–139. [Google Scholar] [CrossRef] [PubMed]
- Ferraris, C.J. Checklist of catfishes, recent and fossil (Osteichthyes: Siluriformes), and catalogue of siluriform primary types. Zootaxa 2007, 1418, 1–628. [Google Scholar] [CrossRef]
- Sczepanski, T.S.; Noleto, R.B.; Cestari, M.M.; Artoni, R.F. A comparative study of two marine catfish (Siluriformes, Ariidae): Cytogenetic tools for determining cytotaxonomy and karyotype evolution. Micron 2010, 41, 193–197. [Google Scholar] [CrossRef]
- Betancur, R.R.; Arturo, A.P.; Bermingham, E.; Cooke, R. Systematics and biogeography of New World sea catfishes (Siluriformes: Ariidae) as inferred from mitochondrial, nuclear, and morphological evidence. Mol. Phylogenet. Evol. 2007, 45, 339–357. [Google Scholar] [CrossRef]
- Betancur, R.R. Molecular phylogenetics and evolutionary history of ariid catfishes revisited: A comprehensive sampling. BMC Evol Biol. 2009, 9, 175. [Google Scholar] [CrossRef]
- Covain, R.; Fisch-Muller, S.; Oliveira, C.; Mol, J.H.; Montoya-Burgos, J.I.; Dray, S. Molecular phylogeny of the highly diversiied catish subfamily Loricariinae (Siluriformes 5421, Loricariidae) reveals incongruences with morphological classification. Mol. Phylogenet. Evol. 2016, 94, 492–517. [Google Scholar] [CrossRef]
- Llera-Herrera, R.; Ramirez-Perez, J.S.; Saavedra-Sotelo, N.C. Complete mitochondrial genome of cominate sea catfish Occidentarius platypogon (Siluriformes: Ariidae). Mitochondrial DNA B 2017, 2, 337–338. [Google Scholar] [CrossRef] [PubMed]
- Ramirez-Perez, J.S.; Saavedra-Sotelo, N.C.; Llera-Herrera, R.; Abadia-Chanona, Q.Y. Complete mitochondrial genome of the Chihuil sea catfish Bagre panamensis (Siluriformes: Ariidae). Mitochondrial DNA B 2017, 2, 341–343. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.F.; Ou, Y.J.; Wen, J.F.; Li, J.E. The complete mitochondrial genome of Arius arius (Siluriformes: Ariidae). Mitochondrial DNA B 2016, 1, 551–552. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tang, M.; Hardman, C.J.; Ji, Y.; Meng, G.; Liu, S.; Tan, M.; Yang, S.; Moss, E.D.; Wang, J.; Yang, C.; et al. High-throughput monitoring of wild bee diversity and abundance via mitogenomics. Methods Ecol. Evol. 2015, 6, 1034–1043. [Google Scholar] [CrossRef] [PubMed]
- Iwasaki, W.; Fukunaga, T.; Isagozawa, R.; Nishida, M.W. MitoFish and MitoAnnotator: A mitochondrial genome database of fish with an accurate and automatic annotation pipeline. Mol. Biol. Evol. 2013, 30, 2531–2540. [Google Scholar] [CrossRef]
- Lowe, T.M.; Eddy, S.R. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997, 25, 955–964. [Google Scholar] [CrossRef]
- Schattner, P.; Brooks, A.N.; Lowe, T.M. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 2007, 33, 686–689. [Google Scholar] [CrossRef]
