Selection and Validation of Reliable Reference Genes for Gene Expression Studies in Different Genotypes and TRV-Infected Fruits of Peach (Prunus persica L. Batsch) during Ripening
Abstract
:1. Introduction
2. Materials and Methods
2.1. Different Genetypes Fruits
2.2. TRV-Infected Fruits
2.3. Design for RT-qPCR Primers
2.4. RNA Isolation and cDNA Preparation
2.5. RT-qPCR with SYBR Green
2.6. Normalization of PpACO1, PpEIN2, and PpPL
2.7. Data Analysis
3. Result
3.1. Detection of Amplification Specificity of Reference Gene Primers
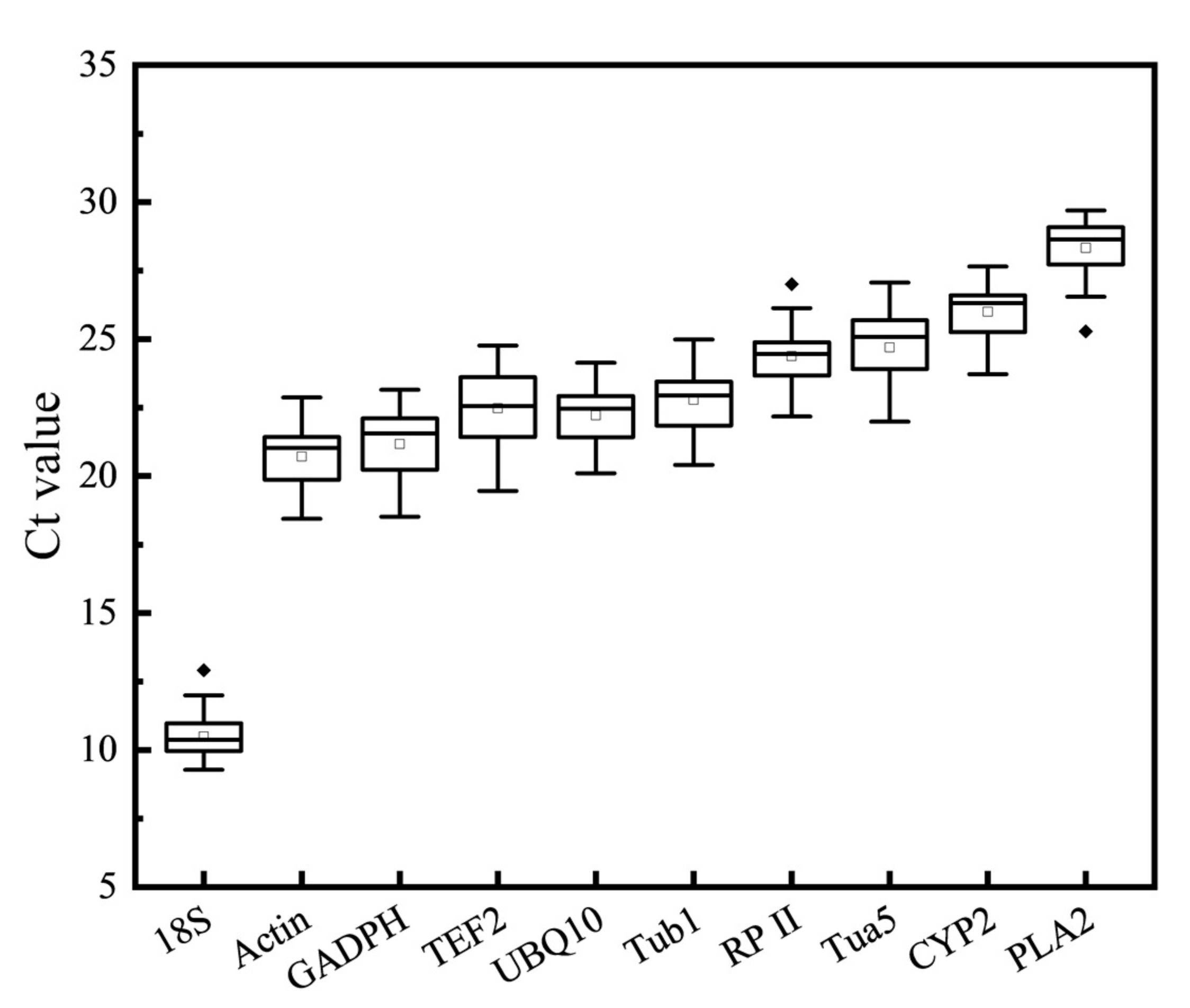
3.2. Expression Level of Reference Genes
3.3. Stability of Reference Genes
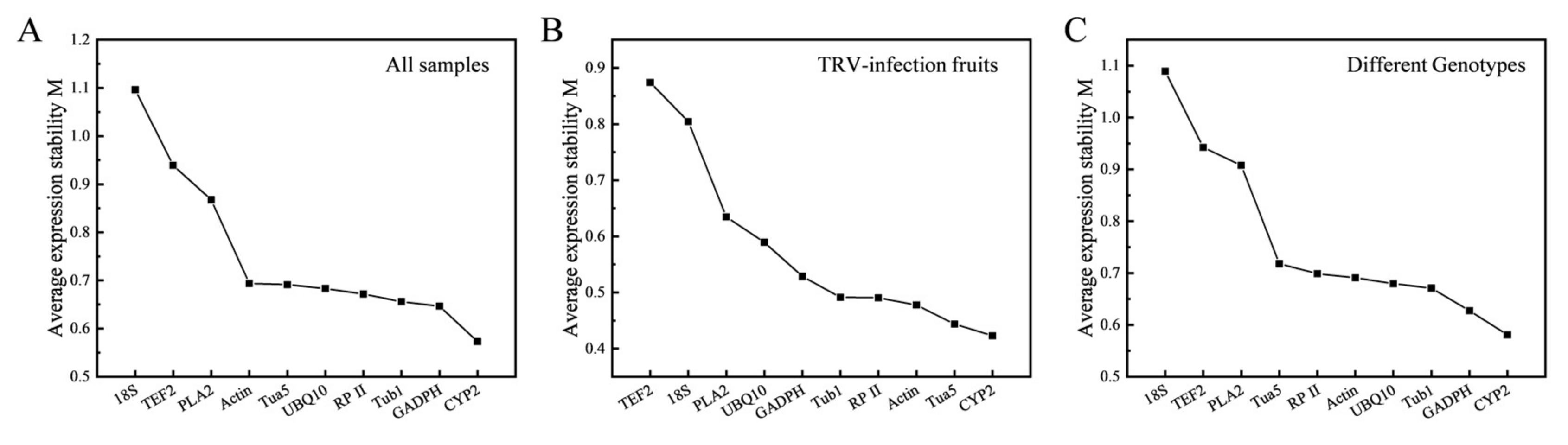
3.3.1. geNorm Analysis
3.3.2. NormFinder Analysis
3.3.3. BestKeeper Analysis
3.3.4. RefFinder Analysis
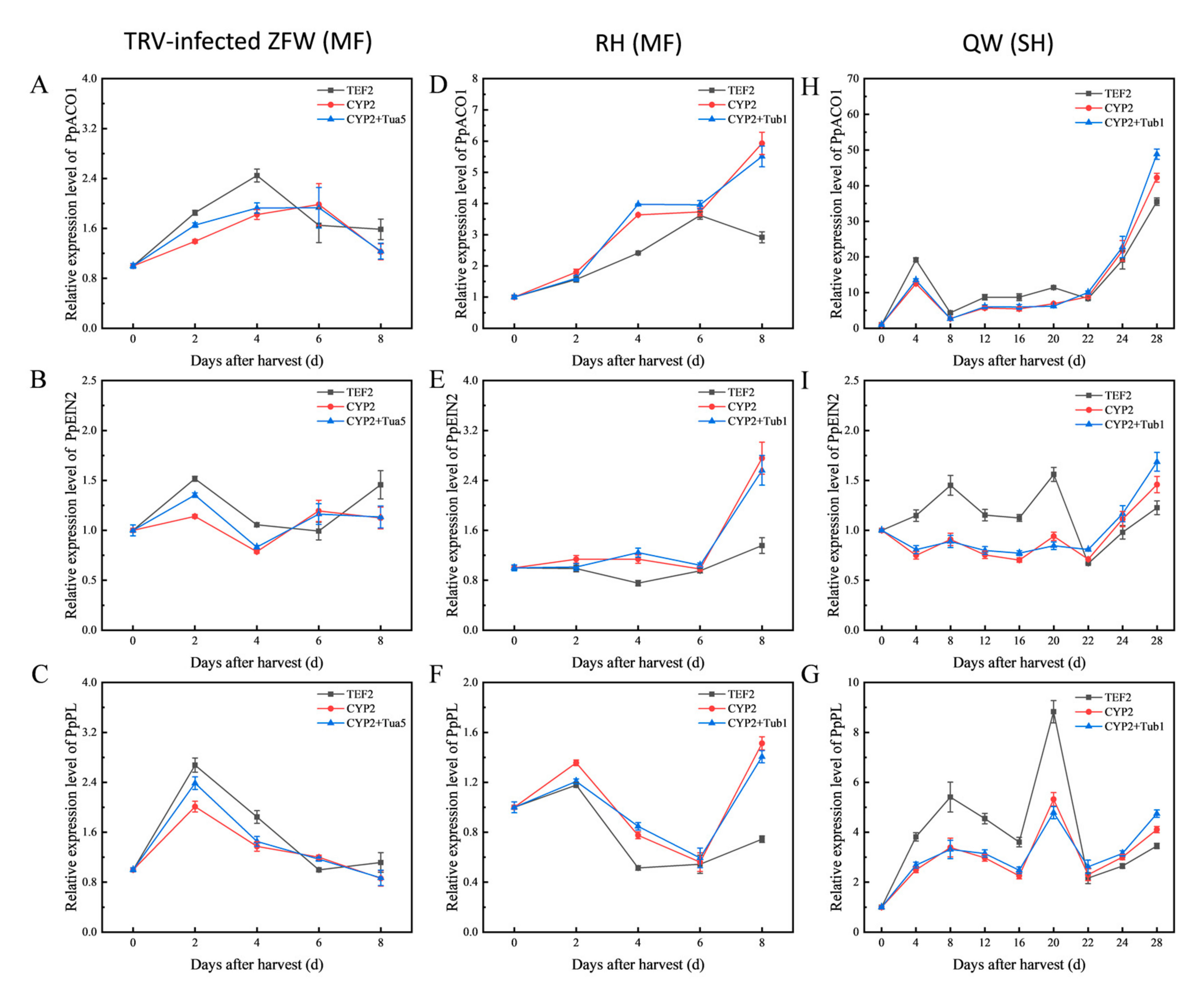
3.4. Validation of Reference Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cao, S.; Liang, M.; Shi, L.; Shao, J.; Song, C.; Bian, K.; Chen, W.; Yang, Z. Accumulation of carotenoids and expression of carotenogenic genes in peach fruit. Food Chem. 2016, 214, 137–146. [Google Scholar] [CrossRef]
- Vimolmangkang, S.; Zheng, H.; Peng, Q.; Jiang, Q.; Wang, H.; Fang, T.; Liao, L.; Wang, L.; He, H.; Han, Y. Assessment of Sugar Components and Genes Involved in the Regulation of Sucrose Accumulation in Peach Fruit. J. Agric. Food Chem. 2016, 64, 6723–6729. [Google Scholar] [CrossRef] [PubMed]
- Engel, H.; Kueppers, C.; Koenig, M.; Loeffert, D. Successful gene expression analysis by multiplex, real-time, one-step RT-PCR, irrespective of the targets amplified. BioTechniques 2007, 43, 230–231. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bustin, S.A.; Benes, V.; Nolan, T.; Pfaffl, M.W. Quantitative real-time RT-PCR—A perspective. J. Mol. Endocrinol. 2005, 34, 597–601. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garson, J.A.; Grant, P.R.; Ayliffe, U.; Ferns, R.B.; Tedder, R.S. Real-time PCR quantitation of hepatitis B virus DNA using automated sample preparation and murine cytomegalovirus internal control. J. Virol. Methods 2005, 126, 207–213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guénin, S.; Mauriat, M.; Pelloux, J.; Van Wuytswinkel, O.; Bellini, C.; Gutierrez, L. Normalization of qRT-PCR data: The necessity of adopting a systematic, experimental conditions-specific, validation of references. J. Exp. Bot. 2009, 60, 487–493. