Seropositivity-Dependent Association between LINE-1 Methylation and Response to Methotrexate Therapy in Early Rheumatoid Arthritis Patients
Abstract
1. Introduction
2. Materials and Methods
3. Results
3.1. Patient Characteristics and Therapy Response
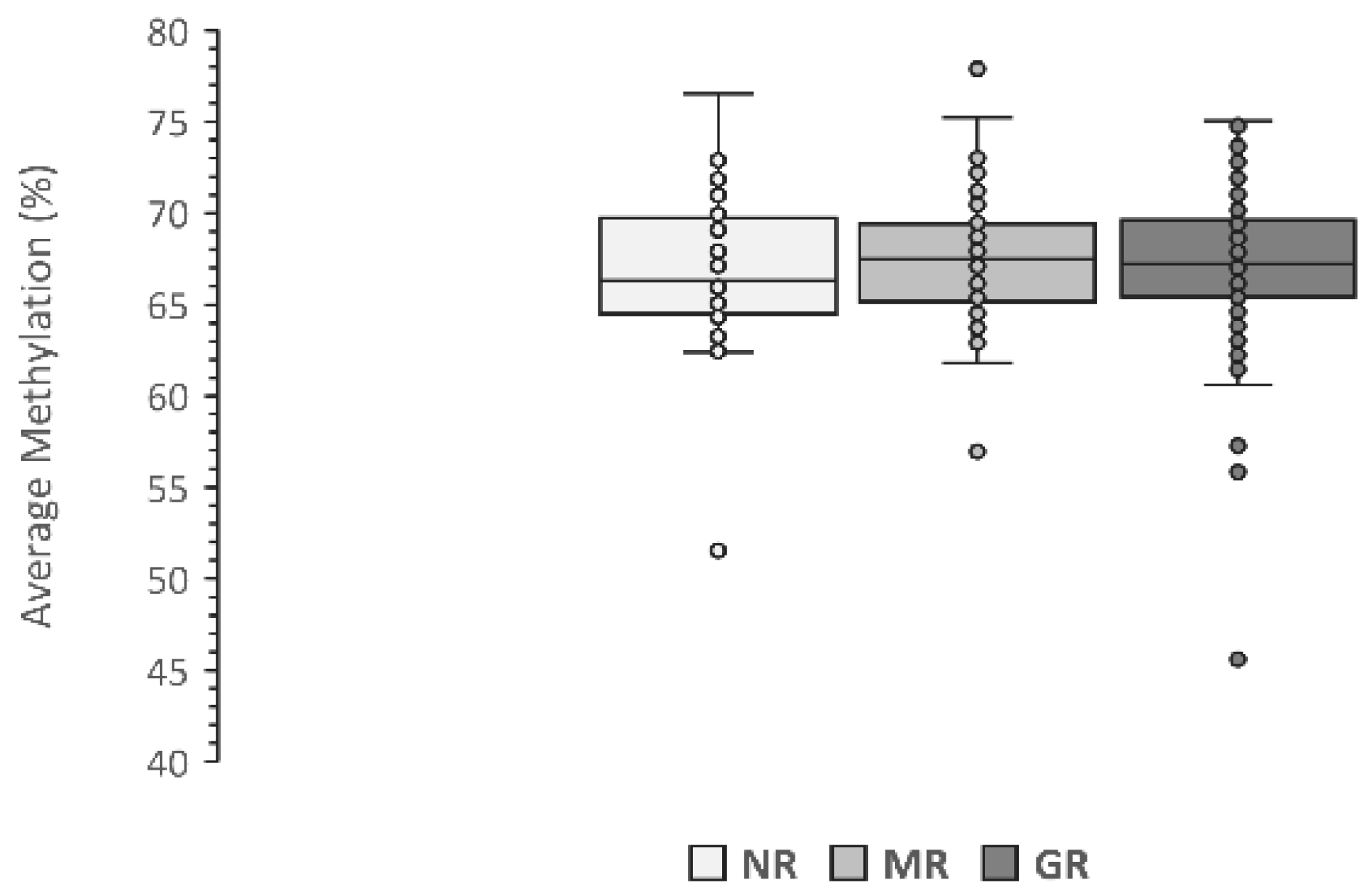
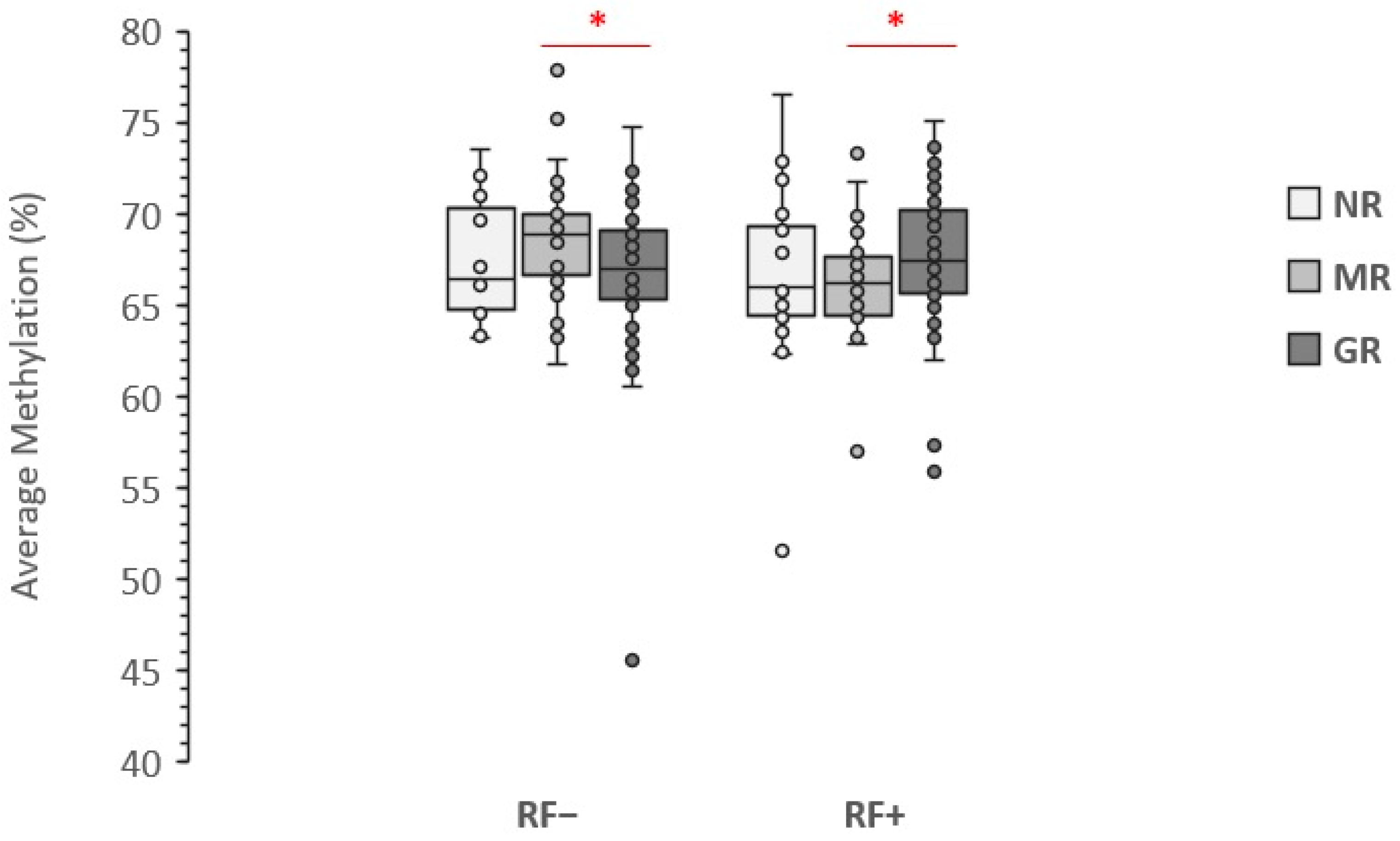
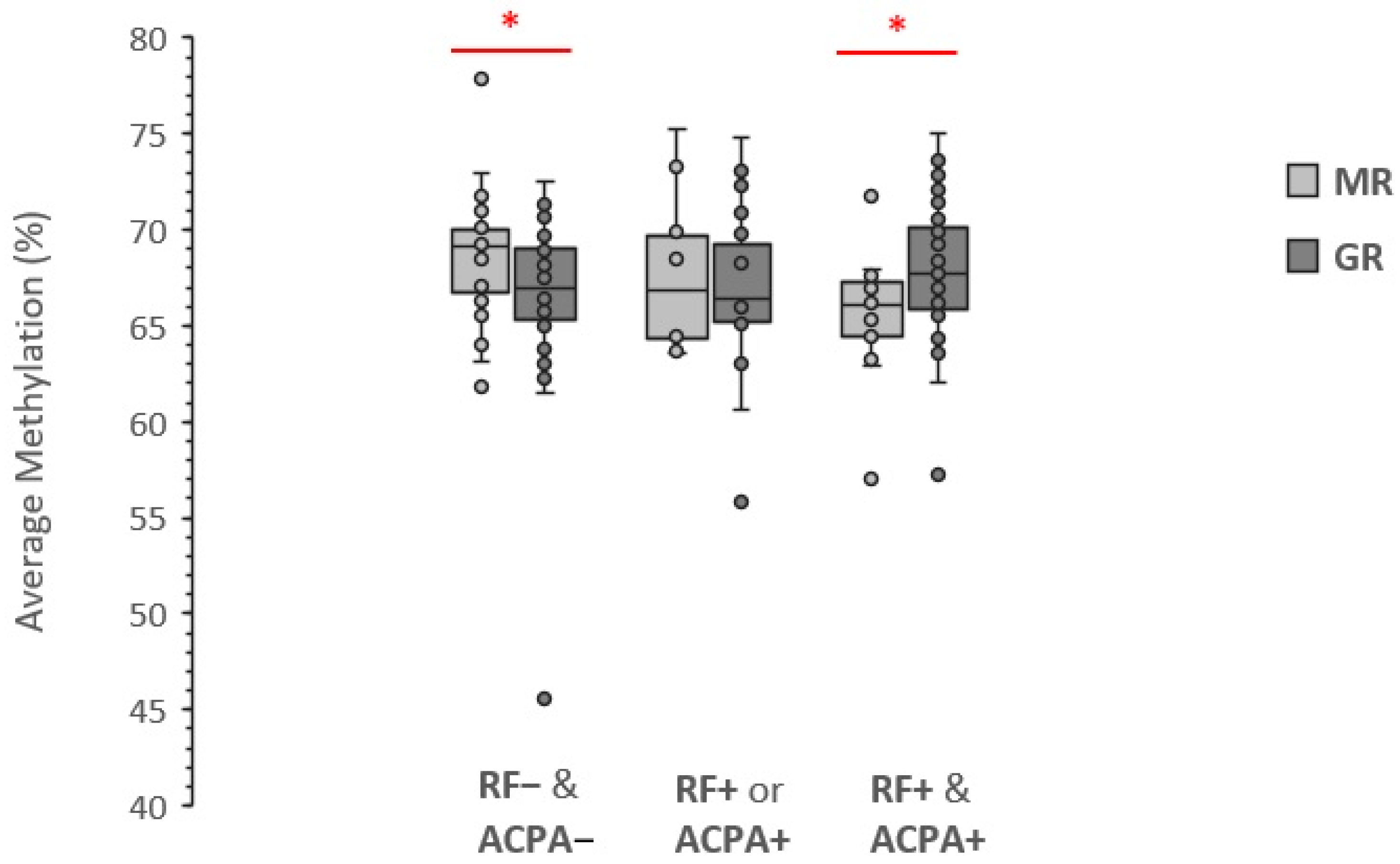
3.2. Association with LINE-1 Methylation Level
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Aletaha, D.; Neogi, T.; Silman, A.J.; Funovits, J.; Felson, D.T.; Bingham, C.O.; Birnbaum, N.S.; Burmester, G.R.; Bykerk, V.P.; Cohen, M.D.; et al. 2010 Rheumatoid Arthritis Classification Criteria: An American College of Rheumatology/European League Against Rheumatism Collaborative Initiative. Ann. Rheum. Dis. 2010, 69, 1580–1588. [Google Scholar] [CrossRef] [PubMed]
- Gibofsky, A. Overview of Epidemiology, Pathophysiology, and Diagnosis of Rheumatoid Arthritis. Am. J. Manag. Care 2012, 18, S295–S302. [Google Scholar] [PubMed]
- Okada, Y.; Wu, D.; Trynka, G.; Raj, T.; Terao, C.; Ikari, K.; Kochi, Y.; Ohmura, K.; Suzuki, A.; Yoshida, S.; et al. Genetics of Rheumatoid Arthritis Contributes to Biology and Drug Discovery. Nature 2014, 506, 376–381. [Google Scholar] [CrossRef] [PubMed]
- Källberg, H.; Ding, B.; Padyukov, L.; Bengtsson, C.; Rönnelid, J.; Klareskog, L.; Alfredsson, L. EIRA Study Group Smoking Is a Major Preventable Risk Factor for Rheumatoid Arthritis: Estimations of Risks after Various Exposures to Cigarette Smoke. Ann. Rheum. Dis. 2011, 70, 508–511. [Google Scholar] [CrossRef]
- Klareskog, L.; Malmström, V.; Lundberg, K.; Padyukov, L.; Alfredsson, L. Smoking, Citrullination and Genetic Variability in the Immunopathogenesis of Rheumatoid Arthritis. Semin. Immunol. 2011, 23, 92–98. [Google Scholar] [CrossRef]
- Ciechomska, M.; Roszkowski, L.; Maslinski, W. DNA Methylation as a Future Therapeutic and Diagnostic Target in Rheumatoid Arthritis. Cells 2019, 8, 953. [Google Scholar] [CrossRef]