- Lohse, M.; Drechsel, O.; Kahlau, S.; Bock, R. OrganellarGenomeDRAW a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 2013, 41, 575–581. [Google Scholar] [CrossRef]
- Perna, N.T.; Kocher, T.D. Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. J. Mol. Evol. 1995, 41, 353–358. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Rozas, J. DNA sequence polymorphism analysis using DnaSP. In Bioinformatics for DNA Sequence Analysis; Methods in Molecular Biology Series; Posada, D., Ed.; Humana Press: Totowa, NJ, USA, 2009; Volume 537, pp. 337–350. [Google Scholar]
- Junqueira, A.C.M.; Lessinger, A.C.; Torres, T.T.; da Silva, F.R.; Vettore, A.L.; Arruda, P.; Espin, A.M.L.A. The mitochondrial genome of the blowfly Chrysomya chloropyga (Diptera: Calliphoridae). Gene 2004, 339, 7–15. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Posada, D. jModelTest: Phylogenetic Model Averaging. Mol. Biol. Evol. 2008, 25, 1253–1256. [Google Scholar] [CrossRef] [PubMed]
- Diego, D.; Taboada, G.L.; Doallo, R.; Posada, D. ProtTest 3: Fast selection of best-fit models of protein evolution. Bioinformatics 2011, 27, 1164–1165. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 2006, 22, 2688–2690. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Nakatani, M.; Miya, M.; Mabuchi, K.; Saitoh, K.; Nishida, M. Evolutionary history of Otophysi (Teleostei), a major clade of the modern freshwater fishes: Pangaean origin and Mesozoic radiation. BMC Evol. Biol. 2011, 11, 177. [Google Scholar] [CrossRef]
- Wang, J.L.; Shen, T.; Ju, J.F.; Yang, G.A. The complete mitochondrial genome of the Chinese longsnout catfish Leiocassis longirostris (Siluriformes: Bagridae) and a time-calibrated phylogeny of ostariophysan fishes. Mol. Biol. Rep. 2011, 38, 2507–2516. [Google Scholar] [CrossRef]
- Zhou, C.J.; Wang, X.Z.; Guan, L.H.; He, S.P. The complete mitochondrial genome of Clarias fuscus (Teleostei, Siluriformes: Clariidae). Mitochondrial DNA 2015, 26, 270–271. [Google Scholar] [CrossRef]
- Han, C.; Li, Q.; Xu, J.Q.; Li, X.F.; Huang, J.R. Characterization of Clarias gariepinus mitochondrial genome sequence and a comparative analysis with other catfishes. Biologia 2015, 70, 1245–1253. [Google Scholar] [CrossRef]
- Yang, W.Z.; Zhang, Y.; Feng, S.Q.; Liu, L.J.; Li, Z.H. The first complete mitochondrial genome of the Japanese beetle Popillia japonica (Coleoptera: Scarabaeidae) and its phylogenetic implications for the superfamily Scarabaeoidea. Int. J Biol. Macromol. 2018, 118, 1406–1413. [Google Scholar] [CrossRef] [PubMed]
- Yu, P.; Zhou, L.; Zhou, X.Y.; Yang, W.T.; Zhang, X.J.; Wang, Y.; Gui, J.F. Unusual AT-skew of Sinorhodeus microlepis mitogenome provides new insights into mitogenome features and phylogenetic implications of bitterling fishes. Int. J Biol. Macromol. 2019, 129, 339–350. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.M.; Zhu, K.H.; Jiang, H.; Lu, X.T.; Liu, B.J.; Ye, Y.Y.; Jiang, L.H.; Liu, L.Q.; Gong, L. Complete mitochondrial genome of Ophichthus brevicaudatus reveals novel gene order and phylogenetic relationships of Anguilliformes. Int. J. Biol. Macromol. 2019, 135, 609–618. [Google Scholar] [CrossRef] [PubMed]
