Comparative Transcriptomic Analyses of Different Jujube Cultivars Reveal the Co-Regulation of Multiple Pathways during Fruit Cracking
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Measurements of Fruit Cracking and Fruit Size
2.3. RNA Extraction and Sequencing
2.4. Sequence Data Analysis
2.5. Differential Expression Analysis
2.6. Functional Annotation, Enrichment Analysis and Transcription Factor Identification
2.7. Co-Expression Analysis
2.8. Validation by qRT-PCR Analysis
2.9. Statistical Analysis
3. Results
3.1. Analysis of Fruit Characteristics in Three Jujube Cultivars
3.2. Transcriptome Sequencing Results
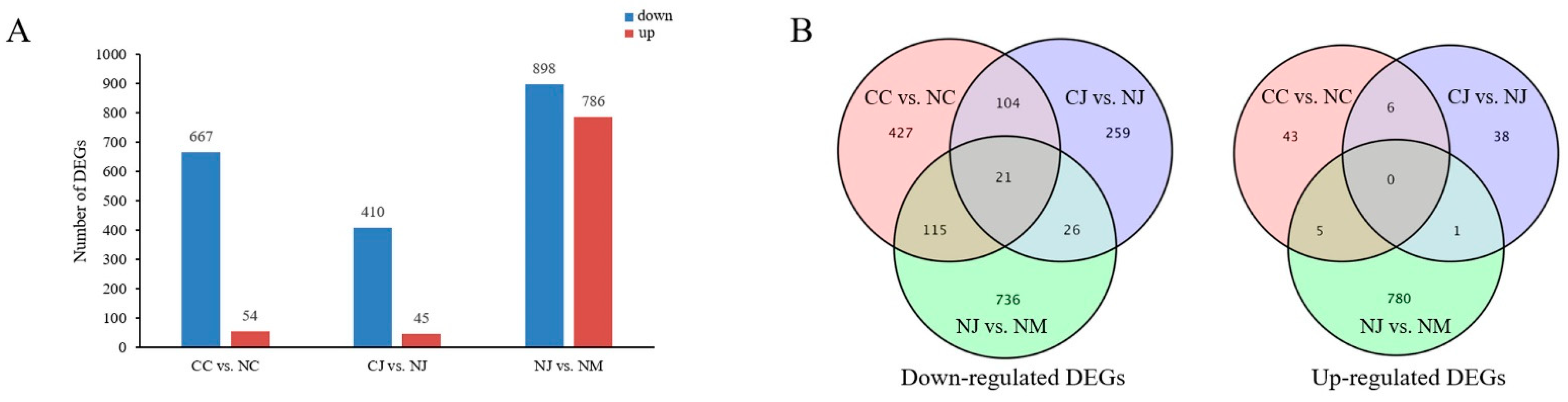
3.3. Analysis of DEGs among Different Jujube Cultivars
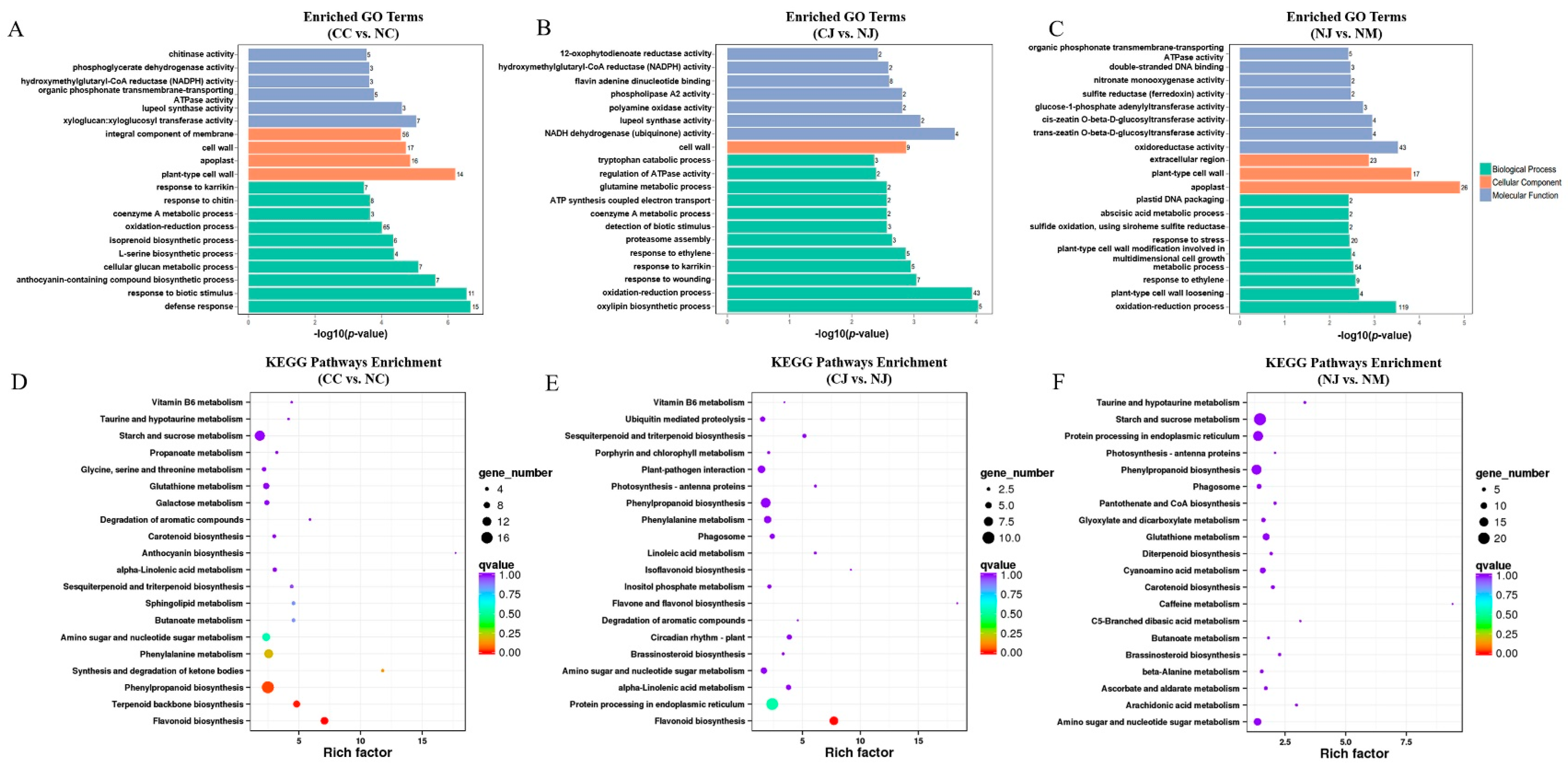
3.4. Functional Annotation of DEGs
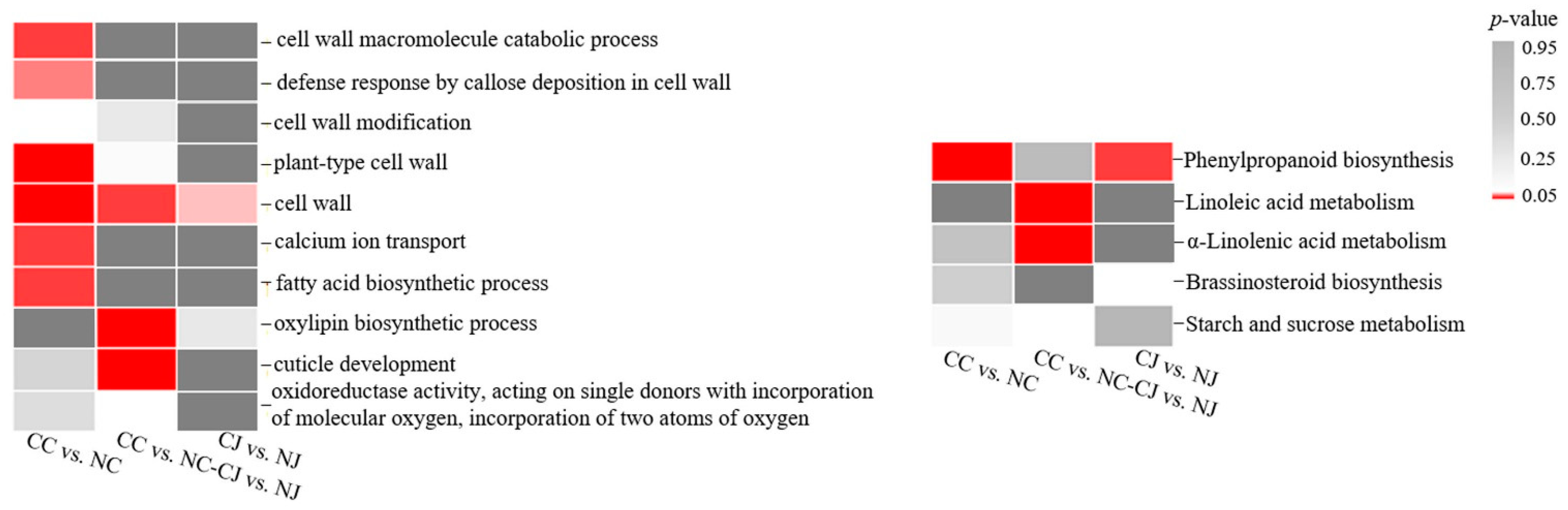
3.5. Venn Diagram and Trend Analyses
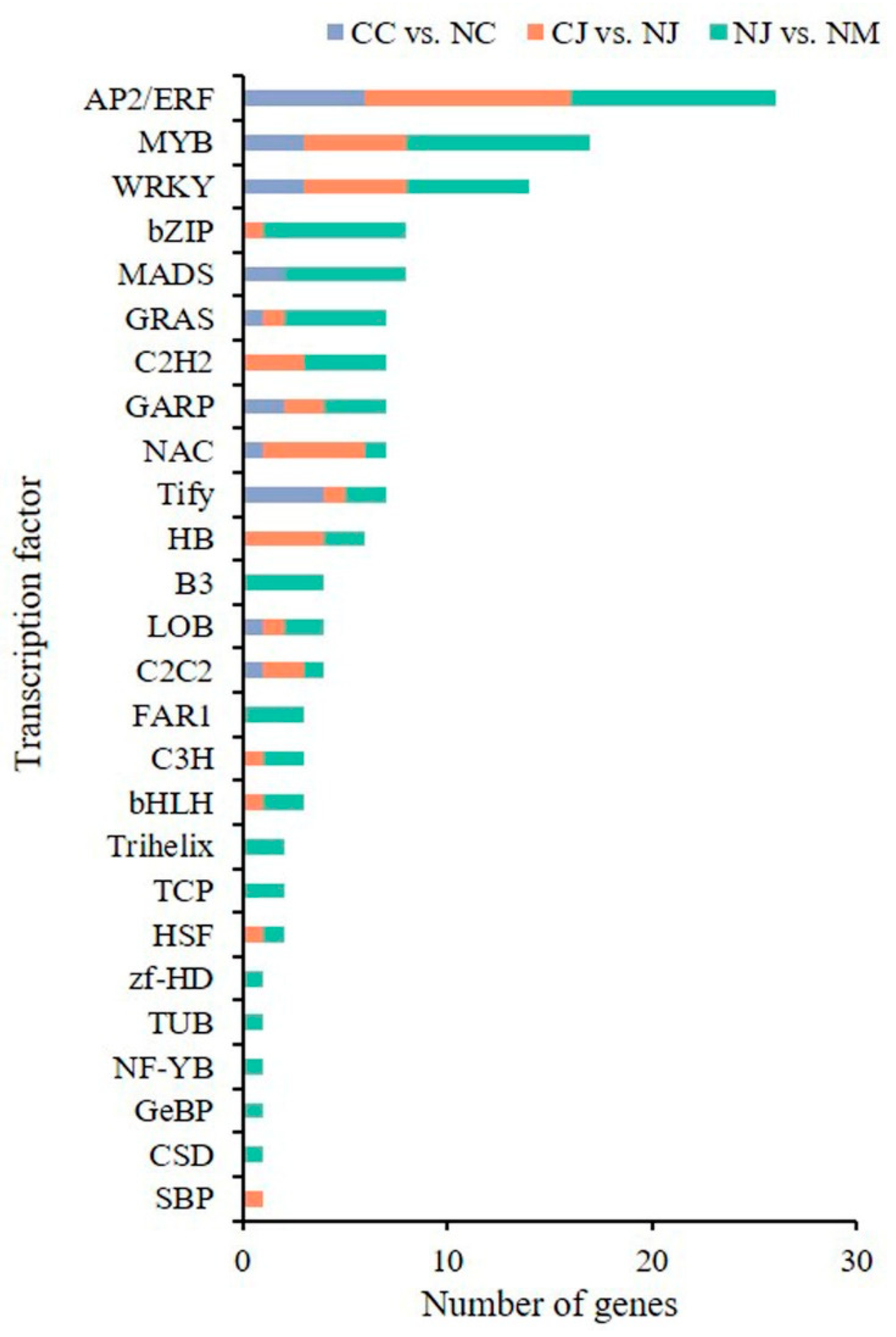
3.6. Identification of Differentially Expressed Transcription Factors
3.7. Co-Expression Analysis of Cracking-Related Genes
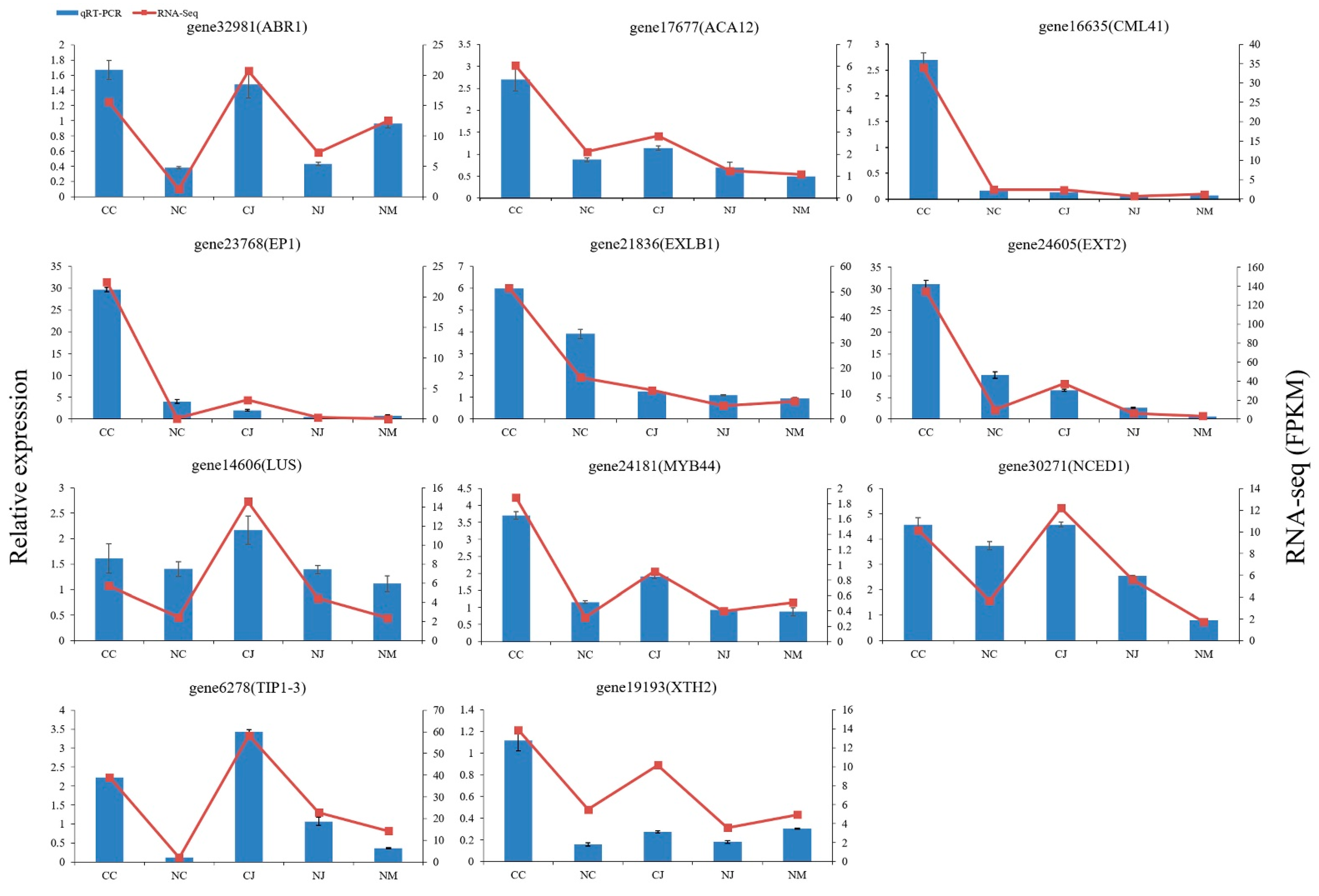
3.8. qRT-PCR Validation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Liu, M. Handbook of High Quality Production of Chinese Jujube; China Agriculture Press: Beijing, China, 2004; pp. 1–22, 275–323. [Google Scholar]
- Liu, M. The challenges and countermeasures of jujube industry during transition period. China Fruits 2018, 1, 1–4. [Google Scholar]
- Wang, Z.L.; Tian, Y.P.; Liu, M.J.; Han, H.Z.; Shao, X.H.; Li, K.S. Crack resistance and its mechanism of different Chinese jujube cultivars. Non-Wood For. Res. 2011, 3, 74–77. [Google Scholar]
- Wang, Y.; Guo, L.; Zhao, X.; Zhao, Y.; Hao, Z.; Luo, H.; Yuan, Z. Advances in Mechanisms and Omics Pertaining to Fruit Cracking in Horticultural Plants. Agronomy 2021, 11, 1045. [Google Scholar] [CrossRef]
