Different Mechanisms Cause Hypomethylation of Both H19 and KCNQ1OT1 Imprinted Differentially Methylated Regions in Two Cases of Silver–Russell Syndrome Spectrum
Abstract
1. Introduction
2. Material and Methods
2.1. Patients
2.2. DNA Extraction
2.3. Methylation Analysis
2.4. SNP-Array
2.5. FISH
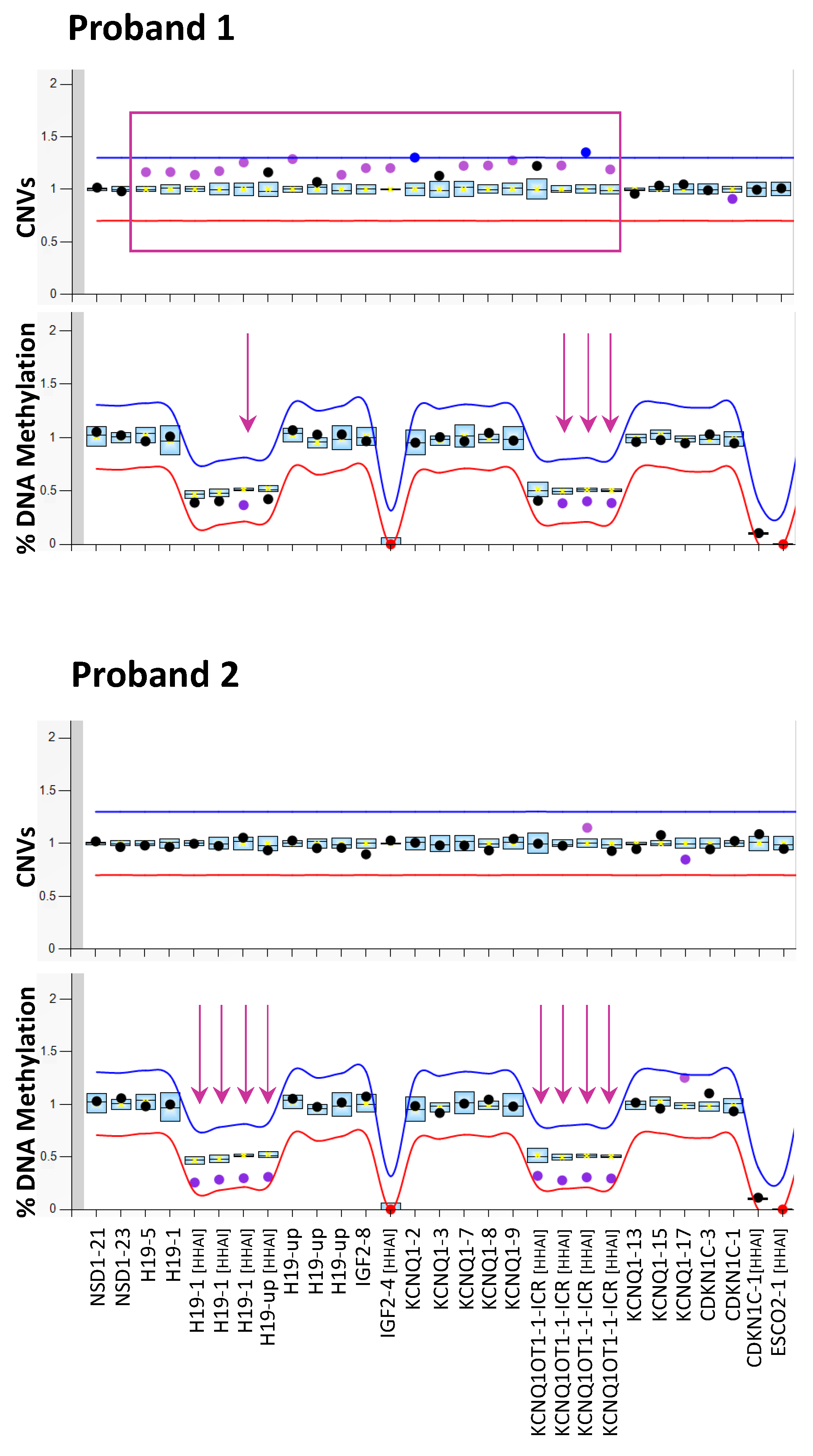
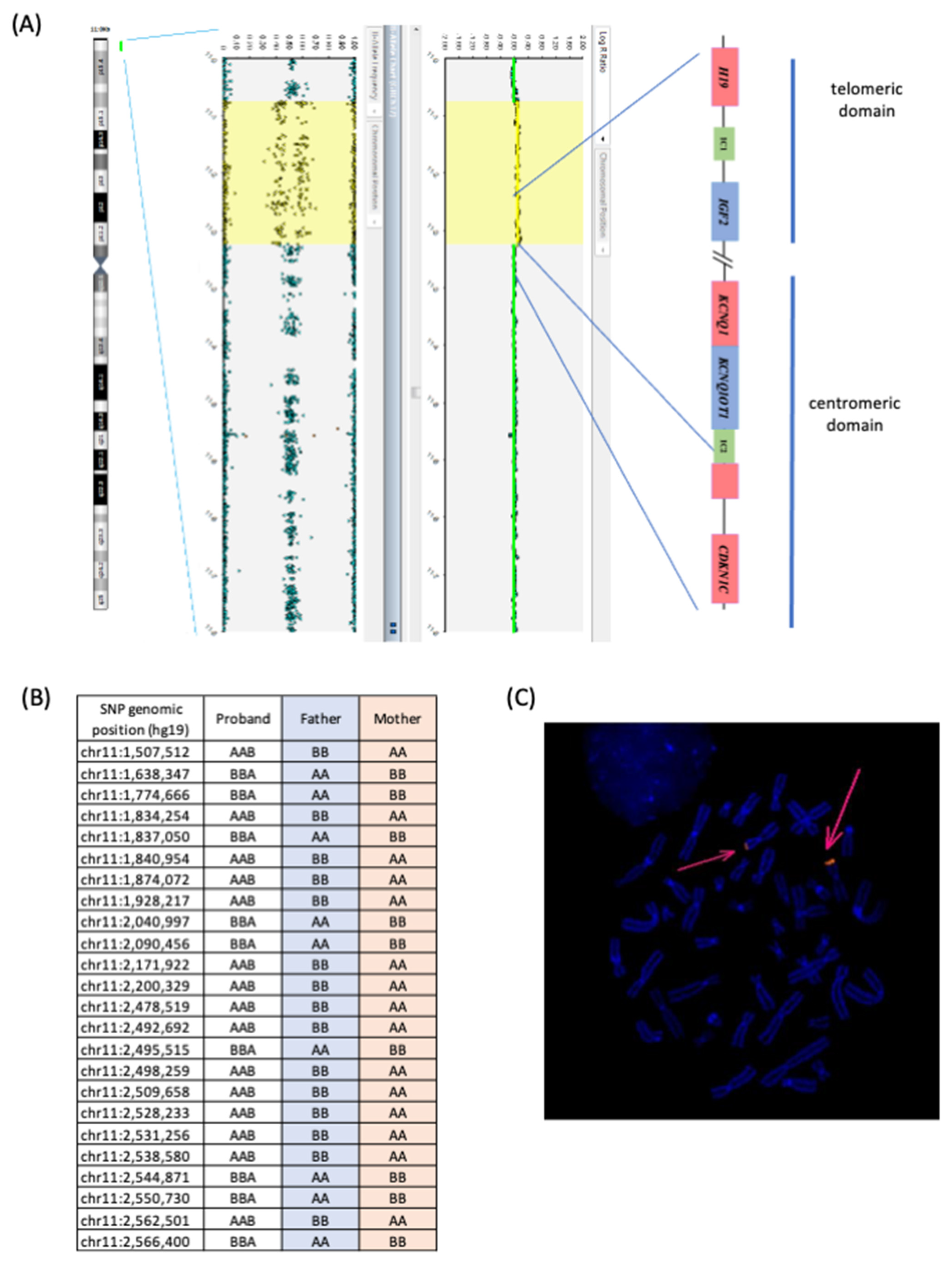
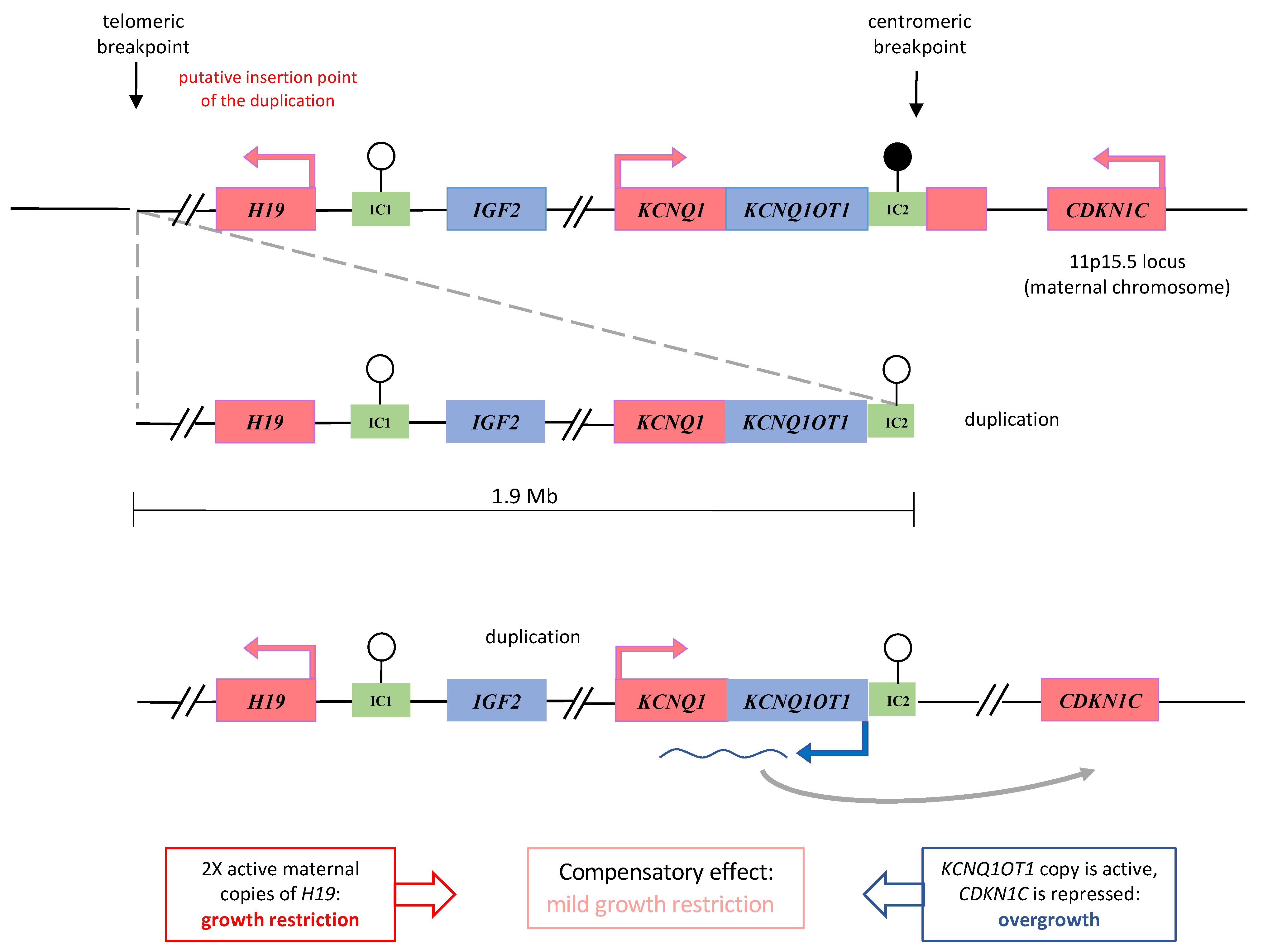
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wakeling, E.L.; Brioude, F.; Lokulo-Sodipe, O.; O’Connell, S.M.; Salem, J.; Bliek, J.; Canton, A.P.M.; Chrzanowska, K.H.; Davies, J.H.; Dias, R.P.; et al. Diagnosis and management of Silver–Russell syndrome: First international consensus statement. Nat. Rev. Endocrinol. 2016, 13, 105–124. [Google Scholar] [CrossRef] [PubMed]
- Mackay, D.J.G.; Temple, I.K. Ongoing Challenges in the Diagnosis of 11p15.5-Associated Imprinting Disorders. Mol. Diagn. Ther. 2022, 26, 263–272. [Google Scholar] [CrossRef]
- Monk, D.; Mackay, D.J.; Eggermann, T.; Maher, E.R.; Riccio, A. Genomic imprinting disorders: Lessons on how genome, epigenome and environment interact. Nat. Rev. Genet. 2019, 20, 235–248. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.C.; Choufani, S.; Ferreira, J.C.; Weksberg, R. Growth Regulation, Imprinted Genes, and Chromosome 11p15.5. Pediatr. Res. 2007, 61, 43R–47R. [Google Scholar] [CrossRef] [PubMed]
- Pignata, L.; Sparago, A.; Palumbo, O.; Andreucci, E.; Lapi, E.; Tenconi, R.; Carella, M.; Riccio, A.; Cerrato, F. Mosaic Segmental and Whole-Chromosome Upd(11)mat in Silver-Russell Syndrome. Genes 2021, 12, 581. [Google Scholar] [CrossRef] [PubMed]
