Abstract
The year 2021 marks the 50th anniversary of the National Cancer Act, signed by President Nixon, which declared a national “war on cancer.” Powered by enormous financial support, this past half-century has witnessed remarkable progress in understanding the individual molecular mechanisms of cancer, primarily through the characterization of cancer genes and the phenotypes associated with their pathways. Despite millions of publications and the overwhelming volume data generated from the Cancer Genome Project, clinical benefits are still lacking. In fact, the massive, diverse data also unexpectedly challenge the current somatic gene mutation theory of cancer, as well as the initial rationales behind sequencing so many cancer samples. Therefore, what should we do next? Should we continue to sequence more samples and push for further molecular characterizations, or should we take a moment to pause and think about the biological meaning of the data we have, integrating new ideas in cancer biology? On this special anniversary, we implore that it is time for the latter. We review the Genome Architecture Theory, an alternative conceptual framework that departs from gene-based theories. Specifically, we discuss the relationship between genes, genomes, and information-based platforms for future cancer research. This discussion will reinforce some newly proposed concepts that are essential for advancing cancer research, including two-phased cancer evolution (which reconciles evolutionary contributions from karyotypes and genes), stress-induced genome chaos (which creates new system information essential for macroevolution), the evolutionary mechanism of cancer (which unifies diverse molecular mechanisms to create new karyotype coding during evolution), and cellular adaptation and cancer emergence (which explains why cancer exists in the first place). We hope that these ideas will usher in new genomic and evolutionary conceptual frameworks and strategies for the next 50 years of cancer research.
1. Introduction
The year 2021 is the National Cancer Act’s 50th birthday [1]. The Act had a profound impact on scientific progress and status, shifting the landscape of cancer research. According to leading cancer gene researcher Robert Weinberg, “The molecular cancer story really began early in the decade—1971 to be precise—when an enormous pot of money suddenly became available for cancer research. President Nixon’s War on Cancer, as it came to be called, was fueled by the conviction that cancer was ultimately a disease of infectious tumor viruses.” [2].
However, as Weinberg describes, it soon turned out that the scientific conviction to start the war was wrong.
“Looking back, it’s clear that the scramble to find human retroviruses represented the major irony of the War on Cancer: it had been launched for the wrong reason, since cancer-causing human retroviruses were never found (with the exception of rare leukemias in the Caribbean and southern Japan).”[2]
Nevertheless, as with many other complex adaptive systems, the war on cancer found its own life. Supported by renewed funding, new convictions arose and became new targeted battles with new rationalizations: from oncogenes to tumor suppressors, differentiation genes, DNA repair genes, cell cycle genes, cell death genes, metabolic genes, stress response genes, genome instability genes, and immune genes; from a handful of cancer genes to hundreds classified by “cancer hallmarks” to thousands of genes and beyond; from DNA to RNA to proteins, lipids, and sugars; and from cellular organelles to gap junctions to tissue structure. Although each battle promised to end cancer once and for all, the war goes on, seemingly endlessly [3,4].
These battles have faced highly diverse targets, although all share the same assumption: cancer is a disease of uncontrollable growth, with a causation that can be identified by studying gene mutation and epigenetic regulation. Then, when all these individual battles failed to deliver, a bold idea emerged as the final battle: can we sequence every single gene mutation in cancer? Therefore, the Cancer Genome Atlas (TCGA) was born. It promised to leave no stone unturned and finally understand and cure all cancers [3,5].
Meanwhile, after conducting “watching cancer evolution in action” experiments [6], we proposed that karyotype-mediated macroevolution, not gene mutation, is the common driving force for most cancers. Our genome-based theory (which would formally develop into the Genome Architecture Theory) predicted that large-scale sequencing would reveal high levels of data heterogeneity—in other words, a larger sample size could not solve our problems [5,7]. Unfortunately, despite efforts made by pioneers such as Richard Goldschmidt and Barbara McClintock, few researchers have appreciated the ultimate importance of genome-level changes [3,8]. According to the dogma of molecular genetics, the chromosome is just a carrier of genes. Most genetic errors are rare events due to multiple failures in error correction mechanisms such as DNA repair, cell cycle checkpoints, ER stress responses, information feedback, and apoptosis [8]. Such elegant and precise regulatory mechanisms resulted from billions of years of natural selection accumulating small changes over time; thus, punctuated, large-scale genomic aberrations must be harmful and cannot fit into the evolutionary process. Additionally, molecular genetics claims that drastically altered karyotypes, which are often observed among dying cells, cannot become clonal populations. Moreover, compared with genes, chromosome aberrations are harder to quantitatively measure for mathematical models, and efforts to identify common chromosomal aberrations for a majority of cancers have, thus far, failed [9].