- Zhu, M.; Yu, J.; Zhao, M.; Wang, M.; Yang, G. Transcriptome analysis of metabolisms related to fruit cracking during ripening of a cracking-susceptible grape berry cv. Xiangfei (Vitis vinifera L.). Genes Genom. 2020, 42, 639–650. [Google Scholar] [CrossRef] [PubMed]
- Cuartero, J.; Palomares, G.; Balasch, S.; Nuez, F. Tomato fruit cracking under plastic-house and in the open air. II. General and specific combining abilities. In Genetics and Breeding of Tomato, Proceeding of the Meeting Eucarpia Tomato Working Group, France, Avignon, 18–21 May 1981; Institut National de la Recherche Agronomique: Versailles, France, 1981; pp. 91–98. [Google Scholar]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef]
- Li, W.C.; Wu, J.Y.; Zhang, H.N.; Shi, S.Y.; Liu, L.Q.; Shu, B.; Liang, Q.Z.; Xie, J.H.; Wei, Y.Z. De novo assembly and characterization of pericarp transcriptome and identification of candidate genes mediating fruit cracking in Litchi chinensis Sonn. Int. J. Mol. Sci. 2014, 15, 17667–17685. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Gao, X.; Ma, Z.; Chen, J.; Liu, Y. Analysis of the molecular basis of fruit cracking susceptibility in Litchi chinensis cv. Baitangying by transcriptome and quantitative proteome profiling. J. Plant Physiol. 2019, 234–235, 106–116. [Google Scholar] [CrossRef]
- Li, H.; Li, W.; Zhang, T.; Zhong, J.; Liu, J.; Yuan, C.; Liu, K. Comparative transcriptomic analysis of split and non-split atemoya (Annona cherimola Mill. × Annona squamosa L.) fruit to identify potential genes involved in the fruit splitting process. Sci. Hortic. 2019, 248, 216–224. [Google Scholar] [CrossRef]