- Chiesa, N.; De Crescenzo, A.; Mishra, K.; Perone, L.; Carella, M.; Palumbo, O.; Mussa, A.; Sparago, A.; Cerrato, F.; Russo, S.; et al. The KCNQ1OT1 imprinting control region and non-coding RNA: New properties derived from the study of Beckwith–Wiedemann syndrome and Silver–Russell syndrome cases. Hum. Mol. Genet. 2012, 21, 10–25. [Google Scholar] [CrossRef] [PubMed]
- Schönherr, N.; Meyer, E.; Roos, A.; Schmidt, A.; Wollmann, H.A.; Eggermann, T. The centromeric 11p15 imprinting centre is also involved in Silver-Russell syndrome. J. Med. Genet. 2006, 44, 59–63. [Google Scholar] [CrossRef]
- De Crescenzo, A.; Sparago, A.; Cerrato, F.; Palumbo, O.; Carella, M.; Miceli, M.; Bronshtein, M.; Riccio, A.; Yaron, Y. Paternal deletion of the 11p15.5 centromeric-imprinting control region is associated with alteration of imprinted gene expression and recurrent severe intrauterine growth restriction. J. Med. Genet. 2012, 50, 99–103. [Google Scholar] [CrossRef] [PubMed]
- Begemann, M.; Rezwan, F.I.; Beygo, J.; Docherty, L.E.; Kolarova, J.; Schroeder, C.; Buiting, K.; Chokkalingam, K.; Degenhardt, F.; Wakeling, E.L.; et al. Maternal variants in NLRP and other maternal effect proteins are associated with multilocus imprinting disturbance in offspring. J. Med. Genet. 2018, 55, 497–504. [Google Scholar] [CrossRef] [PubMed]
- Elbracht, M.; Mackay, D.J.; Begemann, M.; Kagan, K.O.; Eggermann, T. Disturbed genomic imprinting and its relevance for human reproduction: Causes and clinical consequences. Hum. Reprod. Update 2020, 26, 197–213. [Google Scholar] [CrossRef] [PubMed]
- Azzi, S.; Rossignol, S.; Steunou, V.; Sas, T.; Thibaud, N.; Danton, F.; Le Jule, M.; Heinrichs, C.; Cabrol, S.; Gicquel, C.; et al. Multilocus methylation analysis in a large cohort of 11p15-related foetal growth disorders (Russell Silver and Beckwith Wiedemann syndromes) reveals simultaneous loss of methylation at paternal and maternal imprinted loci. Hum. Mol. Genet. 2009, 18, 4724–4733. [Google Scholar] [CrossRef]