- Zhu, T.B.; He, Y.F.; Yang, D.G. The complete mitochondrial genome of the Pseudecheneis immaculatus (Siluriformes:Sisoridae). Mitochondrial DNA B 2019, 4, 3120–3121. [Google Scholar] [CrossRef]
- Alam, M.J.; Andriyono, S.; Lee, S.R.; Hossain, M.A.R.; Eunus, A.T.M.; Hassan, M.T.; Kim, H.W. Characterization of the complete mitochondrial genome of Gangetic ailia, Ailia coila (Siluriformes: Ailiidae). Mitochondrial DNA B 2019, 4, 2258–2259. [Google Scholar] [CrossRef]
- Barman, A.S.; Singh, M.; Pandey, P.K. Complete mitochondrial genome of near threatened butter Catfish Ompok bimaculatus (Siluriformes: Siluridae). Mitochondrial DNA B 2017, 2, 313–314. [Google Scholar] [CrossRef][Green Version]
- Zhou, C.L.; Tian, J.; Yang, C.G. The complete mitochondrial genome sequence of Hemibagrus sp. (Siluriformes: Bagridae), molecular data for species identification. Mitochondrial DNA A 2016, 27, 1914–1915. [Google Scholar] [CrossRef]
- Lian, Z.Q.; Wu, X.D.; Xiao, W.; Sai, Q.Y.; Gun, S.B. Complete sequence and characterization of the Silurus lanzhouensis (Siluriformes: Siluridae) mitochondrial genome. Mitochondrial DNA A 2016, 27, 2483–2484. [Google Scholar] [CrossRef]
- Kim, N.K.; Gietbong, F.Z.; Andriyono, S.; Kim, A.R.; Kim, H.W. The complete mitogenome of Bagrid catfish Chrysichthys nigrodigitatus (Siluriformes: Claroteidae). Mitochondrial DNA B 2018, 3, 1239–1240. [Google Scholar] [CrossRef]
- Janke, A.; Pääbo, S. Editing of a tRNA anticodon in marsupial mitochondria changes its codon recognition. Nucleic. Acids. Res. 1993, 21, 1523–1525. [Google Scholar] [CrossRef]
- Shen, X.; Ren, J.; Cui, Z.; Sha, Z.; Wang, B.; Xiang, J.; Liu, B. The complete mitochondrial genomes of two common shrimps (Litopenaeus vannamei and Fenneropenaeus chinensis) and their phylogenomic considerations. Gene 2007, 403, 98–109. [Google Scholar] [CrossRef] [PubMed]
- Li, X.J.; Huang, Y.; Lei, F.M. Comparative mitochondrial genomics and phylogenetic relationships of the Crossoptilon species (Phasianidae, Galliformes). BMC Genom. 2015, 16, 42. [Google Scholar] [CrossRef] [PubMed]
- Nei, M.; Kumar, S. Molecular Evolution and Phylogenetics; Oxford University Press: New York, NY, USA, 2000. [Google Scholar]
- Yang, Z.; Bielawski, J.P. Statistical methods for detecting molecular adaptation. Trends Ecol Evol. 2000, 15, 496–503. [Google Scholar] [CrossRef]
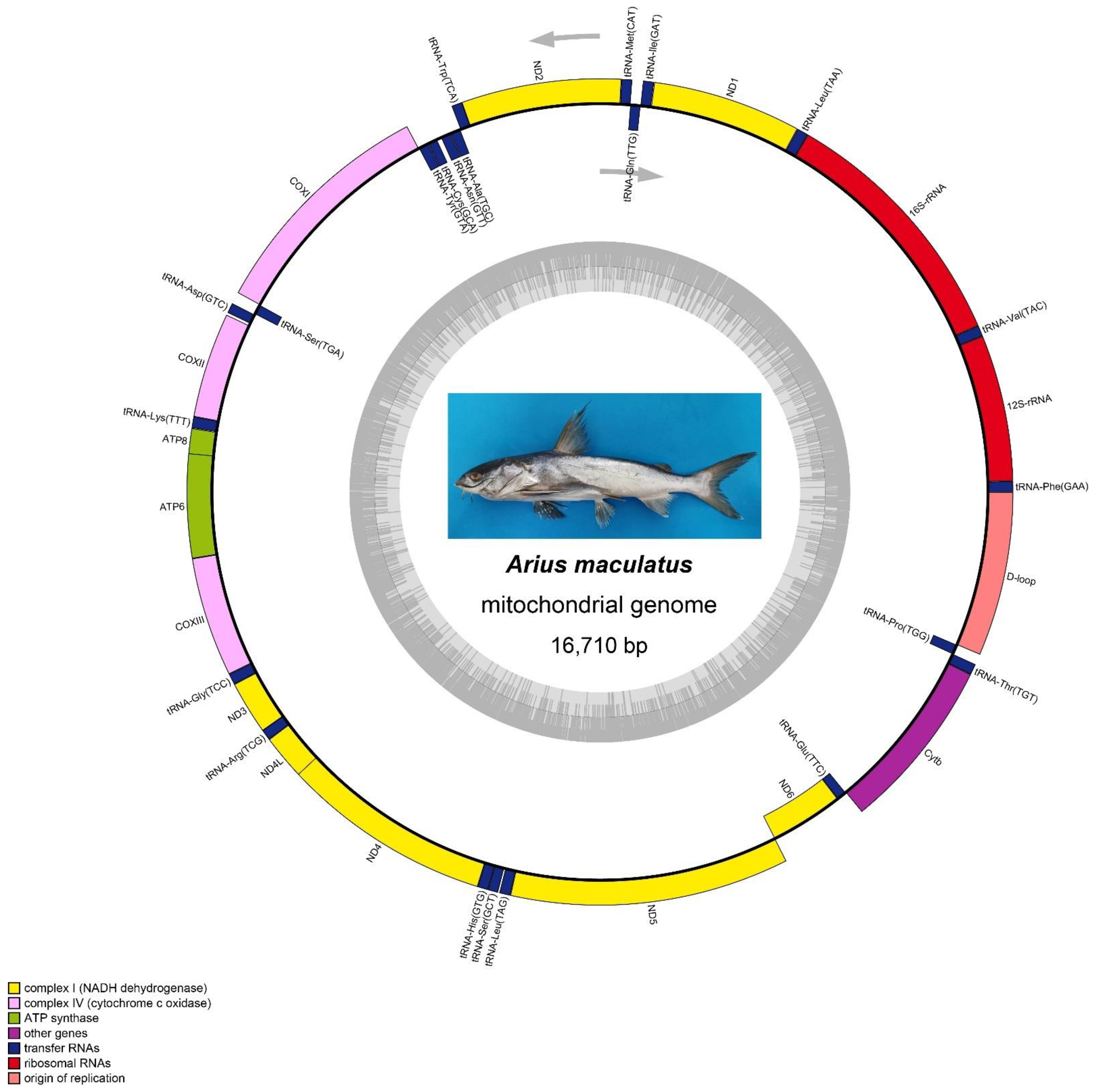

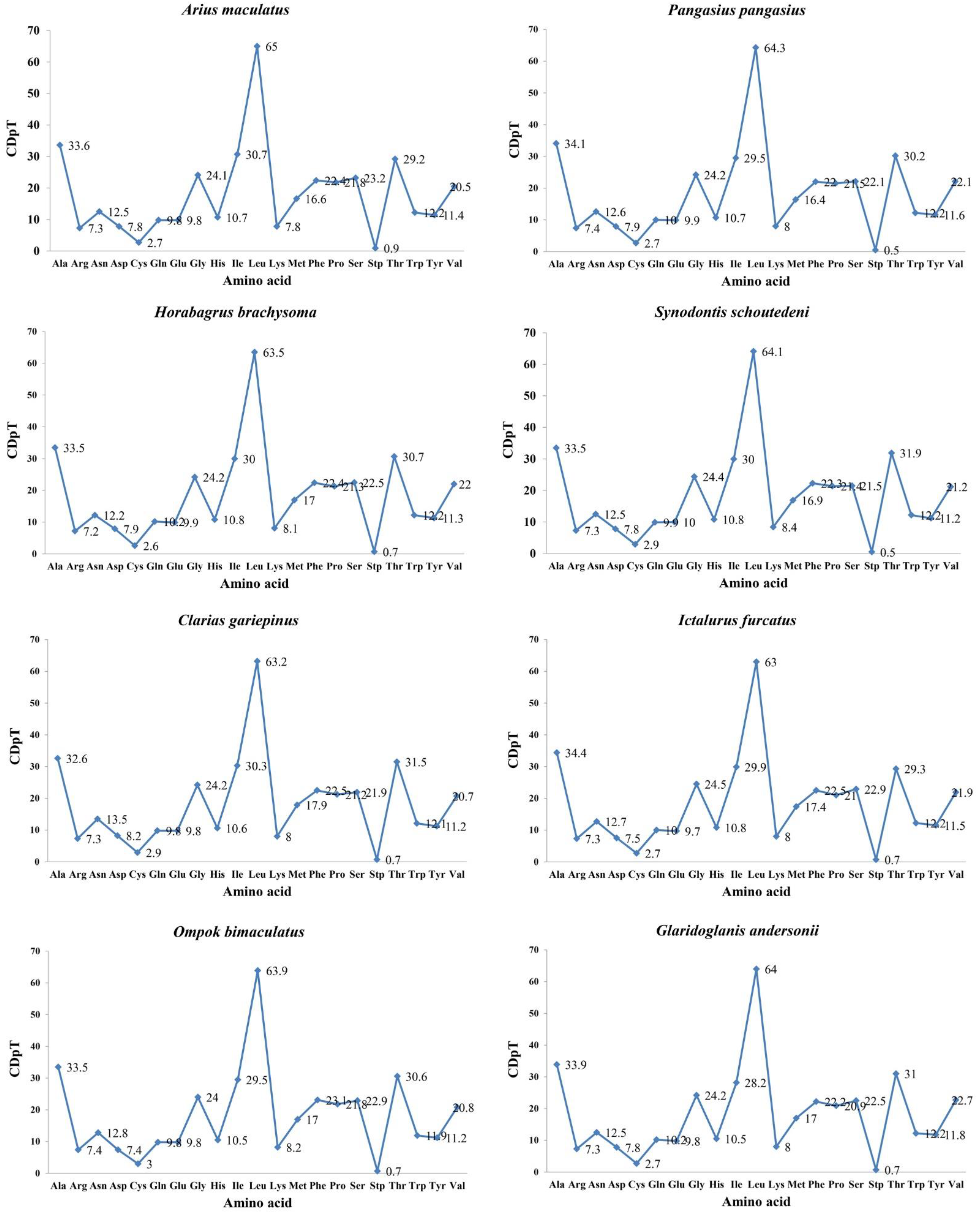

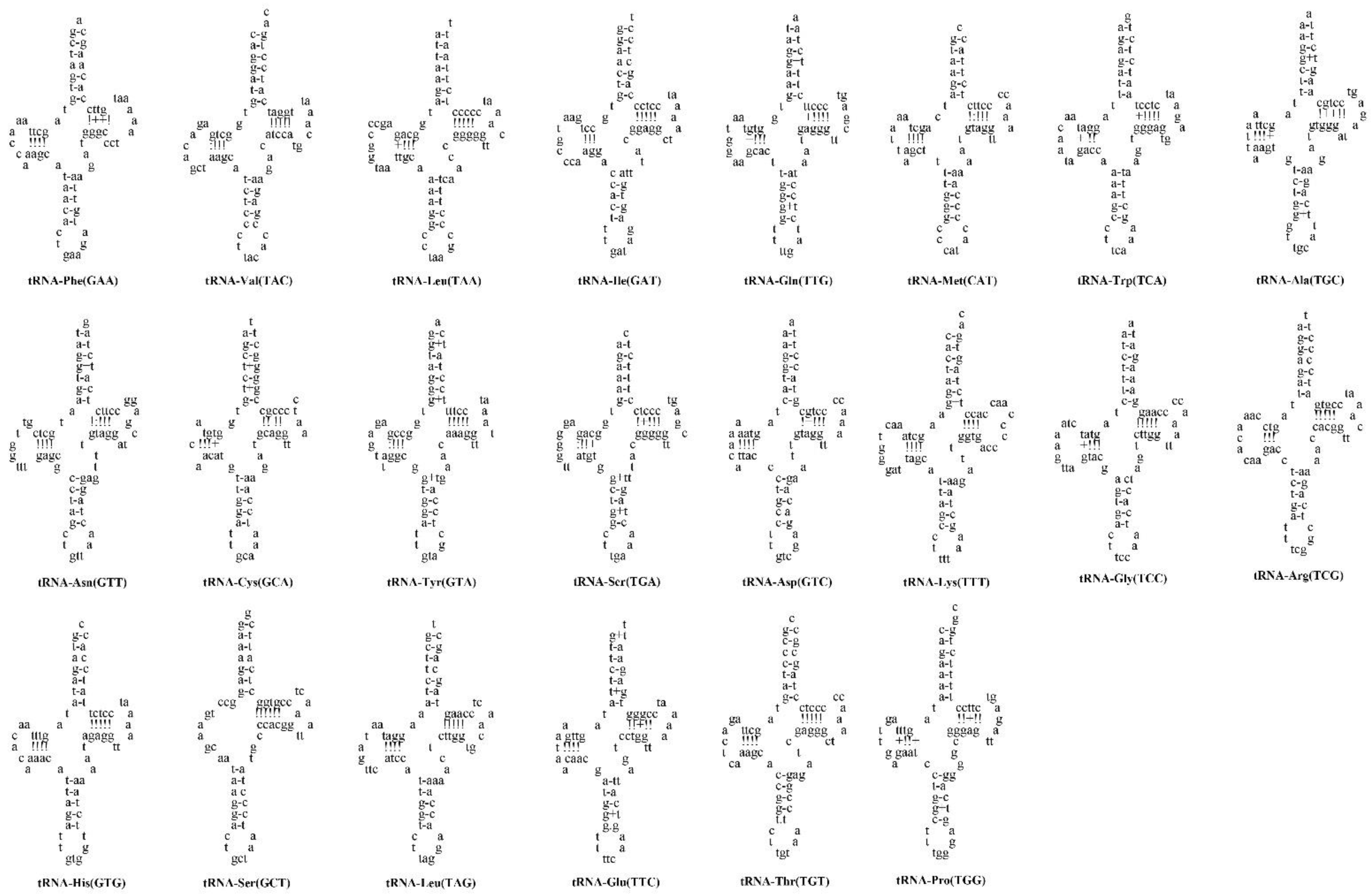
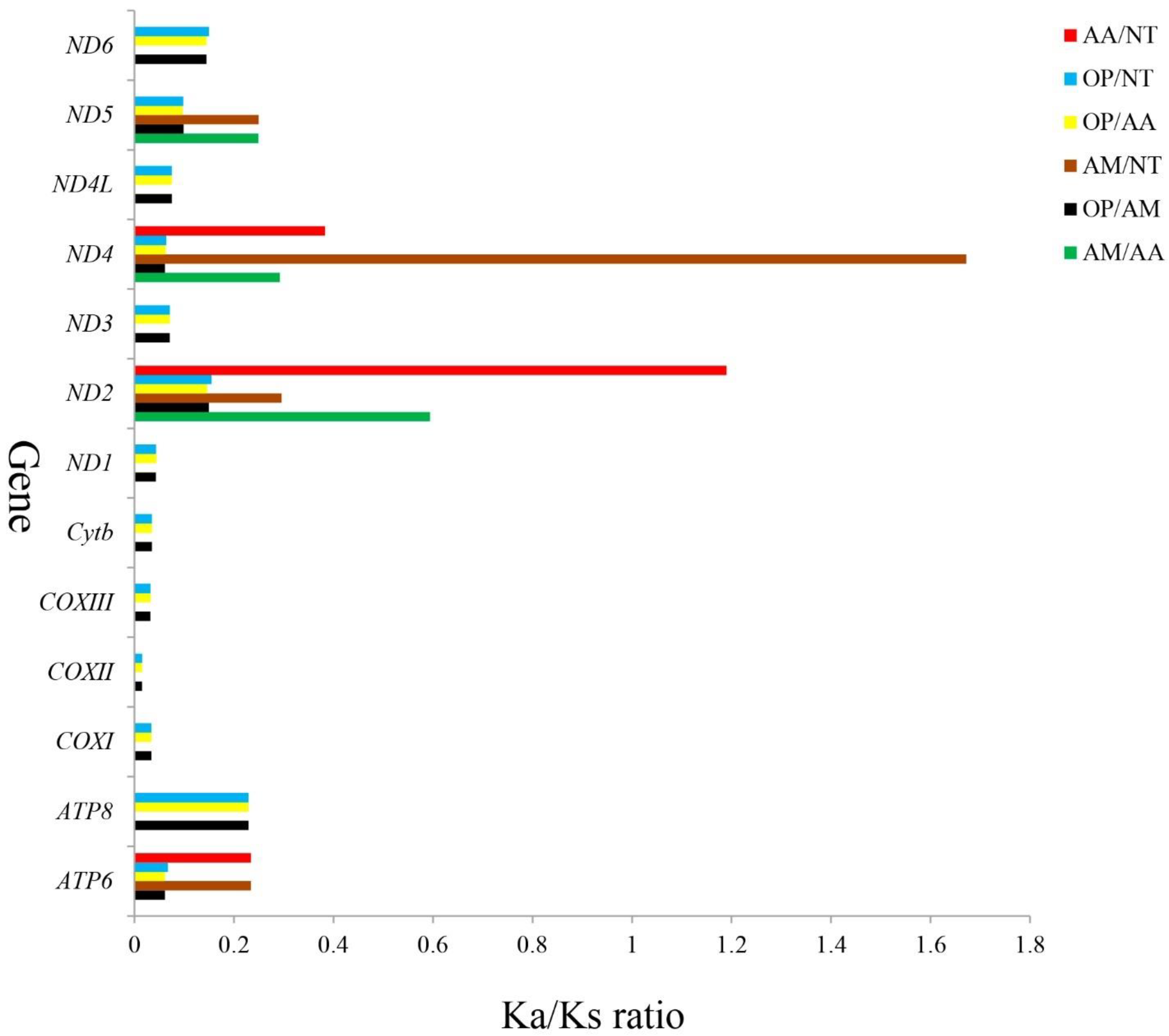