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thellin, O.; Zorzi, W.; Lakaye, B.; De Borman, B.; Coumans, B.; Hennen, G.; Grisar, T.; Igout, A.; Heinen, E. Housekeeping genes as internal standards: Use and limits. J. Biotechnol. 1999, 75, 291–295. [Google Scholar] [CrossRef] [Green Version]
- Schmittgen, T.D.; Zakrajsek, B.A. Effect of experimental treatment on housekeeping gene expression: Validation by real-time, quantitative RT-PCR. J. Biochem. Biophys. Methods 2000, 46, 69–81. [Google Scholar] [CrossRef]
- Tong, Z.; Gao, Z.; Wang, F.; Zhou, J.; Zhang, Z. Selection of reliable reference genes for gene expression studies in peach using real-time PCR. BMC Mol. Biol. 2009, 10, 71. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, X.; Shi, T.; Sun, H.; Song, J.; Ni, Z.; Gao, Z. Selection of suitable inner reference genes for normalisation of microRNA expression response to abiotic stresses by RT-qPCR in leaves, flowers and young stems of peach. Sci. Hortic. 2014, 165, 281–287. [Google Scholar] [CrossRef]
- Kou, X.; Zhang, L.; Yang, S.; Li, G.; Ye, J. Selection and validation of reference genes for quantitative RT-PCR analysis in peach fruit under different experimental conditions. Sci. Hortic. 2017, 225, 195–203. [Google Scholar] [CrossRef]
- Marini, N.; Bevilacqua, C.B.; Büttow, M.V.; Raseira, M.C.B.; Bonow, S. Identification of reference genes for RT-qPCR analysis in peach genotypes with contrasting chilling requirements. Genet. Mol. Res. 2017, 16, gmr16029666. [Google Scholar] [CrossRef] [PubMed]
- You, S.; Cao, K.; Chen, C.; Li, Y.; Wu, J.; Zhu, G.; Fang, W.; Wang, X.; Wang, L. Selection and validation reference genes for qRT-PCR normalization in different cultivars during fruit ripening and softening of peach (Prunus persica). Sci. Rep. 2021, 11, 7302. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Hongjian, W.; Yuejian, Y. Characterization and Selection of Reference Genes for Real-time Quantitative RT-PCR of Plants. Chin. Bull. Bot. 2012, 47, 427–436. [Google Scholar] [CrossRef]