Fast forward, the avalanche of sequencing data we have is at odds with the initial goal of TCGA, to identify a few shared cancer gene mutations. TCGA has exposed a highly heterogeneous genomic landscape that challenges the somatic gene mutation theory of cancer [3]: interestingly, (1) chromosomal alterations are overwhelming in most cancer types, and the profiles of chromosomal changes have much better clinical prediction power than those of gene mutations [10,11,12,13,14]; (2) for many patients, driver gene mutations are hard to identify, although benign tissue could have even more gene mutations than its malignant counterparts [15,16]; and (3) every gene is a relevant “cancer gene”, undermining TCGA’s original mission [17]. The disappointment of gene-based prediction is not limited to TCGA, however. GWASs have revealed many involved loci and few dominant positions [18,19]; experiments have illustrated that the lost function of specific genes can be recovered by aneuploidy [20]; high levels of genomic mosaicism are detected in somatic cells [21,22,23]; and somatic genomic and/or non-genomic dynamics are common during development, aging, and stress response processes. Altogether, the gap between gene profile and phenotype becomes wider, and the power of gene-based prediction is drastically reducing [8].
Obviously, genetics and genomics have now entered uncharted waters. Genes are still considered not only the basis of current genetic and evolutionary theories, but also fundamentally important for medical genetics, the driving force for diseases as well as organismal evolution. If the power of genes is less certain than we have imagined, how will the most dominant theories in biology and their biotechnological and medical implications be impacted? Additionally, equally importantly, how should we practice biology?
Recently, Nobel Laureate Paul Nurse suggested that biology is generating data without ideas to match [24]. In his opinion piece, Nurse quoted Sydney Brenner’s earlier warning for biology that “We are drowning in a sea of data and starving for knowledge.” Clearly, the gap between data and explanation has increased rapidly due to various large-scale -omics platforms, including single-cell technologies, and this gap will only be exacerbated in the future. Nurse recommends a shift in the culture of scientific research by emphasizing ideas and theories in addition to data generation, because current biology lacks the theories to explain or even generate data, as well as worthwhile ideas to inspire new generations. Although the piece is a timely reminder for molecular biologists in the era of Big Data that data and ideas should go hand in hand, Nurse did not address the issue of why technically orientated ideas differ from key theories of biology. Moreover, we must ask ourselves why and when more data means less understanding. What must we do when current theories and data conflict [5,8]?
Supposedly, more data helps us understand more. However, this statement is only true if the data are relevant to the question being studied and the data are explainable within our scientific theory. For example, we usually do not try to classify different species based on the behavior of their atoms. According to Thomas Kuhn, during a routine phase of science, when most data make sense, science progresses. However, when data obviously conflict with the theories that guide the efforts of data generation and collection, alternative theories may be necessary. Quickly reviewing some major cancer genome sequencing data published from the top journals, much of the field’s data can be explained although by neither the somatic gene mutation theory nor neo-Darwinian evolution. Interestingly, researchers often avoid pointing out such inconsistencies or seem altogether unkeen in discussing ideas and theories.
Interestingly, alternative theories that better explain our “surprising” cancer gene mutation data exist, though there are no common frameworks to unify them (Table 1).

Table 1.
Examples of alternative theories/concepts for explaining cancer.
Nevertheless, many of these different ideas are suppressed by the mainstream research community. This suggests that we do not lack ideas/observations, but lack new frameworks to hone and mature these alternative ideas. In other words, we might have reached a moment of scientific revolution. The current lack of new ideas and the difficulties in solving key anomalies represent key features of a crisis, a necessary stage of a paradigm shift: “One of the key preconditions and signals of a paradigm shift is the transition from a routine progression stage to a crisis stage of a given scientific field. In a real crisis stage, the dominating paradigm is losing its capability to explain fundamental facts (most of which are newly discovered), despite that there are many superficial technical achievements being made and a large amount of data being collected. In other words, the more data that are collected, the more confusion there is and the less we can comprehend it, as these new discoveries contradict the expectations of the current paradigm. Such increased anomalies are highly unfit between the existing theory and reality.” [8].