- Liu, Y.; Aryee, M.J.; Padyukov, L.; Fallin, M.D.; Hesselberg, E.; Runarsson, A.; Reinius, L.; Acevedo, N.; Taub, M.; Ronninger, M.; et al. Epigenome-Wide Association Data Implicate DNA Methylation as an Intermediary of Genetic Risk in Rheumatoid Arthritis. Nat. Biotechnol. 2013, 31, 142–147. [Google Scholar] [CrossRef]
- Epigenome-Wide Association Study of Rheumatoid Arthritis Identifies Differentially Methylated Loci in B Cells|Human Molecular Genetics|Oxford Academic. Available online: https://academic.oup.com/hmg/article/26/14/2803/3798734?login=true (accessed on 2 August 2022).
- Zhu, H.; Wu, L.-F.; Mo, X.-B.; Lu, X.; Tang, H.; Zhu, X.-W.; Xia, W.; Guo, Y.-F.; Wang, M.-J.; Zeng, K.-Q.; et al. Rheumatoid Arthritis–Associated DNA Methylation Sites in Peripheral Blood Mononuclear Cells. Ann. Rheum. Dis. 2019, 78, 36–42. [Google Scholar] [CrossRef]
- Heidari, B. Rheumatoid Arthritis: Early Diagnosis and Treatment Outcomes. Casp. J. Intern. Med. 2011, 2, 161–170. [Google Scholar]
- Salliot, C.; van der Heijde, D. Long-Term Safety of Methotrexate Monotherapy in Patients with Rheumatoid Arthritis: A Systematic Literature Research. Ann. Rheum. Dis. 2009, 68, 1100–1104. [Google Scholar] [CrossRef]
- Horsburgh, S.; Ciechomska, M.; O’Reilly, S. CpG-Specific Methylation at Rheumatoid Arthritis Diagnosis as a Marker of Treatment Response. Epigenomics 2017, 9, 595–597. [Google Scholar] [CrossRef] [PubMed]
- Burgers, L.E.; Raza, K.; van der Helm-van Mil, A.H. Window of Opportunity in Rheumatoid Arthritis-Definitions and Supporting Evidence: From Old to New Perspectives. RMD Open 2019, 5, e000870. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.A. Functions of DNA Methylation: Islands, Start Sites, Gene Bodies and Beyond. Nat. Rev. Genet. 2012, 13, 484–492. [Google Scholar] [CrossRef] [PubMed]
- Ballestar, E.; Sawalha, A.H.; Lu, Q. Clinical Value of DNA Methylation Markers in Autoimmune Rheumatic Diseases. Nat. Rev. Rheumatol. 2020, 16, 514–524. [Google Scholar] [CrossRef] [PubMed]
- Lin, R.-K.; Wang, Y.-C. Dysregulated Transcriptional and Post-Translational Control of DNA Methyltransferases in Cancer. Cell Biosci. 2014, 4, 46. [Google Scholar] [CrossRef]