- Rickes, L.N.; Klumb, E.K.; Benitez, L.C.; Braga, E.J.B.; Bianchi, V.J. Stable reference genes for studies of gene expression in Prunus persica under water stress. Aust. J. Crop Sci. 2016, 10, 1348–1356. [Google Scholar] [CrossRef]
- Wu, J.; Zhang, H.; Liu, L.; Li, W.; Wei, Y.; Shi, S. Validation of Reference Genes for RT-qPCR Studies of Gene Expression in Preharvest and Postharvest Longan Fruits under Different Experimental Conditions. Front. Plant Sci. 2016, 7, 780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shukla, P.; Reddy, R.A.; Ponnuvel, K.M.; Rohela, G.K.; Shabnam, A.A.; Ghosh, M.K.; Mishra, R.K. Selection of suitable reference genes for quantitative real-time PCR gene expression analysis in Mulberry (Morus alba L.) under different abiotic stresses. Mol. Biol. Rep. 2019, 46, 1809–1817. [Google Scholar] [CrossRef]
- Hayama, H.; Shimada, T.; Fujii, H.; Ito, A.; Kashimura, Y. Ethylene-regulation of fruit softening and softening-related genes in peach. J. Exp. Bot. 2006, 57, 4071–4077. [Google Scholar] [CrossRef] [PubMed]
- Yoshioka, H.; Hayama, H.; Tatsuki, M.; Nakamura, Y. Cell wall modification during development of mealy texture in the stony-hard peach “Odoroki” treated with propylene. Postharvest Biol. Technol. 2010, 55, 1–7. [Google Scholar] [CrossRef]
- Hayama, H.; Tatsuki, M.; Nakamura, Y. Combined treatment of aminoethoxyvinylglycine (AVG) and 1-methylcyclopropene (1-MCP) reduces melting-flesh peach fruit softening. Postharvest Biol. Technol. 2008, 50, 228–230. [Google Scholar] [CrossRef]
- Lu, M.-T.; Song, C.-W.; Huang, C.-C.; Ou, S.-K. Changes in Flesh Firmness and Ethylene Production of Different Peach Types during Fruit Ripening. Acta Hortic. 2008, 153–159. [Google Scholar] [CrossRef]
- Haji, T.; Yaegaki, H.; Yamaguchi, M. Changes in ethylene production and flesh firmness of melting, nonmelting and stony hard peaches after harvest. J. Jpn. Soc. Hortic. Sci. 2001, 70, 458–459. [Google Scholar] [CrossRef] [Green Version]