- Xue, L.; Sun, M.; Wu, Z.; Yu, L.; Yu, Q.; Tang, Y.; Jiang, F. LncRNA regulates tomato fruit cracking by coordinating gene expression via a hormone-redox-cell wall network. BMC Plant Biol. 2020, 20, 162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Wu, X.F.; Tang, Y.; Li, J.G.; Zhao, M.L. RNA-Seq Provides New Insights into the Molecular Events Involved in “Ball-Skin versus Bladder Effect” on Fruit Cracking in Litchi. Int. J. Mol. Sci. 2021, 22, 454. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Fu, L.; Song, Y.; Li, J.; Xue, X.; Li, S.; Li, L. Water entry in jujube fruit and its relationship with cracking. Acta Physio. Plant. 2019, 41, 162. [Google Scholar] [CrossRef]
- Li, N.; Fu, L.; Song, Y.; Li, J.; Xue, X.; Li, S.; Li, L. Wax composition and concentration in jujube (Ziziphus jujuba Mill.) cultivars with differential resistance to fruit cracking. J. Plant Physiol. 2019, 255, 153294. [Google Scholar] [CrossRef] [PubMed]
- Lichter, A.; Dvir, O.; Fallik, E.; Cohen, S.; Golan, R.; Shemer, Z.; Sagi, M. Cracking of cherry tomatoes in solution. Postharvest Biol. Technol. 2002, 26, 305–312. [Google Scholar] [CrossRef]
- Ozturk, B. Cracking and quality attributes of jujube fruits as affected by covering and pre-harvest Parka and GA3 treatments. Sci. Hortic. 2018, 240, 65–71. [Google Scholar] [CrossRef]
- Hou, L.; Chen, W.; Zhang, Z.; Pang, X.; Li, Y. Genome-wide association studies of fruit quality traits in jujube germplasm collections using genotyping-by-sequencing. Plant Genome 2020, 13, e20036. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, P.; Geng, Y.; Xie, X.; Wen, P. Cracking of jujube fruits is associated with differential expression of metabolic genes. FEBS Open Bio. 2020, 10, 1765–1773. [Google Scholar] [CrossRef]
- Chen, W.; Kong, D.; Cui, Y.; Ming, C.; Pang, X.; Li, Y. Phenotypic genetic diversity of a core collection of Ziziphus jujuba and correlation analysis of dehiscent characters. J. Beijing For. Univ. 2017, 39, 78–84. [Google Scholar]
- Du, W.; Li, X.G.; Wang, C.Z.; Gao, W.H.; Wang, Y.Q. Mechanism of fruit dehiscent in Zizyphus jujube. J. Fruit Sci. 2012, 29, 374–381. [Google Scholar]