| Locus Name | One Letter Code | From | To | Size | Strand | No. of Amino Acids | Anticodon | Inferred Initiation Codon | Inferred Termination Codon | GC_Percent | Intergenic Nucleotides |
|---|---|---|---|---|---|---|---|---|---|---|---|
| tRNA-Phe | F | 1 | 70 | 70 | H | GAA | 40.00% | 0 | |||
| 12S-rRNA | 71 | 1028 | 958 | H | 49.16% | 0 | |||||
| tRNA-Val | V | 1029 | 1100 | 72 | H | TAC | 48.61% | 0 | |||
| 16S-rRNA | 1101 | 2775 | 1675 | H | 46.15% | 0 | |||||
| tRNA-Leu | L | 2776 | 2850 | 75 | H | TAA | 48.00% | 1 | |||
| ND1 | 2852 | 3826 | 975 | H | 324 | ATG | TAA | 47.08% | 1 | ||
| tRNA-Ile | I | 3828 | 3899 | 72 | H | GAT | 50.00% | −1 | |||
| tRNA-Gln | Q | 3899 | 3969 | 71 | L | TTG | 43.66% | −1 | |||
| tRNA-Met | M | 3969 | 4038 | 70 | H | CAT | 40.00% | 0 | |||
| ND2 | 4039 | 5085 | 1047 | H | 348 | ATG | TAG | 45.75% | −2 | ||
| tRNA-Trp | W | 5084 | 5154 | 71 | H | TCA | 38.03% | 2 | |||
| tRNA-Ala | A | 5157 | 5225 | 69 | L | TGC | 40.58% | 1 | |||
| tRNA-Asn | N | 5227 | 5299 | 73 | L | GTT | 47.95% | 31 | |||
| tRNA-Cys | C | 5331 | 5396 | 66 | L | GCA | 50.00% | 1 | |||
| tRNA-Tyr | Y | 5398 | 5467 | 70 | L | GTA | 47.14% | 1 | |||
| COXI | 5469 | 7019 | 1551 | H | 516 | GTG | TAA | 44.36% | 0 | ||
| tRNA-Ser | S | 7020 | 7090 | 71 | L | TGA | 52.11% | 4 | |||
| tRNA-Asp | D | 7095 | 7163 | 69 | H | GTC | 42.03% | 14 | |||
| COXII | 7178 | 7868 | 691 | H | 230 | ATG | T | 42.11% | 0 | ||
| tRNA-Lys | K | 7869 | 7942 | 74 | H | TTT | 44.59% | 1 | |||
| ATP8 | 7944 | 8111 | 168 | H | 55 | ATG | TAA | 38.69% | −10 | ||
| ATP6 | 8102 | 8785 | 684 | H | 227 | ATG | TAA | 42.69% | −1 | ||
| COXIII | 8785 | 9568 | 784 | H | 261 | ATG | T | 47.19% | 0 | ||
| tRNA-Gly | G | 9569 | 9641 | 73 | H | TCC | 36.99% | 0 | |||
| ND3 | 9642 | 9992 | 351 | H | 116 | ATG | TAG | 45.87% | −2 | ||
| tRNA-Arg | R | 9991 | 10,061 | 71 | H | TCG | 46.48% | 0 | |||
| ND4L | 10,062 | 10,358 | 297 | H | 98 | ATG | TAA | 49.83% | −7 | ||
| ND4 | 10,352 | 11,732 | 1381 | H | 460 | ATG | T | 45.84% | 0 | ||
| tRNA-His | H | 11,733 | 11,802 | 70 | H | GTG | 30.00% | 0 | |||
| tRNA-Ser | S | 11,803 | 11,869 | 67 | H | GCT | 50.75% | 8 | |||
| tRNA-Leu | L | 11,878 | 11,950 | 73 | H | TAG | 41.10% | 0 | |||
| ND5 | 11,951 | 13,777 | 1827 | H | 608 | ATG | TAA | 43.13% | −4 | ||
| ND6 | 13,774 | 14,286 | 513 | L | 170 | ATG | TAG | 47.76% | 0 | ||
| tRNA-Glu | E | 14,287 | 14,355 | 69 | L | TTC | 36.23% | 1 | |||
| Cytb | 14,357 | 15,494 | 1138 | H | 379 | ATG | T | 46.40% | 0 | ||
| tRNA-Thr | T | 15,495 | 15,566 | 72 | H | TGT | 56.94% | −2 | |||
| tRNA-Pro | P | 15,565 | 15,634 | 70 | L | TGG | 47.14% | 0 | |||
| D-loop | □ | 15,635 | 16,710 | 1076 | H | □ | □ | □ | □ | 37.45% | 0 |
| Species | Size (bp) | A% | T% | G% | C% | A + T % | AT Skewness | GC Skewness |