- Eggermann, T.; Yapici, E.; Bliek, J.; Pereda, A.; Begemann, M.; Russo, S.; Tannorella, P.; Calzari, L.; de Nanclares, G.P.; Lombardi, P.; et al. Trans-acting genetic variants causing multilocus imprinting disturbance (MLID): Common mechanisms and consequences. Clin. Epigenet. 2022, 14, 41. [Google Scholar] [CrossRef] [PubMed]
- Pignata, L.; Palumbo, O.; Cerrato, F.; Acurzio, B.; De Álava, E.; Roma, J.; Gallego, S.; Mora, J.; Carella, M.; Riccio, A.; et al. Both Epimutations and Chromosome Aberrations Affect Multiple Imprinted Loci in Aggressive Wilms Tumors. Cancers 2020, 12, 3411. [Google Scholar] [CrossRef] [PubMed]
- Heide, S.; Chantot-Bastaraud, S.; Keren, B.; Harbison, M.D.; Azzi, S.; Rossignol, S.; Michot, C.; Lys, M.L.-P.; Demeer, B.; Heinrichs, C.; et al. Chromosomal rearrangements in the 11p15 imprinted region: 17 new 11p15.5 duplications with associated phenotypes and putative functional consequences. J. Med. Genet. 2018, 55, 205–213. [Google Scholar] [CrossRef]
- Demars, J.; Rossignol, S.; Netchine, I.; Lee, K.S.; Shmela, M.; Faivre, L.; Weill, J.; Odent, S.; Azzi, S.; Callier, P.; et al. New insights into the pathogenesis of beckwith-wiedemann and silver-russell syndromes: Contribution of small copy number variations to 11p15 imprinting defects. Hum. Mutat. 2011, 32, 1171–1182. [Google Scholar] [CrossRef]
- Keniry, A.; Oxley, D.; Monnier, P.; Kyba, M.; Dandolo, L.; Smits, G.; Reik, W. The H19 lincRNA is a developmental reservoir of miR-675 that suppresses growth and Igf1r. Nat. Cell Biol. 2012, 14, 659–665. [Google Scholar] [CrossRef]
- Gabory, A.; Ripoche, M.-A.; Le Digarcher, A.; Watrin, F.; Ziyyat, A.; Forné, T.; Jammes, H.; Ainscough, J.F.X.; Surani, M.A.; Journot, L.; et al. H19 acts as a trans regulator of the imprinted gene network controlling growth in mice. Development 2009, 136, 3413–3421. [Google Scholar] [CrossRef] [PubMed]