- Yuan, Z.; Lu, Y.Q.; Zhao, J.; Liu, M.J. Screening and evaluation of germplasms with high resistance to fruit dehiscent in Ziziphus jujuba Mill. Sci Agric. Sin. 2013, 46, 4968–4976. [Google Scholar]
- Ewing, B.; Hillier, L.; Wendl, M.C.; Green, P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 1998, 8, 175–185. [Google Scholar] [CrossRef] [Green Version]
- Liu, M.; Zhao, J.; Cai, Q.; Liu, G.; Wang, J.; Zhao, Z.; Liu, P.; Dai, L.; Yan, G.; Wang, W.; et al. The complex jujube genome provides insights into fruit tree biology. Nat. Commun. 2014, 5, 5315. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013, 14, R36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [Green Version]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, 14. [Google Scholar] [CrossRef] [Green Version]
- Mao, X.; Cai, T.; Olyarchuk, J.G.; Wei, L. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 2005, 21, 3787–3793. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Jiao, C.; Sun, H.; Rosli, H.; Pombo, M.A.; Zhang, P.; Banf, M.; Dai, X.; Martin, G.; Giovannoni, J.J.; et al. iTAK: A Program for Genome-wide Prediction and Classification of Plant Transcription Factors, Transcriptional Regulators, and Protein Kinases. Mol. Plant 2016, 9, 1667–1670. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga1, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Bu, J.; Zhao, J.; Liu, M. Expression stabilities of candidate reference genes for RT-qPCR in Chinese jujube (Ziziphus jujuba Mill.) under a variety of conditions. PLoS ONE 2016, 11, e0154212. [Google Scholar] [CrossRef] [Green Version]
- Alexandersson, E.; Fraysse, L.; Sjövall-Larsen, S.; Gustavsson, S.; Fellert, M.; Karlsson, M.; Johanson, U.; Kjellbom, P. Whole gene family expression and drought stress regulation of aquaporins. Plant Mol. Biol. 2005, 59, 469–484. [Google Scholar] [CrossRef]