- van der Maarel, S.M. Epigenetic Mechanisms in Health and Disease. Ann. Rheum. Dis. 2008, 67 (Suppl. S3), iii97–iii100. [Google Scholar] [CrossRef]
- Burmester, G.R.; Feist, E.; Dörner, T. Emerging Cell and Cytokine Targets in Rheumatoid Arthritis. Nat. Rev. Rheumatol. 2014, 10, 77–88. [Google Scholar] [CrossRef]
- Karouzakis, E.; Gay, R.E.; Michel, B.A.; Gay, S.; Neidhart, M. DNA Hypomethylation in Rheumatoid Arthritis Synovial Fibroblasts. Arthritis Rheum. 2009, 60, 3613–3622. [Google Scholar] [CrossRef]
- de Andres, M.C.; Perez-Pampin, E.; Calaza, M.; Santaclara, F.J.; Ortea, I.; Gomez-Reino, J.J.; Gonzalez, A. Assessment of Global DNA Methylation in Peripheral Blood Cell Subpopulations of Early Rheumatoid Arthritis before and after Methotrexate. Arthritis Res. Ther. 2015, 17, 233. [Google Scholar] [CrossRef]
- Klein, K.; Gay, S. Epigenetic Modifications in Rheumatoid Arthritis, a Review. Curr. Opin. Pharmacol. 2013, 13, 420–425. [Google Scholar] [CrossRef]
- Neidhart, M.; Rethage, J.; Kuchen, S.; Künzler, P.; Crowl, R.M.; Billingham, M.E.; Gay, R.E.; Gay, S. Retrotransposable L1 Elements Expressed in Rheumatoid Arthritis Synovial Tissue: Association with Genomic DNA Hypomethylation and Influence on Gene Expression. Arthritis Rheum. 2000, 43, 2634–2647. [Google Scholar] [CrossRef]
- Karouzakis, E.; Rengel, Y.; Jüngel, A.; Kolling, C.; Gay, R.E.; Michel, B.A.; Tak, P.P.; Gay, S.; Neidhart, M.; Ospelt, C. DNA Methylation Regulates the Expression of CXCL12 in Rheumatoid Arthritis Synovial Fibroblasts. Genes Immun. 2011, 12, 643–652. [Google Scholar] [CrossRef] [PubMed]
- Nakano, K.; Whitaker, J.W.; Boyle, D.L.; Wang, W.; Firestein, G.S. DNA Methylome Signature in Rheumatoid Arthritis. Ann. Rheum. Dis. 2013, 72, 110–117. [Google Scholar] [CrossRef] [PubMed]
- Nile, C.J.; Read, R.C.; Akil, M.; Duff, G.W.; Wilson, A.G. Methylation Status of a Single CpG Site in the IL6 Promoter Is Related to IL6 Messenger RNA Levels and Rheumatoid Arthritis. Arthritis Rheum. 2008, 58, 2686–2693. [Google Scholar] [CrossRef]