After 50 years of war, does current cancer research need a new paradigm? If yes, what should it look like? How much do cancer theories and research strategies need to be altered? Our key predictions about the outcome of the Cancer Genome Project have proven to be correct [5,7], and we believe it is time to renew the call to invoke a new genomic and evolutionary framework [8]: the Genome Architecture Theory (GAT). The GAT (previously referred to as the Genome Theory or Genome System Theory) departs from the somatic gene mutation theory of cancer [5,7,8,84,86]. In this perspective, genetic and genomic concepts are briefly compared, and some newly emergent principles of the GAT are clarified in the context of inheritance, information, and evolution patterns. Finally, some lessons are offered from the war on cancer.
2. Newly Emergent Genomic and Evolutionary Concepts
2.1. Genes vs. Karyotypes (Chromosome Sets): Redefining Inheritance
The transition from studying karyotypes to genes once represented a technological advancement in cancer research [54]. Based on reductionist practice, the higher the resolution, the better the experimental approach. As soon as fusion genes could be isolated from translated chromosomal regions, karyotype analyses became less important. Compared with gene studies, karyotype studies were often dismissed as “not mechanistic enough” and relegated to the less fundable “descriptive studies” category.
However, following 50 years of gene-based cancer research, especially after the Cancer Genome Project, increased gene data have unexpectedly illustrated the complexity and uncertainty of cancer, which forcefully challenges some of the key assumptions of the somatic gene mutation theory. For example, the gene was assumed to be an independent information unit, where key genes are responsible for key traits. It followed that cancer genes would be common drivers for cancer evolution. Why, then, were sequencing results data and theoretical predictions so far off? Among many explanations (see [3,5,8]), two key conceptual misunderstandings of genetics and evolution are worth re-emphasizing:
(1). The gene’s predictive power has been artificially overestimated by researchers through their selective experimental systems and data interpretations.
As we have previously discussed [7,8], clear-cut relationships between genes and phenotypes are often only observable in exceptional cases. Artificial experimental systems and selective datasets have been key to establishing the power of single genetic elements or genes, ever since Mendel’s foundational experiments. Not only it is difficult to repeat Mendel’s experiments using different species, but he also only reported seven traits among the many he had examined. When studying each trait, his extreme selection of individuals played a major role, choosing uncommon stem lengths for example:
“Third, Mendel had a strict selection criterion for each sample. He had purposely avoided collecting “average data” by using exceptional samples in his experiments. For example, to compare the difference in the stem length (one of his 7 traits), a long axis of 6–7 ft was always crossed with a short one of 0.75–1.5 ft. By pushing extreme cases rather than using average long and short populations, the certainty of data becomes much more impressive. Paradoxically, however, the pattern he discovered based on selection will not represent the majority of the data he ignored.
Fourth, Mendel had tried his best to reduce environmental variations that could influence the data, such as growth conditions, the timing of experiments, and the effect of all foreign pollen, which invariably created ideal systems with minimal environmental influences.
Together, Mendel had created a perfect yet highly exceptional system. Perfect for a manipulated linear model with reduced variants, exceptional for the reality of genetics where most genetic traits do not contribute by a single gene and heterogeneity dominates within a population.” [8].
Knowing how Mendel studied the genetic basis of the length of pea stems, we can understand why it has been so difficult for current researchers to study the genetic basis of traits such as human height despite better technologies and sample sizes. Mendel likely would have faced similar difficulties had he used the entire set of stem lengths in the pea plant population. The genetic basis of human height could be much simpler if we only examine extreme “giant” and “dwarf” phenotypes, because an excess or deficiency of growth hormone would be the obvious cause. However, such a simple answer will not explain the majority of differences in individuals’ heights. Recently, more than 100,000 genomic variants affecting human height were identified, most of which were previously considered statistical noise and ignored; these variants are less useful for pattern identification because they are distributed across the entire genome [18].
Even so-called Mendelian diseases, which are purportedly caused by a single gene, involve many other genes, such as multiple levels of modifiers, and genes related to system function, including homeostasis. The same “causative” mutation can cause different phenotypes across individuals.