- Khadivi-Khub, A. Physiological and genetic factors influencing fruit cracking. Acta Physio. Plant. 2015, 37, 1718. [Google Scholar] [CrossRef]
- Li, X.; Chen, X.; Wen, X.; Zhao, Y.; Ma, H. Correlation Analysis between the Sugar Components and Fruit Cracking in Easily Cracked and Resistant Jujube. Mol. Plant Breed. 2020, 18, 6180–6186. [Google Scholar]
- Wang, J.G.; Gao, X.M.; Ma, Z.L.; Chen, J.; Liu, Y.N.; Shi, W.Q. Metabolomic and transcriptomic profiling of three types of litchi pericarps reveals that changes in the hormone balance constitute the molecular basis of the fruit cracking susceptibility of Litchi chinensis cv. Baitangying. Mol. Biol. Rep. 2019, 46, 5295–5308. [Google Scholar] [CrossRef] [PubMed]
- Wójcik, P.; Akgül, H.; Demirtaş, I.; Sarısu, C.; Aksu, M.; Gubbuk, H. Effect of preharvest spays of calcium chloride and sucrose on cracking and quality of ‘Burlat’ sweet cherry fruit. J. Plant Nutr. 2013, 36, 1453–1465. [Google Scholar] [CrossRef]
- Correia, S.; Schouten, R.; Silva, A.P.; Gonçalves, B. Sweet cherry fruit cracking mechanisms and prevention strategies: A review. Sci. Hortic. 2018, 240, 369–377. [Google Scholar] [CrossRef]
- Limonta, M.; Romanowsky, S.; Olivari, C.; Bonza, M.C.; Luoni, L.; Rosenberg, A.; Harper, J.F.; De Michelis, M.I. ACA12 is a deregulated isoform of plasma membrane Ca2+-ATPase of Arabidopsis thaliana. Plant Mol. Biol. 2014, 84, 387–397. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, N.; Wang, G.; Zong, M.; Han, S.; Liu, F. Genome-wide identification, and phylogenetic and expression profiling analyses of CaM and CML genes in Brassica rapa and Brassica oleracea. Gene 2018, 677, 232–244. [Google Scholar] [CrossRef]
- Yang, S.; Zheng, G.; Zhang, Y.; Zang, Y. Ultracytochemical localization of Ca2+ in the normal fruits and cracking ones of Lingwu Long-jujube. Acta Bot. Bor-Occid. Sin. 2011, 31, 84–88. [Google Scholar]