- Haji, T.; Yaegaki, H.; Yamaguchi, M. Varietal Differences in the Relationship between Maturation Characteristics, Storage Life and Ethylene Production in Peach Fruit. J. Jpn. Soc. Hortic. Sci. 2004, 73, 97–104. [Google Scholar] [CrossRef] [Green Version]
- Lu, R.; Martin-Hernandez, A.M.; Peart, J.R.; Malcuit, I.; Baulcombe, D.C. Virus-induced gene silencing in plants. Methods 2003, 30, 296–303. [Google Scholar] [CrossRef]
- Senthil-Kumar, M.; Mysore, K.S. New dimensions for VIGS in plant functional genomics. Trends Plant Sci. 2011, 16, 656–665. [Google Scholar] [CrossRef] [PubMed]
- Burch-Smith, T.M.; Anderson, J.C.; Martin, G.B.; Dinesh-Kumar, S.P. Applications and advantages of virus-induced gene silencing for gene function studies in plants. Plant J. 2004, 39, 734–746. [Google Scholar] [CrossRef] [PubMed]
- Bai, S.; Tuan, P.A.; Tatsuki, M.; Yaegaki, H.; Ohmiya, A.; Yamamizo, C.; Moriguchi, T. Knockdown of Carotenoid Cleavage Dioxygenase 4 (CCD4) via Virus-Induced Gene Silencing Confers Yellow Coloration in Peach Fruit: Evaluation of Gene Function Related to Fruit Traits. Plant Mol. Biol. Rep. 2015, 34, 257–264. [Google Scholar] [CrossRef]
- Li, J.; Li, F.; Qian, M.; Han, M.; Liu, H.; Zhang, D.; Ma, J.; Zhao, C. Characteristics and regulatory pathway of the PrupeSEP1 SEPALLATA gene during ripening and softening in peach fruits. Plant Sci. 2017, 257, 63–73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, Y.; Zhang, L.; Ma, R. Functional characterization and mapping of two MADS box genes from peach (Prunus persica). Chin. Sci. Bull. 2008, 53, 853–859. [Google Scholar] [CrossRef] [Green Version]
- Tatsuki, M.; Nakajima, N.; Fujii, H.; Shimada, T.; Nakano, M.; Hayashi, K.-I.; Hayama, H.; Yoshioka, H.; Nakamura, Y. Increased levels of IAA are required for system 2 ethylene synthesis causing fruit softening in peach (Prunus persica L. Batsch). J. Exp. Bot. 2013, 64, 1049–1059. [Google Scholar] [CrossRef] [PubMed]