|---|---|---|---|---|---|---|---|---|
| Whole Mitogenome | ||||||||
| A. maculatus | 16,710 | 29.63 | 25.42 | 29.65 | 15.3 | 55.05 | 0.0764 | −0.3194 |
| S. seemanni | 16,830 | 30.21 | 26.68 | 27.88 | 14.63 | 56.89 | 0.0619 | −0.3115 |
| P. ondon | 16,534 | 31.06 | 25.72 | 27.97 | 15.24 | 56.78 | 0.094 | −0.2946 |
| P. pangasius | 16,476 | 30.48 | 25.09 | 28.74 | 15.68 | 55.57 | 0.097 | −0.294 |
| P. ussuriensis | 16,536 | 31.79 | 26.84 | 26.5 | 14.87 | 58.63 | 0.0845 | −0.2811 |
| B. panamensis | 16,718 | 30.75 | 26.64 | 28.17 | 14.34 | 57.39 | 0.0716 | −0.3253 |
| C. gariepinus | 16,508 | 32.53 | 24.68 | 27.96 | 14.84 | 57.21 | 0.1372 | −0.3066 |
| I. furcatus | 16,499 | 29.35 | 25.37 | 29.17 | 16.1 | 54.72 | 0.0728 | −0.2886 |
| G. andersonii | 16,532 | 31.24 | 24.73 | 28.51 | 15.52 | 55.97 | 0.1163 | −0.2951 |
| O. bimaculatus | 16,482 | 31.67 | 25.13 | 28.3 | 14.9 | 56.8 | 0.1151 | −0.3101 |
| A. occidentalis | 16,535 | 31.01 | 25.23 | 28.91 | 14.85 | 56.24 | 0.1029 | −0.3212 |
| O. platypogon | 16,714 | 30.73 | 26.37 | 28.54 | 14.35 | 57.1 | 0.0765 | −0.3308 |
| P. gigas | 16,533 | 30.42 | 25.5 | 28.42 | 15.66 | 55.92 | 0.0879 | −0.2895 |
| N. thalassina | 16,711 | 29.65 | 25.38 | 29.69 | 15.28 | 55.03 | 0.0775 | −0.3204 |
| S. soldatovi | 16,527 | 30.45 | 25.49 | 28.2 | 15.86 | 55.94 | 0.0886 | −0.2799 |
| S. schoutedeni | 16,540 | 31.44 | 24.53 | 28.98 | 15.05 | 55.97 | 0.1235 | −0.3162 |
| P. fulvidraco | 16,527 | 30.83 | 25.53 | 28.23 | 15.41 | 56.36 | 0.0941 | −0.2937 |
| M. cavasius | 16,554 | 31.93 | 25.73 | 27.4 | 14.95 | 57.66 | 0.1075 | −0.2942 |
| H. brachysoma | 16,567 | 31.16 | 25.35 | 28.15 | 15.34 | 56.51 | 0.1028 | −0.2944 |
| Protein-Coding Genes | ||||||||
| A. maculatus | 11,407 | 28.78 | 26.10 | 31.33 | 13.79 | 54.88 | 0.0489 | −0.3888 |
| S. seemanni | 11,403 | 29.59 | 27.84 | 29.44 | 13.13 | 57.43 | 0.0304 | −0.3832 |
| P. ondon | 11,406 | 30.18 | 26.38 | 29.58 | 13.86 | 56.56 | 0.0671 | −0.3619 |
| P. pangasius | 11,407 | 29.44 | 25.77 | 30.58 | 14.21 | 55.21 | 0.0664 | −0.3654 |
| P. ussuriensis | 11,406 | 31.12 | 27.85 | 27.67 | 13.37 | 58.96 | 0.0555 | −0.3484 |
| B. panamensis | 11,397 | 29.91 | 27.66 | 29.68 | 12.74 | 57.58 | 0.039 | −0.3994 |
| C. gariepinus | 11,409 | 32.24 | 25.23 | 29.34 | 13.19 | 57.47 | 0.1219 | −0.3796 |
| I. furcatus | 11,403 | 28.23 | 26.06 | 30.90 | 14.81 | 54.29 | 0.0399 | −0.3519 |
| G. andersonii | 11,409 | 30.42 | 25.16 | 30.41 | 14.00 | 55.59 | 0.0946 | −0.3696 |
| O. bimaculatus | 11,403 | 31.04 | 25.93 | 29.93 | 13.09 | 56.98 | 0.0897 | −0.3914 |
| A. occidentalis | 11,405 | 30.04 | 25.76 | 30.86 | 13.34 | 55.8 | 0.0767 | −0.3965 |
| O. platypogon | 11,403 | 30.01 | 27.19 | 30.13 | 12.66 | 57.2 | 0.0492 | −0.4082 |
| P. gigas | 11,411 | 29.52 | 26.11 | 30.16 | 14.21 | 55.63 | 0.0614 | −0.3597 |