The real challenges for future genomics are the facts that the power of any gene is limited and that genes do not function individually but within complex networks. Furthermore, gene-coded information is fuzzy, and environments can “select” outcomes among many coded potentials [8]. By introducing linear, causative experimental systems, researchers often can “prove” the predictive power of a gene under study, but these linear relationships fall apart in a natural setting, where uncertainty is much higher. It is thus no surprise that so many genes—nearly all genes—can be linked to cancer formation and growth [17]. Now, the more serious question becomes: if (almost) every gene affects (almost) everything, how does genetics actually work [8]?;
(2). The mechanisms of inheritance in macro- and microevolution have traditionally been misunderstood, resulting in much confusion in cancer research
Since the gene was identified as a basic unit of inheritance, the research landscape of modern biology has been dominated by studying how genes program cellular phenotypes through various bioprocesses. Successful efforts include characterization of the gene itself (structure, replication, repair) and its relationship with RNA, proteins, and other biological components/pathways; understanding gene regulation and function in developmental as well as normal physiological processes; gene frequency dynamics within a given population; and the cloning of genes responsible for inherited diseases, including some familial cancers (however, for most sporadic cancers, gene theory has lost much of its explanatory power).
Confusion about the power of genes arises from misunderstandings of gene-defined inheritance and cancer. First, Mendelian genetics is objectively constrained to study genetic processes occurring within a given species. Second, since the beginning of molecular cancer research, cancer has been defined as a gene mutation problem, where the oncogene can promote cellular proliferation. Both these assumptions fail to realize that cancer is a new system emergent from multiple levels of system constraints. Theodor Boveri linked cancer to chromosomal aberrations, the only genetic mechanism he knew, in 1902 [87]; since then, molecular geneticists have concluded that the gene is a more advanced concept than the chromosome, because the chromosome is just a vehicle carrying genes. As a result of the dominance of genes, few molecular geneticists are interested in examining the impact of an altered karyotype on their favored gene pathways. However, crucially, the karyotype plays a key role in organizing gene interactions and preserving system information (for more details, refer to later sections). In other words, karyotype change generates a new system. Furthermore, our decades-long research has linked karyotype change with “system inheritance”, which mainly involves macroevolution, and gene mutation with “parts inheritance”, which involves microevolution, including normal developmental processes [7,8,88].
The importance of the karyotype was also ignored by most biologists who use model systems to study gene function. In these systems, the karyotype is often maintained; therefore, the function of a gene can be observed across various experimental settings. In cancer evolution, in contrast, the function of a gene is often indecipherable amid karyotype changes that impact many genes and the networks among them [3,8].
In essence [3,8], current confusions in cancer research are rooted in misunderstandings of inheritance and evolution. The purpose of this piece is to call for the re-examination of basic biological theories to rethink the cancer issue. For example, if the gene and karyotype represent two different types of inheritance, and cancer is an issue of system emergence with a new genome, monitoring the karyotype is an obvious necessity, especially when exorbitant gene-based research has failed to deliver its promises.
2.2. Molecular vs. Evolutionary Mechanisms: The Parts/Pathways, the System, and the Selective Processes
Research strategies are heavily influenced by theories. Since the identification of the first oncogene, the characterization of parts (e.g., identifying and analyzing cancer genes and proteins) has been the main focus of cancer research. When many components of the accumulated data conflicted with each other, various -omics approaches and systems biology were introduced. However, although overwhelming complexity in cancer has been confirmed, there are few specific suggestions of how to proceed. Researchers favor different approaches, including sequencing more samples of genomic and epigenetic landscapes for different cancer types, targeting various pathways, analyzing micro-RNA/non-coding RNA, studying metabolic contribution and influence of microbiota, and examining options of immunotherapy.
Many factors, genomic and environmental alike (from gene to lifestyle), are involved in cancer [3,4]. Under different experimental systems and conditions, any and all factors can promote, slow, or stochastically alter the cancer progression landscape; a given gene can switch from a tumor suppressor to an oncogene function or vice versa; the outliers can multiply into a dominant population or a key population can be eliminated; and a defined, specific pathway can soon become unpredictable when further variants are introduced. These data, saturated with complexity and uncertainty, force researchers to ask some important questions: What is the clinical value of studying one specific gene mutation within a highly adaptive system?; When data components conflict with each other, how do we choose which component to prioritize?
To address these questions, one first needs to understand the relationship between different molecular mechanisms and the common mechanism of cancer [89,90]. Cancer is an evolutionary process; therefore, it must involve the generation of variations in the cellular population (which involves information creation at genome and gene-level); these variations display different degrees of reproduction, survival, and/or evolvability. These variations must be heritable. Furthermore, these altered cellular systems need to respond to evolutionary stress and must ultimately break environmental constraints. Then, any individual molecular mechanism that can contribute to the cancer evolutionary process can be linked to cancer.
However, although there are many specific and highly diverse gene-mediated molecular mechanisms, these can be unified by the common mechanism of genome-mediated cancer evolution under the evolutionary mechanism of cancer.