- Leida, C.A.; Terol, J.; Martí, G.; Agustí, M.; Llácer, G.; Badenes, M.L.; Ríos, G. Identification of genes associated with bud dormancy release in Prunus persica by suppression subtractive hybridization. Tree Physiol. 2010, 30, 655–666. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Z.; Reighard, G.L.; Abbott, A.G.; Bielenberg, D.G. Dormancy-associated MADS genes from the EVG locus of peach [Prunus persica (L.) Batsch] have distinct seasonal and photoperiodic expression patterns. J. Exp. Bot. 2009, 60, 3521–3530. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, research0034.1–research0034.11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andersen, C.L.; Jensen, J.L.; Ørntoft, T.F. Normalization of Real-Time Quantitative Reverse Transcription-PCR Data: A Model-Based Variance Estimation Approach to Identify Genes Suited for Normalization, Applied to Bladder and Colon Cancer Data Sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper—Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef] [PubMed]
- Xie, F.; Xiao, P.; Chen, D.; Xu, L.; Zhang, B. miRDeepFinder: A miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol. Biol. 2012, 80, 75–84. [Google Scholar] [CrossRef] [PubMed]
- Valasek, M.A.; Repa, J.J. The power of real-time PCR. Adv. Physiol. Educ. 2005, 29, 151–159. [Google Scholar] [CrossRef] [PubMed]
- Løvdal, T.; Lillo, C. Reference gene selection for quantitative real-time PCR normalization in tomato subjected to nitrogen, cold, and light stress. Anal. Biochem. 2009, 387, 238–242. [Google Scholar] [CrossRef]
- Mascia, T.; Santovito, E.; Gallitelli, D.; Cillo, F. Evaluation of reference genes for quantitative reverse-transcription polymerase chain reaction normalization in infected tomato plants. Mol. Plant Pathol. 2010, 11, 805–816. [Google Scholar] [CrossRef] [PubMed]
- Remans, T.; Smeets, K.; Opdenakker, K.; Mathijsen, D.; Vangronsveld, J.; Cuypers, A. Normalisation of real-time RT-PCR gene expression measurements in Arabidopsis thaliana exposed to increased metal concentrations. Planta 2008, 227, 1343–1349. [Google Scholar] [CrossRef] [Green Version]