- Bui, C.; Barter, M.J.; Scott, J.L.; Xu, Y.; Galler, M.; Reynard, L.N.; Rowan, A.D.; Young, D.A. CAMP Response Element-Binding (CREB) Recruitment Following a Specific CpG Demethylation Leads to the Elevated Expression of the Matrix Metalloproteinase 13 in Human Articular Chondrocytes and Osteoarthritis. FASEB J. 2012, 26, 3000–3011. [Google Scholar] [CrossRef]
- Guderud, K.; Sunde, L.H.; Flåm, S.T.; Mæhlen, M.T.; Mjaavatten, M.D.; Lillegraven, S.; Aga, A.-B.; Evenrød, I.M.; Norli, E.S.; Andreassen, B.K.; et al. Rheumatoid Arthritis Patients, Both Newly Diagnosed and Methotrexate Treated, Show More DNA Methylation Differences in CD4+ Memory Than in CD4+ Naïve T Cells. Front. Immunol. 2020, 11, 194. [Google Scholar] [CrossRef]
- Guderud, K.; Sunde, L.H.; Flåm, S.T.; Mæhlen, M.T.; Mjaavatten, M.D.; Norli, E.S.; Evenrød, I.M.; Andreassen, B.K.; Franzenburg, S.; Franke, A.; et al. Methotrexate Treatment of Newly Diagnosed RA Patients Is Associated with DNA Methylation Differences at Genes Relevant for Disease Pathogenesis and Pharmacological Action. Front. Immunol. 2021, 12, 713611. [Google Scholar] [CrossRef]
- Nair, N.; Plant, D.; Verstappen, S.M.; Isaacs, J.D.; Morgan, A.W.; Hyrich, K.L.; Barton, A.; Wilson, A.G.; MATURA Investigators. Differential DNA Methylation Correlates with Response to Methotrexate in Rheumatoid Arthritis. Rheumatol. Oxf. Engl. 2020, 59, 1364–1371. [Google Scholar] [CrossRef]
- Liebold, I.; Grützkau, A.; Göckeritz, A.; Gerl, V.; Lindquist, R.; Feist, E.; Zänker, M.; Häupl, T.; Poddubnyy, D.; Zernicke, J.; et al. Peripheral Blood Mononuclear Cells Are Hypomethylated in Active Rheumatoid Arthritis and Methylation Correlates with Disease Activity. Rheumatol. Oxf. Engl. 2021, 60, 1984–1995. [Google Scholar]
- Gosselt, H.R.; Vallerga, C.L.; Mandaviya, P.R.; Lubberts, E.; Hazes, J.M.W.; de Jonge, R.; Heil, S.G. Epigenome Wide Association Study of Response to Methotrexate in Early Rheumatoid Arthritis Patients. PLoS ONE 2021, 16, e0247709. [Google Scholar] [CrossRef]