During comparative studies of tumorigenicity, in which five different well-characterized model systems were used, the degree of karyotypic heterogeneity (population diversity) was directly linked to tumorigenicity. The tumorigenicity of each model has been linked with different and specific molecular pathways and there is no common molecular mechanism shared among them; therefore, we realized that the common link of tumorigenicity between these diverse models is elevated genome diversity. Based on the concept that genome-level heterogeneity is a key to cancer evolution and that stress can induce increased system dynamics, reflected as increased NCCA frequencies, we proposed that the evolutionary mechanism of cancer is equal to the sum of all individual molecular mechanisms:
Evolutionary Mechanism = ∑ Individual Molecular Mechanisms
Although there are many individual molecular mechanisms, there are four key components for understanding how the evolutionary mechanism works: (1) stress-induced system dynamics (e.g., genomic, epigenomic, and increased stochastic changes); (2) population diversity (genome heterogeneity, which can be triggered by diverse gene mutations or molecular pathways when the stress is sufficiently high); (3) selection based on the genome package (macrocellular evolution); and (4) new genome systems capable of breaking down of higher levels of constraints and becoming the dominant population [8].
With the above realization, we now understand the limitation of focusing on the partial characterization of individual gene mutations, because there are so many, and most are trivial compared with karyotype alterations. Instead, we need to focus on evolutionary selection based on new emergent systems (cellular populations with new karyotypes). In addition, the unity of cellular macroevolution occurs at the genome level, rather than at the gene level. Gene mutations are mainly involved in the microevolutionary phase.
Changes in research strategy also reflect general trends in biological research. At the initial stage of molecular research, researchers only knew to study isolated parts. Currently, researchers are struggling to integrate these well-studied parts, which requires holistic concepts and tools. In the future, researchers need to focus on profiling the evolutionary selection process, perhaps through watching-evolution-in-action experiments [6]. For example, over the course of a cancer treatment, it could be tolerable not knowing specific potential pathways, which can be quickly replaced by another pathway when the system undergoes a highly dynamic adaptation. Instead, we ought to focus on better predicting a system’s behavior and modifying its general trends.
2.3. Genome Chaos: The Essential Process of System Information Self-Creation
The main reason why cancer often wins battles in the war on cancer is its incredible evolvability. Ironically, this evolvability is greatly enhanced by our medical treatment strategies. In other words, in the name of killing monsters, we may be propagating them more, because treatment options designed for the maximal killing of cancer cells can result in the formation of more aggressive cancers via genome chaos.
Genome chaos, rapid and massive genome re-organization under crisis, was initially systematically described during watching-evolution-in-action experiments [6,70,91]. It was soon realized that genome chaos is essential for key phase transitions in cancer evolution, including immortalization, transformation, metastasis, and drug resistance [3,8]. Chaotic karyotypes had occasionally been observed by cytogenetics, but they were largely ignored without the foundation of specific mechanisms and experimental systems to reproducibly generate them. Even after our initial report, suspicions remained high, based on the general view that: (1) there is no chance for these drastically altered structures to survive, and they have no evolutionary significance because the evolutionary selection is based on the accumulation of the small changes, and (2) high genetic integrity will not tolerate such structures. Thus, any chaotic genomes belong to genetic noise, which will certainly be eliminated by evolutionary selection.
In contrast, unexpectedly, chaotic genomes have commonly been detected from cancers in the Cancer Genome Project [92,93,94]. Although many different new terms were employed to describe the phenomena, previous cytogenetic discoveries of genome chaos were confirmed. As described in our previous publication:
“Recently, the cancer genome sequencing project has generated large amounts of data, which inevitably confirmed the importance of studying genome chaos [95,96,97]. Various chaotic genomes were detected within nearly all types of cancers. In some cancer types such as prostate cancer, chaotic genomes were detected in a majority of cases. Interestingly, these fragmented and stitched chromosomes were given many different names by different investigators including “chromothripsis”, “chromoplexy”, “chromoanagenesis”, “chromoanasynthesis”, “chromosome catastrophes”, and “structural mutations” [98,99,100,101,102,103,104,105,106,107,108]. Based on their descriptions chromothripsis refers to the chaotic genome mainly involving local re-organization (within a single chromosome), while chromoplexy refers to a more whole genome re-organization involving many individual chromosomes. We prefer using “genome chaos”, “karyotype chaos”, or “chromosomal chaos” to refer these structures due to their broad coverage (from local to global re-organization, from structural to numerical changes) and the simplicity of terminology [3,109,110,111].