- Nicot, N.; Hausman, J.-F.; Hoffmann, L.; Evers, D. Housekeeping gene selection for real-time RT-PCR normalization in potato during biotic and abiotic stress. J. Exp. Bot. 2005, 56, 2907–2914. [Google Scholar] [CrossRef] [PubMed]
- Libault, M.; Thibivilliers, S.; Bilgin, D.D.; Radwan, O.; Benitez, M.; Clough, S.J.; Stacey, G. Identification of Four Soybean Reference Genes for Gene Expression Normalization. Plant Genome 2008, 1, 44–54. [Google Scholar] [CrossRef]
- Hu, R.; Fan, C.; Li, H.; Zhang, Q.; Fu, Y.-F. Evaluation of putative reference genes for gene expression normalization in soybean by quantitative real-time RT-PCR. BMC Mol. Biol. 2009, 10, 93. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, L.; Zhong, H.-Y.; Kuang, J.-F.; Li, J.-G.; Lu, W.-J.; Chen, J.-Y. Validation of reference genes for RT-qPCR studies of gene expression in banana fruit under different experimental conditions. Planta 2011, 234, 377–390. [Google Scholar] [CrossRef] [PubMed]
- Jain, M.; Nijhawan, A.; Tyagi, A.K.; Khurana, J.P. Validation of housekeeping genes as internal control for studying gene expression in rice by quantitative real-time PCR. Biochem. Biophys. Res. Commun. 2006, 345, 646–651. [Google Scholar] [CrossRef] [PubMed]
- Joseph, J.T.; Poolakkalody, N.J.; Shah, J.M. Plant reference genes for development and stress response studies. J. Biosci. 2018, 43, 173–187. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Peng, X.; Liu, Y.; Li, Y.; Luo, Y.; Wang, X.; Tang, H. Evaluation of suitable reference genes for qRT-PCR normalization in strawberry (Fragaria × ananassa) under different experimental conditions. BMC Mol. Biol. 2018, 19, 8. [Google Scholar] [CrossRef]
- Feng, K.; Liu, J.-X.; Xing, G.-M.; Sun, G.; Li, S.; Duan, A.-Q.; Wang, F.; Li, M.-Y.; Xu, Z.-S.; Xiong, A.-S. Selection of appropriate reference genes for RT-qPCR analysis under abiotic stress and hormone treatment in celery. PeerJ 2019, 7, e7925. [Google Scholar] [CrossRef]
- Migocka, M.; Papierniak, A. Identification of suitable reference genes for studying gene expression in cucumber plants subjected to abiotic stress and growth regulators. Mol. Breed. 2010, 28, 343–357. [Google Scholar] [CrossRef]