- Gosselt, H.R.; van Zelst, B.D.; de Rotte, M.C.F.J.; Hazes, J.M.W.; de Jonge, R.; Heil, S.G. Higher Baseline Global Leukocyte DNA Methylation Is Associated with MTX Non-Response in Early RA Patients. Arthritis Res. Ther. 2019, 21, 157. [Google Scholar] [CrossRef] [PubMed]
- Lander, E.S.; Linton, L.M.; Birren, B.; Nusbaum, C.; Zody, M.C.; Baldwin, J.; Devon, K.; Dewar, K.; Doyle, M.; FitzHugh, W.; et al. Initial Sequencing and Analysis of the Human Genome. Nature 2001, 409, 860–921. [Google Scholar] [PubMed]
- Rodriguez, J.; Frigola, J.; Vendrell, E.; Risques, R.-A.; Fraga, M.F.; Morales, C.; Moreno, V.; Esteller, M.; Capellà, G.; Ribas, M.; et al. Chromosomal Instability Correlates with Genome-Wide DNA Demethylation in Human Primary Colorectal Cancers. Cancer Res. 2006, 66, 8462–9468. [Google Scholar] [CrossRef] [PubMed]
- Beck, C.R.; Collier, P.; Macfarlane, C.; Malig, M.; Kidd, J.M.; Eichler, E.E.; Badge, R.M.; Moran, J.V. LINE-1 Retrotransposition Activity in Human Genomes. Cell 2010, 141, 1159–1170. [Google Scholar] [CrossRef] [PubMed]
- Brouha, B.; Schustak, J.; Badge, R.M.; Lutz-Prigge, S.; Farley, A.H.; Moran, J.V.; Kazazian, H.H. Hot L1s Account for the Bulk of Retrotransposition in the Human Population. Proc. Natl. Acad. Sci. USA 2003, 100, 5280–5285. [Google Scholar] [CrossRef] [PubMed]
- Lopes, L.L.; Bressan, J.; Peluzio, M.D.C.G.; Hermsdorff, H.H.M. LINE-1 in Obesity and Cardiometabolic Diseases: A Systematic Review. J. Am. Coll. Nutr. 2019, 38, 478–484. [Google Scholar] [CrossRef]
- van Gestel, A.M.; Prevoo, M.L.; van’t Hof, M.A.; van Rijswijk, M.H.; van de Putte, L.B.; van Riel, P.L. Development and Validation of the European League Against Rheumatism Response Criteria for Rheumatoid Arthritis. Comparison with the Preliminary American College of Rheumatology and the World Health Organization/International League Against Rheumatism Criteria. Arthritis Rheum. 1996, 39, 34–40. [Google Scholar]
- van Gestel, A.M.; Haagsma, C.J.; van Riel, P.L. Validation of Rheumatoid Arthritis Improvement Criteria That Include Simplified Joint Counts. Arthritis Rheum. 1998, 41, 1845–1850. [Google Scholar] [CrossRef]