- Yang, W.; Zeng, H.; Zou, M.; Chaozhong, L.U. An overview of the roles of cell wall modification in fruit pericarp cracking. Chin. J. Trop. Crops 2011, 32, 1995–1999. [Google Scholar]
- Jiang, F.; Lopez, A.; Jeon, S.; de Freitas, S.T.; Yu, Q.; Wu, Z.; Labavitch, J.M.; Tian, S.; Powell, A.L.T.; Mitcham, E. Disassembly of the fruit cell wall by the ripening-associated polygalacturonase and expansin influences tomato cracking. Hortic. Res. 2019, 6, 17. [Google Scholar] [CrossRef] [Green Version]
- Petit, J.; Bres, C.; Mauxion, J.P.; Jun, W.; Tai, F.; Martin, L.B.B.; Fich, E.A.; Joubès, J.; Rose, J.K.C.; Domergue, F.; et al. The glycerol-3-phosphate acyltransferase GPAT6 from tomato plays a central role in fruit cutin biosynthesis. Plant Physiol. 2016, 171, 894–913. [Google Scholar] [CrossRef] [PubMed]
- Domínguez, E.; Cuartero, J.; Heredia, A. An overview on plant cuticle biomechanics. Plant Sci. 2011, 181, 77–84. [Google Scholar] [CrossRef]
- Mutuku, J.M.; Cui, S.; Hori, C.; Takeda, Y.; Tobimatsu, Y.; Nakabayashi, R.; Mori, T.; Saito, K.; Demura, T.; Umezawa, T.; et al. The structural integrity of lignin is crucial for resistance against Striga hermonthica parasitism in rice. Plant Physiol. 2019, 179, 1796–1809. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marmon, S.K.; Sturtevant, D.; Herrfurth, C.; Chapman, K.D.; Stymne, S.; Feussner, I. Two acyltransferases contribute differently to linolenic acid levels in seed oil. Plant Physiol. 2017, 173, 2081. [Google Scholar] [CrossRef] [Green Version]
- Marboh, E.; Singh, S.K.; Swapnil, P.; Nath, V.; Gupta, A.K.; Pongener, A. Fruit cracking in litchi (Litchi chinensis): An overview. Indian J. Agric. Res. 2017, 87, 3–11. [Google Scholar]