- Bliek, J.; Snijder, S.; Maas, S.; Polstra, A.; van der Lip, K.; Alders, M.; Knegt, A.; Mannens, M. Phenotypic discordance upon paternal or maternal transmission of duplications of the 11p15 imprinted regions. Eur. J. Med. Genet. 2009, 52, 404–408. [Google Scholar] [CrossRef] [PubMed]
- Begemann, M.; Spengler, S.; Gogiel, M.; Grasshoff, U.; Bonin, M.; Betz, R.C.; Dufke, A.; Spier, I.; Eggermann, T. Clinical significance of copy number variations in the 11p15.5 imprinting control regions: New cases and review of the literature. J. Med. Genet. 2012, 49, 547–553. [Google Scholar] [CrossRef]
- Maier, F.; Frühwald, M.; Heinrich, U.; Schimmel, M.; Wahl, D.; Eggermann, T. Overgrowth-associated partial trisomy 15q24.3-qter and mosaic 11p15.5 duplication involving Silver-Russell region in a patient with lateralized asymmetry and developmental delay. Clin. Dysmorphol. 2021, 30, 189–193. [Google Scholar] [CrossRef] [PubMed]
- Sano, S.; Matsubara, K.; Nagasaki, K.; Kikuchi, T.; Nakabayashi, K.; Hata, K.; Fukami, M.; Kagami, M.; Ogata, T. Beckwith–Wiedemann syndrome and pseudohypoparathyroidism type Ib in a patient with multilocus imprinting disturbance: A female-dominant phenomenon? J. Hum. Genet. 2016, 61, 765–769. [Google Scholar] [CrossRef] [PubMed]
- Sparago, A.; Verma, A.; Patricelli, M.G.; Pignata, L.; Russo, S.; Calzari, L.; De Francesco, N.; Del Prete, R.; Palumbo, O.; Carella, M.; et al. The phenotypic variations of multi-locus imprinting disturbances associated with maternal-effect variants of NLRP5 range from overt imprinting disorder to apparently healthy phenotype. Clin. Epigenet. 2019, 11, 190. [Google Scholar] [CrossRef] [PubMed]