- Ravaei, A.; Rubini, M. Rescuing Effect of Folates on Methotrexate Cytotoxicity in Human Trophoblast Cells. Clin. Exp. Rheumatol. 2022, 40, 1403–1410. [Google Scholar] [CrossRef]
- Salma, K.; Nessrine, A.; Krystel, E.; Khaoula, E.K.; Noura, N.; Khadija, E.; Taoufik, H. Rheumatoid Arthritis: Seropositivity versus Seronegativity; A Comparative Cross-Sectional Study Arising from Moroccan Context. Curr. Rheumatol. Rev. 2020, 16, 143–148. [Google Scholar] [CrossRef]
- Romão, V.C.; Canhão, H.; Fonseca, J.E. Old Drugs, Old Problems: Where Do We Stand in Prediction of Rheumatoid Arthritis Responsiveness to Methotrexate and Other Synthetic DMARDs? BMC Med. 2013, 11, 17. [Google Scholar] [CrossRef] [PubMed]
- Rönnelid, J.; Wick, M.C.; Lampa, J.; Lindblad, S.; Nordmark, B.; Klareskog, L.; van Vollenhoven, R.F. Longitudinal Analysis of Citrullinated Protein/Peptide Antibodies (Anti-CP) during 5 Year Follow up in Early Rheumatoid Arthritis: Anti-CP Status Predicts Worse Disease Activity and Greater Radiological Progression. Ann. Rheum. Dis. 2005, 64, 1744–1749. [Google Scholar] [CrossRef] [PubMed]
- Kokkonen, H.; Mullazehi, M.; Berglin, E.; Hallmans, G.; Wadell, G.; Rönnelid, J.; Rantapää-Dahlqvist, S. Antibodies of IgG, IgA and IgM Isotypes against Cyclic Citrullinated Peptide Precede the Development of Rheumatoid Arthritis. Arthritis Res. Ther. 2011, 13, R13. [Google Scholar] [CrossRef] [PubMed]
- Sokolove, J.; Johnson, D.S.; Lahey, L.J.; Wagner, C.A.; Cheng, D.; Thiele, G.M.; Michaud, K.; Sayles, H.; Reimold, A.M.; Caplan, L.; et al. Rheumatoid Factor as a Potentiator of Anti-Citrullinated Protein Antibody-Mediated Inflammation in Rheumatoid Arthritis. Arthritis Rheumatol. 2014, 66, 813–821. [Google Scholar] [CrossRef] [PubMed]
- Clavel, C.; Nogueira, L.; Laurent, L.; Iobagiu, C.; Vincent, C.; Sebbag, M.; Serre, G. Induction of Macrophage Secretion of Tumor Necrosis Factor α through Fcgamma Receptor IIa Engagement by Rheumatoid Arthritis-Specific Autoantibodies to Citrullinated Proteins Complexed with Fibrinogen. Arthritis Rheum. 2008, 58, 678–688. [Google Scholar] [CrossRef]