- Stern, R.A.; Flaishman, M.; Applebaum, S.; Ben-Arie, R. Effect of synthetic auxins on fruit development of ‘Bing’ cherry (Prunus avium L.). Sci. Hortic. 2007, 114, 275–280. [Google Scholar] [CrossRef]
- De Freitas, S.T.; Shackel, K.A.; Mitcham, E.J. Abscisic acid triggers whole-plant and fruit-specific mechanisms to increase fruit calcium uptake and prevent blossom end rot development in tomato fruit. J. Exp. Bot. 2011, 62, 2645–2656. [Google Scholar] [CrossRef] [Green Version]
- Napier, R.M.; Venis, M.A. From auxin-binding protein to plant hormone receptor? Trends Biochem. Sci. 1991, 16, 72–75. [Google Scholar]
- Sun, L.; Sun, Y.; Zhang, M.; Wang, L.; Ren, J.; Cui, M.; Wang, Y.; Ji, K.; Li, P.; Li, Q.; et al. Suppression of 9-cis-epoxycarotenoid dioxygenase, which encodes a key enzyme in abscisic acid biosynthesis, alters fruit texture in transgenic tomato. Plant Physiol. 2012, 158, 283–298. [Google Scholar] [CrossRef] [Green Version]
- Tran, L.S.P.; Mochida, K. Identification and prediction of abiotic stress responsive transcription factors involved in abiotic stress signaling in soybean. Plant Signal. Behav. 2010, 5, 255–257. [Google Scholar] [CrossRef] [Green Version]
- Liao, N.; Hu, Z.; Li, Y.; Hao, J.; Chen, S.; Xue, Q.; Ma, Y.; Zhang, K.; Mahmoud, A.; Ali, A.; et al. Ethylene-responsive factor 4 is associated with the desirable rind hardness trait conferring cracking resistance in fresh fruits of watermelon. Plant Biotechnol. J. 2020, 18, 1066–1077. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, H.; Yang, W.; Liu, D.; Han, Y.; Zhang, A.; Li, S. Ectopic expression of a grapevine transcription factor VvWRKY11 contributes to osmotic stress tolerance in Arabidopsis. Mol. Biol. Rep. 2011, 38, 417–427. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Jia, J.; Zhao, P.; Guo, X.; Chen, S.; Qi, D.; Cheng, L.; Liu, G. LcMYB4, an unknown function transcription factor gene from sheepgrass, as a positive regulator of chilling and freezing tolerance in transgenic Arabidopsis. BMC Plant Biol. 2020, 20, 238. [Google Scholar] [CrossRef] [PubMed]