- Pignata, L.; Cecere, F.; Verma, A.; Mele, B.H.; Monticelli, M.; Acurzio, B.; Giaccari, C.; Sparago, A.; Mora, J.R.H.; Monteagudo-Sánchez, A.; et al. Novel genetic variants of KHDC3L and other members of the subcortical maternal complex associated with Beckwith–Wiedemann syndrome or Pseudohypoparathyroidism 1B and multi-locus imprinting disturbances. Clin. Epigenet. 2022, 14, 71. [Google Scholar] [CrossRef] [PubMed]
- Docherty, L.E.; Rezwan, F.I.; Poole, R.L.; Turner, C.L.S.; Kivuva, E.; Maher, E.R.; Smithson, S.F.; Hamilton-Shield, J.; Patalan, M.; Giżewska, M.; et al. Mutations in NLRP5 are associated with reproductive wastage and multilocus imprinting disorders in humans. Nat. Commun. 2015, 6, 8086. [Google Scholar] [CrossRef] [PubMed]
- Cubellis, M.V.; Pignata, L.; Verma, A.; Sparago, A.; Del Prete, R.; Monticelli, M.; Calzari, L.; Antona, V.; Melis, D.; Tenconi, R.; et al. Loss-of-function maternal-effect mutations of PADI6 are associated with familial and sporadic Beckwith-Wiedemann syndrome with multi-locus imprinting disturbance. Clin. Epigenet. 2020, 12, 139. [Google Scholar] [CrossRef]
- Eggermann, T.; Kadgien, G.; Begemann, M.; Elbracht, M. Biallelic PADI6 variants cause multilocus imprinting disturbances and miscarriages in the same family. Eur. J. Hum. Genet. 2020, 29, 575–580. [Google Scholar] [CrossRef]
- Tannorella, P.; Calzari, L.; Daolio, C.; Mainini, E.; Vimercati, A.; Gentilini, D.; Soli, F.; Pedrolli, A.; Bonati, M.T.; Larizza, L.; et al. Germline variants in genes of the subcortical maternal complex and Multilocus Imprinting Disturbance are associated with miscarriage/infertility or Beckwith–Wiedemann progeny. Clin. Epigenet. 2022, 14, 43. [Google Scholar] [CrossRef]
| ID | Clinical Features | Hypo/Hyper Methylated Loci | References |
|---|---|---|---|