- Aletaha, D.; Alasti, F.; Smolen, J.S. Rheumatoid Factor, Not Antibodies against Citrullinated Proteins, Is Associated with Baseline Disease Activity in Rheumatoid Arthritis Clinical Trials. Arthritis Res. Ther. 2015, 17, 229. [Google Scholar] [CrossRef]
- Derksen, V.F.A.M.; Ajeganova, S.; Trouw, L.A.; van der Helm-van Mil, A.H.M.; Hafström, I.; Huizinga, T.W.J.; Toes, R.E.M.; Svensson, B.; van der Woude, D. Rheumatoid Arthritis Phenotype at Presentation Differs Depending on the Number of Autoantibodies Present. Ann. Rheum. Dis. 2017, 76, 716–720. [Google Scholar] [CrossRef]
- Miriovsky, B.J.; Michaud, K.; Thiele, G.M.; O’Dell, J.R.; Cannon, G.W.; Kerr, G.; Richards, J.S.; Johnson, D.; Caplan, L.; Reimold, A.; et al. Anti-CCP Antibody and Rheumatoid Factor Concentrations Predict Greater Disease Activity in Men with Rheumatoid Arthritis. Ann. Rheum. Dis. 2010, 69, 1292–1297. [Google Scholar] [CrossRef]
- Krishnamurthy, A.; Joshua, V.; Haj Hensvold, A.; Jin, T.; Sun, M.; Vivar, N.; Ytterberg, A.J.; Engström, M.; Fernandes-Cerqueira, C.; Amara, K.; et al. Identification of a Novel Chemokine-Dependent Molecular Mechanism Underlying Rheumatoid Arthritis-Associated Autoantibody-Mediated Bone Loss. Ann. Rheum. Dis. 2016, 75, 721–729. [Google Scholar] [CrossRef]


| Present DAS28 | DAS28 Improvement | ||
|---|---|---|---|
| >1.2 | >0.6 and ≤1.2 | ≤0.6 | |
| ≤3.2 | Good response | Moderate response | No response |
| >3.2 and ≤5.1 | Moderate response | Moderate response | No response |
| >5.1 | Moderate response | No response | No response |
| Variable | Stratum | (%) |
|---|---|---|
| Sex | Female | 73.7 |
| Age | >60 years | 51.6 |
| BMI 1 | >25 kg/m2 | 54.0 |
| Tobacco smoking | Current smokers | 44.7 |
| RF 2 | Positive | 49.8 |
| ACPA 3 | Positive | 47.4 |
| RF/ACPA 4 | Positive | 56.4 |
| MTX response | NR 5 | 13.9 |
| MR 6 | 28.9 | |
| GR 7 | 57.2 |
| NR 1 | MR 2 | GR 3 | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Patients (N) | Average Methylation (%) | SEM | Patients (N) | Average Methylation (%) | SEM | Patients (N) | Average Methylation (%) | SEM | |
| Sex | |||||||||