- Niu, L.; Tao, Y.-B.; Chen, M.-S.; Fu, Q.; Li, C.; Dong, Y.; Wang, X.; He, H.; Xu, Z.-F. Selection of Reliable Reference Genes for Gene Expression Studies of a Promising Oilseed Crop, Plukenetia volubilis, by Real-Time Quantitative PCR. Int. J. Mol. Sci. 2015, 16, 12513–12530. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Rank | All Samples | TRV-Infected Fruits | Different Genotypes | |||||
|---|---|---|---|---|---|---|---|---|
| No Subgroups | 2 Subgroups | |||||||
| 1 | CYP2 | 0.192 | RP II | 0.110 | CYP2 | 0.125 | CYP2 | 0.222 |
| 2 | RP II | 0.381 | CYP2 | 0.133 | Tua5 | 0.211 | UBQ10 | 0.404 |
| 3 | Tub1 | 0.407 | Tub1 | 0.218 | RP II | 0.226 | RP II | 0.426 |
| 4 | UBQ10 | 0.417 | Tua5 | 0.227 | Actin | 0.252 | Tub1 | 0.432 |
| 5 | Tua5 | 0.437 | UBQ10 | 0.240 | Tub1 | 0.267 | Tua5 | 0.470 |
| 6 | Actin | 0.478 | PLA2 | 0.291 | UBQ10 | 0.433 | Actin | 0.476 |
| 7 | GADPH | 0.625 | Actin | 0.312 | PLA2 | 0.471 | GADPH | 0.544 |
| 8 | PLA2 | 0.691 | TEF2 | 0.343 | GADPH | 0.775 | PLA2 | 0.739 |
| 9 | TEF2 | 0.794 | GADPH | 0.350 | TEF2 | 0.815 | TEF2 | 0.785 |
| 10 | 18S | 0.980 | 18S | 0.444 | 18S | 0.816 | 18S | 0.969 |
| Rank | All Samples | TRV-Infected Fruits | Different Genotypes | ||||||
|---|---|---|---|---|---|---|---|---|---|
| CV ± SD | R | CV ± SD | R | CV ± SD | R | ||||
| 1 | CYP2 | 3.10 ± 0.81 | 0.954 | CYP2 | 3.97 ± 1.01 | 0.999 | Tua5 | 3.24 ± 0.81 | 0.934 |
| 2 | Tua5 | 4.02 ± 0.99 | 0.954 | Actin | 5.88 ± 1.20 | 0.991 | CYP2 | 2.40 ± 0.63 | 0.918 |
| 3 | TEF2 | 5.18 ± 1.16 | 0.928 | Tub1 | 5.37 ± 1.20 | 0.989 | Actin | 4.05 ± 0.84 | 0.907 |
| 4 | RP II | 3.23 ± 0.79 | 0.916 | Tua5 | 4.80 ± 1.15 | 0.984 | RP II | 2.71 ± 0.67 | 0.881 |
| 5 | Actin | 4.66 ± 0.97 | 0.915 | TEF2 | 7.64 ± 1.65 | 0.978 | TEF2 | 4.13 ± 0.94 | 0.867 |
| 6 | Tub1 | 3.66 ± 0.84 | 0.896 | RP II | 3.99 ± 0.95 | 0.971 | UBQ10 | 2.64 ± 0.59 | 0.835 |
| 7 | UBQ10 | 3.51 ± 0.78 | 0.891 | UBQ10 | 3.43 ± 0.73 | 0.969 | GADPH | 3.39 ± 0.73 | 0.815 |
| 8 | GADPH | 4.55 ± 0.96 | 0.848 | PLA2 | 3.27 ± 0.90 | 0.966 | Tub1 | 2.83 ± 0.65 | 0.803 |
| 9 | PLA2 | 2.62 ± 0.74 | 0.741 | 18S | 5.72 ± 0.59 | 0.957 | 18S | 5.75 ± 0.60 | 0.437 |