| N. thalassina | 11,403 | 28.76 | 26.08 | 31.37 | 13.79 | 54.84 | 0.0488 | −0.3891 |
| S. soldatovi | 11,409 | 29.29 | 26.62 | 29.63 | 14.46 | 55.91 | 0.0478 | −0.3439 |
| S. schoutedeni | 11,432 | 30.99 | 24.93 | 30.77 | 13.30 | 55.92 | 0.1084 | −0.3963 |
| P. fulvidraco | 11,406 | 29.86 | 26.09 | 30.00 | 14.05 | 55.95 | 0.0674 | −0.3623 |
| M. cavasius | 11,406 | 31.18 | 26.18 | 29.20 | 13.45 | 57.36 | 0.0871 | −0.3692 |
| H. brachysoma | 11,409 | 30.29 | 25.86 | 29.86 | 13.99 | 56.15 | 0.079 | −0.362 |
| tRNA | ||||||||
| A. maculatus | 1558 | 31.32 | 24.20 | 25.87 | 18.61 | 55.52 | 0.1283 | −0.1631 |
| S. seemanni | 1579 | 32.24 | 24.70 | 25.27 | 17.80 | 56.93 | 0.1324 | −0.1735 |
| P. ondon | 1568 | 32.14 | 25.00 | 24.23 | 18.62 | 57.14 | 0.125 | −0.131 |
| P. pangasius | 1564 | 31.71 | 24.87 | 24.42 | 18.99 | 56.59 | 0.1209 | −0.1252 |
| P. ussuriensis | 1566 | 32.82 | 25.10 | 24.07 | 18.01 | 57.92 | 0.1334 | −0.1442 |
| B. panamensis | 1561 | 31.96 | 24.33 | 25.80 | 17.91 | 56.29 | 0.1357 | −0.1806 |
| C. gariepinus | 1560 | 31.60 | 24.94 | 24.68 | 18.78 | 56.54 | 0.1179 | −0.1357 |
| I. furcatus | 1559 | 31.49 | 25.08 | 24.50 | 18.92 | 56.57 | 0.1134 | −0.1285 |
| G. andersonii | 1553 | 31.62 | 24.53 | 24.79 | 19.06 | 56.15 | 0.1261 | −0.1307 |
| O. bimaculatus | 1557 | 32.18 | 24.79 | 24.41 | 18.63 | 56.97 | 0.1297 | −0.1343 |
| A. occidentalis | 1561 | 31.65 | 24.86 | 24.98 | 18.51 | 56.5 | 0.1202 | −0.1487 |
| O. platypogon | 1562 | 31.82 | 24.71 | 25.42 | 18.05 | 56.53 | 0.1257 | −0.1694 |
| P. gigas | 1559 | 30.98 | 25.02 | 24.70 | 19.31 | 56 | 0.1065 | −0.1224 |
| N. thalassina | 1558 | 31.32 | 24.20 | 25.87 | 18.61 | 55.52 | 0.1283 | −0.1631 |
| S. soldatovi | 1562 | 31.24 | 24.20 | 25.54 | 19.01 | 55.44 | 0.127 | −0.1466 |
| S. schoutedeni | 1560 | 31.79 | 24.87 | 25.13 | 18.21 | 56.67 | 0.1222 | −0.1598 |
| P. fulvidraco | 1568 | 32.33 | 24.68 | 24.30 | 18.69 | 57.02 | 0.1342 | −0.1306 |
| M. cavasius | 1561 | 32.03 | 25.30 | 24.22 | 18.45 | 57.34 | 0.1173 | −0.1351 |
| H. brachysoma | 1561 | 32.03 | 24.92 | 24.54 | 18.51 | 56.95 | 0.1249 | −0.1399 |
| rRNA | ||||||||
| A. maculatus | 2633 | 32.62 | 20.13 | 27.08 | 20.17 | 52.75 | 0.2369 | −0.1463 |
| S. seemanni | 2635 | 33.17 | 21.10 | 25.84 | 19.89 | 54.27 | 0.2224 | −0.1303 |
| P. ondon | 2632 | 34.80 | 21.66 | 23.97 | 19.57 | 56.46 | 0.2328 | −0.1012 |
| P. pangasius | 2633 | 33.38 | 20.74 | 25.48 | 20.40 | 54.12 | 0.2337 | −0.1109 |
| P. ussuriensis | 2636 | 34.98 | 21.93 | 23.56 | 19.54 | 56.9 | 0.2293 | −0.0933 |
| B. panamensis | 2636 | 33.69 | 21.48 | 25.36 | 19.47 | 55.17 | 0.2212 | −0.1315 |
| C. gariepinus | 2627 | 34.64 | 20.25 | 25.43 | 19.68 | 54.89 | 0.2621 | −0.1274 |
| I. furcatus | 2614 | 32.44 | 21.12 | 25.71 | 20.73 | 53.56 | 0.2114 | −0.1071 |