| Female | 27 | 67.40 | 0.94 | 65 | 67.19 | 0.41 | 109 | 67.31 | 0.34 |
| Male | 11 | 66.11 | 0.81 | 14 | 68.60 | 0.69 | 47 | 67.09 | 0.53 |
| Age | |||||||||
| >60 | 18 | 67.24 | 1.30 | 43 | 67.78 | 0.55 | 80 | 67.09 | 0.44 |
| ≤60 | 20 | 66.84 | 0.70 | 36 | 67.05 | 0.45 | 76 | 67.40 | 0.37 |
| BMI | |||||||||
| ≤25 | 14 | 66.29 | 1.59 | 31 | 67.57 | 0.62 | 76 | 67.17 | 0.37 |
| >25 | 24 | 67.46 | 0.65 | 44 | 67.31 | 0.47 | 74 | 67.24 | 0.45 |
| Smoking | |||||||||
| Non-smokers | 14 | 67.24 | 0.84 | 51 | 67.80 | 0.43 | 86 | 67.45 | 0.41 |
| Smokers | 24 | 66.90 | 1.02 | 28 | 66.79 | 0.66 | 70 | 66.99 | 0.39 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ravaei, A.; Pulsatelli, L.; Assirelli, E.; Meliconi, R.; Ciaffi, J.; Gremese, E.; Tolusso, B.; Salvarani, C.; Govoni, M.; Rubini, M. Seropositivity-Dependent Association between LINE-1 Methylation and Response to Methotrexate Therapy in Early Rheumatoid Arthritis Patients. Genes 2022, 13, 2012. https://doi.org/10.3390/genes13112012
Ravaei A, Pulsatelli L, Assirelli E, Meliconi R, Ciaffi J, Gremese E, Tolusso B, Salvarani C, Govoni M, Rubini M. Seropositivity-Dependent Association between LINE-1 Methylation and Response to Methotrexate Therapy in Early Rheumatoid Arthritis Patients. Genes. 2022; 13(11):2012. https://doi.org/10.3390/genes13112012
Chicago/Turabian StyleRavaei, Amin, Lia Pulsatelli, Elisa Assirelli, Riccardo Meliconi, Jacopo Ciaffi, Elisa Gremese, Barbara Tolusso, Carlo Salvarani, Marcello Govoni, and Michele Rubini. 2022. "Seropositivity-Dependent Association between LINE-1 Methylation and Response to Methotrexate Therapy in Early Rheumatoid Arthritis Patients" Genes 13, no. 11: 2012. https://doi.org/10.3390/genes13112012
APA StyleRavaei, A., Pulsatelli, L., Assirelli, E., Meliconi, R., Ciaffi, J., Gremese, E., Tolusso, B., Salvarani, C., Govoni, M., & Rubini, M. (2022). Seropositivity-Dependent Association between LINE-1 Methylation and Response to Methotrexate Therapy in Early Rheumatoid Arthritis Patients. Genes, 13(11), 2012. https://doi.org/10.3390/genes13112012