| SRS | SGA, PNGR, protruding forehead, body asymmetry, dolichocephaly, triangular face, nasal hypoplasia, clinodactyly of the V fingers, flat philtrum | H19, KCNQ1OT1, NAP1L5, INPP5F | Proband 2 of present study |
| SRS | SGA, PNGR, relative macrocephaly at birth, protruding forehead, body asymmetry, feeding difficulties | H19, KCNQ1OT1, MEST | [11,12] |
| SRS | Birth weight at 27 wg 465 g, OFC 32 cm. PNGR, respiratory support for 2 months, gastric tube feeding for first year. Microcephaly, precocious puberty, dysmorphism. Developmental delay. 47, XXY | H19, KCNQ1OT1, GRB10, MEST, MEG3, GNAS-AS, GNAS | [11,12] |
| SRS | Discordant monozygotic twin, kidney failure in infancy, bilateral renal dysplasia | H19, KCNQ1OT1, PLAGL1, IGF2R, IGF1R, PEG3, GNAS-AS | [11,12] |
| SRS | PNGR, relative macrocephaly, facial gestalt (prominent forehead, triangular face, downturned corners of the mouth, micrognathia), asymmetry and clinodactyly of the fifth digit | H19, KCNQ1OT1 | [11,12] |
| SRS | N/A | H19, KCNQ1OT1, MEG3 | [11,12] |
| SRS | N/A | H19, KCNQ1OT1, MEG3, IGF2R | [11,12] |
| SRS | N/A | H19, KCNQ1OT1, PLAGL1, MEST, MEG3 | [11,12] |
| BWS | Polyhydramnios, macroglossia, umbilical ernia, hypoglicaemia, naevus flammeus | H19, KCNQ1OT1, MEG3, GNAS-AS, GNAS, PEG3, PLAGL1 GRB10, MEST | [11,12] |
| BWS | Macroglossia, umbilical ernia, hypoglicaemia hemihypertrophy, naevus flammeus, ear creases, low birth weight, hypocalcemia, facial dysmorphism, slight cognitive delay | GRB10, MEST, H19, KCNQ1OT1, MEG3, GNAS-AS, GNAS, PEG3, PLAGL1, SNRPN | [11,12] |
| BWS | Macroglossia, cheek and tongue right-side hemihyperplasia, naevus flammeus, diastasis recti | H19, KCNQ1OT1, PLAGL1, MEST, GNAS-AS, GNAS | [11,12] |
| BWS | BW 90th–97th centile, macrosomia, macroglossia, asymmetry, naevus flammeus, ear creases, developmental delay | H19, KCNQ1OT1, GRB10, MEST, IGF2R, IGF1R | [11,12] |