| G. andersonii | 2630 | 33.95 | 20.68 | 25.32 | 20.04 | 54.64 | 0.2429 | −0.1165 |
| O. bimaculatus | 2616 | 33.98 | 20.68 | 25.38 | 19.95 | 54.66 | 0.2434 | −0.1197 |
| A. occidentalis | 2648 | 34.48 | 20.96 | 25.04 | 19.52 | 55.44 | 0.2439 | −0.1237 |
| O. platypogon | 2627 | 33.12 | 20.94 | 26.19 | 19.76 | 54.05 | 0.2254 | −0.14 |
| P. gigas | 2633 | 33.42 | 21.34 | 24.91 | 20.32 | 54.77 | 0.2205 | −0.1016 |
| N. thalassina | 2633 | 32.66 | 20.02 | 27.19 | 20.13 | 52.68 | 0.2401 | −0.1493 |
| S. soldatovi | 2627 | 34.60 | 20.67 | 24.90 | 19.83 | 55.27 | 0.2521 | −0.1132 |
| S. schoutedeni | 2639 | 33.95 | 20.27 | 25.77 | 20.01 | 54.23 | 0.2523 | −0.1258 |
| P. fulvidraco | 2631 | 34.70 | 21.63 | 24.02 | 19.65 | 56.33 | 0.2321 | −0.1001 |
| M. cavasius | 2626 | 34.69 | 22.54 | 23.15 | 19.61 | 57.24 | 0.2122 | −0.0828 |
| H. brachysoma | 2634 | 34.17 | 21.18 | 25.06 | 19.59 | 55.35 | 0.2346 | −0.1224 |
| Control Region | ||||||||
| A. maculatus | 1076 | 29.46 | 33.09 | 23.05 | 14.41 | 62.55 | −0.0579 | −0.2308 |
| S. seemanni | 1186 | 27.57 | 30.86 | 20.15 | 12.98 | 58.43 | −0.0563 | −0.2163 |
| P. ondon | 891 | 29.97 | 30.98 | 24.92 | 14.14 | 60.94 | −0.0166 | −0.2759 |
| P. pangasius | 87 | 33.33 | 29.89 | 21.84 | 14.94 | 63.22 | 0.0545 | −0.1875 |
| P. ussuriensis | 892 | 29.82 | 31.50 | 23.99 | 14.69 | 61.32 | −0.0274 | −0.2406 |
| B. panamensis | 1080 | 32.04 | 32.31 | 22.13 | 13.52 | 64.35 | −0.0043 | −0.2416 |
| C. gariepinus | 864 | 32.29 | 30.44 | 22.69 | 14.58 | 62.73 | 0.0295 | −0.2174 |
| I. furcatus | 886 | 31.83 | 30.14 | 24.38 | 13.66 | 61.96 | 0.0273 | −0.2819 |
| G. andersonii | 101 | 36.63 | 35.64 | 17.82 | 9.90 | 72.28 | 0.0137 | −0.2857 |
| O. bimaculatus | 864 | 32.52 | 28.82 | 22.11 | 16.55 | 61.34 | 0.0604 | −0.1437 |
| A. occidentalis | 881 | 32.46 | 31.56 | 22.36 | 13.62 | 64.02 | 0.0142 | −0.2429 |
| O. platypogon | 1076 | 31.60 | 33.27 | 21.65 | 13.48 | 64.87 | −0.0258 | −0.2328 |
| P. gigas | 899 | 32.70 | 30.92 | 22.25 | 14.13 | 63.63 | 0.028 | −0.2232 |
| N. thalassina | 1077 | 29.81 | 32.96 | 23.12 | 14.11 | 62.77 | −0.0503 | −0.2419 |
| S. soldatovi | 891 | 32.32 | 27.39 | 24.13 | 16.16 | 59.71 | 0.0827 | −0.1978 |
| S. schoutedeni | 896 | 29.24 | 31.92 | 21.99 | 16.85 | 61.16 | −0.0438 | −0.1322 |
| P. fulvidraco | 888 | 29.96 | 31.31 | 24.21 | 14.53 | 61.26 | −0.0221 | −0.25 |
| M. cavasius | 910 | 33.85 | 29.78 | 22.09 | 14.29 | 63.63 | 0.0639 | −0.2145 |
| H. brachysoma | 925 | 32.65 | 31.35 | 21.73 | 14.27 | 64 | 0.0203 | −0.2072 |
| Amino Acid | Codon | Number | Frequency (%) | RSCU | Amino Acid | Codon | Number | Frequency (%) | RSCU |
|---|---|---|---|---|---|---|---|---|---|
| Ala | GCC | 161 | 4.24 | 1.92 | CAT | 23 | 0.61 | 0.43 | |
| GCA | 100 | 2.63 | 1.19 | Ile | ATC | 156 | 4.10 | 1.02 | |
| GCT | 67 | 1.76 | 0.80 | ATT | 151 | 3.97 | 0.98 | ||