| Sample | Raw Reads | Clean Reads | Clean Bases | GC (%) | Q30 (%) | Total Mapped Reads (%) | Uniquely Mapped Reads (%) |
|---|---|---|---|---|---|---|---|
| CC | 26,363,434 | 25,818,100 | 7,722,697,490 | 44.42 | 91.42 | 73.16 | 64.03 |
| NC | 26,315,272 | 25,807,498 | 7,725,295,650 | 44.86 | 86.92 | 70.40 | 60.43 |
| CJ | 29,154,642 | 28,593,357 | 8,556,528,121 | 44.48 | 86.91 | 70.66 | 62.15 |
| NJ | 27,450,994 | 25,637,345 | 7,667,051,071 | 45.45 | 86.92 | 71.72 | 57.72 |
| NM | 28,834,319 | 28,252,252 | 8,438,681,007 | 44.51 | 87.27 | 71.11 | 62.54 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hou, L.; Li, M.; Zhang, C.; Liu, N.; Liu, X.; Bo, W.; Pang, X.; Li, Y. Comparative Transcriptomic Analyses of Different Jujube Cultivars Reveal the Co-Regulation of Multiple Pathways during Fruit Cracking. Genes 2022, 13, 105. https://doi.org/10.3390/genes13010105
Hou L, Li M, Zhang C, Liu N, Liu X, Bo W, Pang X, Li Y. Comparative Transcriptomic Analyses of Different Jujube Cultivars Reveal the Co-Regulation of Multiple Pathways during Fruit Cracking. Genes. 2022; 13(1):105. https://doi.org/10.3390/genes13010105
Chicago/Turabian StyleHou, Lu, Meng Li, Chenxing Zhang, Ningwei Liu, Xinru Liu, Wenhao Bo, Xiaoming Pang, and Yingyue Li. 2022. "Comparative Transcriptomic Analyses of Different Jujube Cultivars Reveal the Co-Regulation of Multiple Pathways during Fruit Cracking" Genes 13, no. 1: 105. https://doi.org/10.3390/genes13010105
APA StyleHou, L., Li, M., Zhang, C., Liu, N., Liu, X., Bo, W., Pang, X., & Li, Y. (2022). Comparative Transcriptomic Analyses of Different Jujube Cultivars Reveal the Co-Regulation of Multiple Pathways during Fruit Cracking. Genes, 13(1), 105. https://doi.org/10.3390/genes13010105