| BWS | N/A | H19, KCNQ1OT1, MEG3, GNAS-AS, GNAS | [11,12] |
| BWS | N/A | H19, KCNQ1OT1, MEG3, SNRPN, PEG3 | [11,12] |
| BWS | N/A | H19, KCNQ1OT1, PLAGL1, MEST, GNAS-AS, GNAS | [11,12] |
| BWS | N/A | H19, KCNQ1OT1, PLAGL1, MEST, IGF2R | [11,12] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Passaretti, F.; Pignata, L.; Vitiello, G.; Alesi, V.; D’Elia, G.; Cecere, F.; Acquaviva, F.; De Brasi, D.; Novelli, A.; Riccio, A.; et al. Different Mechanisms Cause Hypomethylation of Both H19 and KCNQ1OT1 Imprinted Differentially Methylated Regions in Two Cases of Silver–Russell Syndrome Spectrum. Genes 2022, 13, 1875. https://doi.org/10.3390/genes13101875
Passaretti F, Pignata L, Vitiello G, Alesi V, D’Elia G, Cecere F, Acquaviva F, De Brasi D, Novelli A, Riccio A, et al. Different Mechanisms Cause Hypomethylation of Both H19 and KCNQ1OT1 Imprinted Differentially Methylated Regions in Two Cases of Silver–Russell Syndrome Spectrum. Genes. 2022; 13(10):1875. https://doi.org/10.3390/genes13101875
Chicago/Turabian StylePassaretti, Francesco, Laura Pignata, Giuseppina Vitiello, Viola Alesi, Gemma D’Elia, Francesco Cecere, Fabio Acquaviva, Daniele De Brasi, Antonio Novelli, Andrea Riccio, and et al. 2022. "Different Mechanisms Cause Hypomethylation of Both H19 and KCNQ1OT1 Imprinted Differentially Methylated Regions in Two Cases of Silver–Russell Syndrome Spectrum" Genes 13, no. 10: 1875. https://doi.org/10.3390/genes13101875
APA StylePassaretti, F., Pignata, L., Vitiello, G., Alesi, V., D’Elia, G., Cecere, F., Acquaviva, F., De Brasi, D., Novelli, A., Riccio, A., Iolascon, A., & Cerrato, F. (2022). Different Mechanisms Cause Hypomethylation of Both H19 and KCNQ1OT1 Imprinted Differentially Methylated Regions in Two Cases of Silver–Russell Syndrome Spectrum. Genes, 13(10), 1875. https://doi.org/10.3390/genes13101875