| GCG | 8 | 0.21 | 0.10 | Leu | CTA | 274 | 7.21 | 2.53 | |
| Arg | CGA | 39 | 1.03 | 2.14 | CTC | 138 | 3.63 | 1.27 | |
| CGC | 16 | 0.42 | 0.88 | TTA | 102 | 2.68 | 0.94 | ||
| CGG | 11 | 0.29 | 0.60 | CTT | 78 | 2.05 | 0.72 | ||
| CGT | 7 | 0.18 | 0.38 | CTG | 41 | 1.08 | 0.38 | ||
| Asn | AAC | 79 | 2.08 | 1.26 | TTG | 17 | 0.45 | 0.16 | |
| AAT | 46 | 1.21 | 0.74 | Lys | AAA | 75 | 1.97 | 1.92 | |
| Asp | GAC | 53 | 1.39 | 1.36 | AAG | 3 | 0.08 | 0.08 | |
| GAT | 25 | 0.66 | 0.64 | Met | ATA | 120 | 3.16 | 1.45 | |
| Cys | TGC | 15 | 0.39 | 1.11 | ATG | 46 | 1.21 | 0.55 | |
| TGT | 12 | 0.32 | 0.89 | Phe | TTC | 130 | 3.42 | 1.16 | |
| Gln | CAA | 86 | 2.26 | 1.76 | TTT | 94 | 2.47 | 0.84 | |
| CAG | 12 | 0.32 | 0.24 | Pro | CCC | 90 | 2.37 | 1.65 | |
| GAA | 83 | 2.18 | 1.69 | CCA | 86 | 2.26 | 1.58 | ||
| GAG | 15 | 0.39 | 0.31 | CCT | 38 | 1.00 | 0.70 | ||
| Gly | GGA | 102 | 2.68 | 1.69 | CCG | 4 | 0.11 | 0.07 | |
| GGC | 88 | 2.32 | 1.46 | Ser | TCC | 86 | 2.26 | 2.22 | |
| GGG | 27 | 0.71 | 0.45 | TCA | 70 | 1.84 | 1.81 | ||
| GGT | 24 | 0.63 | 0.40 | AGC | 38 | 1.00 | 0.98 | ||
| His | CAC | 84 | 2.21 | 1.57 | TCT | 27 | 0.71 | 0.70 | |
| AGT | 6 | 0.16 | 0.16 | ACG | 4 | 0.11 | 0.05 | ||
| TCG | 5 | 0.13 | 0.13 | Trp | TGA | 99 | 2.60 | 1.62 | |
| Stp | TAA | 6 | 0.16 | 2.67 | TGG | 23 | 0.61 | 0.38 | |
| TAG | 3 | 0.08 | 1.33 | Tyr | TAC | 72 | 1.89 | 1.26 | |
| AGA | 0 | 0.00 | 0.00 | TAT | 42 | 1.10 | 0.74 | ||
| AGG | 0 | 0.00 | 0.00 | Val | GTA | 77 | 2.03 | 1.50 | |
| Thr | ACC | 121 | 3.18 | 1.66 | GTT | 55 | 1.45 | 1.07 | |
| ACA | 113 | 2.97 | 1.55 | GTC | 51 | 1.34 | 0.99 | ||
| ACT | 54 | 1.42 | 0.74 | GTG | 23 | 0.61 | 0.45 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, M.; Yang, Z.; Liu, C.; Lee, X.; Zhu, K. Characterization of the Complete Mitochondrial Genome of the Spotted Catfish Arius maculatus (Thunberg, 1792) and Its Phylogenetic Implications. Genes 2022, 13, 2128. https://doi.org/10.3390/genes13112128
Yang M, Yang Z, Liu C, Lee X, Zhu K. Characterization of the Complete Mitochondrial Genome of the Spotted Catfish Arius maculatus (Thunberg, 1792) and Its Phylogenetic Implications. Genes. 2022; 13(11):2128. https://doi.org/10.3390/genes13112128
Chicago/Turabian StyleYang, Min, Zimin Yang, Cuiyu Liu, Xuezhu Lee, and Kecheng Zhu. 2022. "Characterization of the Complete Mitochondrial Genome of the Spotted Catfish Arius maculatus (Thunberg, 1792) and Its Phylogenetic Implications" Genes 13, no. 11: 2128. https://doi.org/10.3390/genes13112128
APA StyleYang, M., Yang, Z., Liu, C., Lee, X., & Zhu, K. (2022). Characterization of the Complete Mitochondrial Genome of the Spotted Catfish Arius maculatus (Thunberg, 1792) and Its Phylogenetic Implications. Genes, 13(11), 2128. https://doi.org/10.3390/genes13112128