| 10 | 18S | 5.87 ± 0.61 | 0.554 | GADPH | 5.40 ± 1.09 | 0.145 | PLA2 | 1.66 ± 0.47 | 0.435 |
| Rank | All Samples | TRV-Infected Fruits | Different Genotypes | |||
|---|---|---|---|---|---|---|
| 1 | CYP2 | 1.50 | CYP2 | 2.11 | CYP2 | 1.41 |
| 2 | Tub1 | 2.45 | Tua5 | 3.03 | Tub1 | 2.51 |
| 3 | RPII | 3.13 | Actin | 3.22 | UBQ10 | 2.63 |
| 4 | UBQ10 | 4.12 | Tub1 | 3.56 | RPII | 4.61 |
| 5 | Actin | 5.42 | RPII | 4.16 | PLA2 | 4.76 |
| 6 | 18S | 5.62 | UBQ10 | 4.56 | Actin | 5.05 |
| 7 | PLA2 | 5.66 | 18S | 5.62 | Tua5 | 6.16 |
| 8 | Tua5 | 5.79 | PLA2 | 5.66 | GADPH | 7.00 |
| 9 | GADPH | 7.00 | GADPH | 7.90 | 18S | 7.40 |
| 10 | TEF2 | 9.24 | TEF2 | 8.71 | TEF2 | 9.24 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, Z.; Dai, J.; Su, W.; Wu, H.; Shah, K.; Xing, L.; Ma, J.; Zhang, D.; Zhao, C. Selection and Validation of Reliable Reference Genes for Gene Expression Studies in Different Genotypes and TRV-Infected Fruits of Peach (Prunus persica L. Batsch) during Ripening. Genes 2022, 13, 160. https://doi.org/10.3390/genes13010160
Xu Z, Dai J, Su W, Wu H, Shah K, Xing L, Ma J, Zhang D, Zhao C. Selection and Validation of Reliable Reference Genes for Gene Expression Studies in Different Genotypes and TRV-Infected Fruits of Peach (Prunus persica L. Batsch) during Ripening. Genes. 2022; 13(1):160. https://doi.org/10.3390/genes13010160
Chicago/Turabian StyleXu, Ze, Jieyu Dai, Weijing Su, Haixia Wu, Kamran Shah, Libo Xing, Juanjuan Ma, Dong Zhang, and Caiping Zhao. 2022. "Selection and Validation of Reliable Reference Genes for Gene Expression Studies in Different Genotypes and TRV-Infected Fruits of Peach (Prunus persica L. Batsch) during Ripening" Genes 13, no. 1: 160. https://doi.org/10.3390/genes13010160
APA StyleXu, Z., Dai, J., Su, W., Wu, H., Shah, K., Xing, L., Ma, J., Zhang, D., & Zhao, C. (2022). Selection and Validation of Reliable Reference Genes for Gene Expression Studies in Different Genotypes and TRV-Infected Fruits of Peach (Prunus persica L. Batsch) during Ripening. Genes, 13(1), 160. https://doi.org/10.3390/genes13010160