Limited mechanistic studies mainly focused on specific gene mutations, pathways, and specific cellular mechanisms, such as how the p53 mutation and micro-nucleus formation contribute to genome chaos. For example, it was elegantly illustrated that chromothripsis may directly originate from DNA damage in micronuclei, linking the restriction of chromothriptic rearrangements to a single chromosome. Alternatively, chromothripsis was linked to telomere crisis [112,113]. From a genome evolutionary perspective, however, there should be large numbers of specific molecular mechanisms that can contribute to genome chaos, and only a small portion of them belong to chromothripsis [3,6,114]. Additional studies have now linked many individual factors, such as hyperploidy and radiation, to chromothripsis [115,116], and the list of the contributing factors should be increasing…” [117].
Following the tradition of molecular research, increased attention has focused on the molecular mechanisms for different subtypes of genome chaos. In a recent comprehensive review article, James Shapiro summarized different molecular mechanisms that are responsible for different chaotic genome subtypes [92]. Although it is interesting to study the individual mechanisms for chromothripsis, chromoplexy, micronuclei clusters, and many other structural and numerical chaotic genomes, such mechanisms unhelpfully involve nearly unlimited pathways and trigger factors [4,114]. There is now increased attention on polyploid giant cancer cells (PGCCs), a numerical chaotic genome subtype, in cancer’s aggressiveness and drug resistance [71,72,73,74,75,76,91,109]. Due to the direct relationship of the different phases within the cell cycle (a defect in the S phase can impact the G2, M, and G1 phases), various types of chromosomal abnormalities are linked (replication defects can lead to the error of chromosomal condensation and segregation, or vice versa) [55]. Furthermore, under the stress response, many subtypes of chaotic genomes are intimately linked by a “cause and consequence cycle”, and can constantly alter subtypes [54,70,110,118]. More challengingly, it would be even harder to study or predict the dynamics of multiple variants during active evolutionary selection. A more practical approach is to focus on the evolutionary mechanism of cancer: regardless of the individual trigger factors and mechanisms of this phenomenon, the common evolutionary consequence is the same—to produce the new genomes that are essential for phase transition during the evolution [8,53,118,119].
Two useful concepts can help one truly appreciate the importance of genome chaos: First, chaos theory is a new frontier of science that differs from the layman’s definition of disorder. Although genome chaos represents a highly heterogeneous process, the outcome of this process is predictable. For example, myriad triggers (uncertain factors) can generate high stress, and the cellular program of genome re-organization will be initiated (a certain consequence). However, the cellular system cannot guarantee the production of only winning karyotypes—it always produces massively different karyotypes (with uncertainty). Then, evolution picks the winners. Given enough chance, there will be winners (certainty), but who the winner will be is unknown (uncertain). That is why it is less useful to focus on any specific pathways. As we have stated:
“Chaos as a system behavior does not simply mean random disorder. Why use the term “chaos” to describe genome-based rapid cancer evolution? Genome behavior during crises shares common features with complex systems described by chaos theory. Chaos theory, a non-linear dynamics concept, states that “within the apparent randomness of chaotic complex systems, there are underlying patterns, interconnectedness, constant feedback loops, repetition, self-similarity, fractals, and self-organization” (https://en.wikipedia.org/wiki/Chaos_theory) (accessed on 12 August 2021). It is important to note that, within the collective frameworks of chaos theory, apparently random states of disorder and irregularities of dynamic systems are often governed by deterministic laws. In layman’s terms, unfortunately, the word “chaos” suggests rampant disorder and randomness, coupled with a negative connotation”“…Genome chaos represents a process that can be described by a combination of uncertainty and certainty, disorder and order, stochasticity and determinism—an excellent example of random means to a nonrandom end function. Therefore, analyzing system behavior (how macroevolution is achieved by the success of creating new systems), rather than characterizing all involved genome variation mechanisms, will make the cellular macroevolution phase transition much easier to understand and predict.” [119];
Second: genome chaos research has led to the important concept of System Information Self-Creation Under Crisis, which will have a profound impact on cancer research and evolutionary biology [120]. Genome reorganization and role driving phase transitions in cancer led us to search for “system inheritance”-coded “system information”, distinctive to “parts inheritance”-coded “parts information.” Such efforts have established the concept of karyotype coding:
“‘Karyotype coding’ or ‘chromosomal (set) coding’ functions as an organizer of gene interactions within the entire genome. Its biological effect is not just on individual genes but on the entire genomic network. As opposed to gene coding or vague ideas that chromosomes carry additional information, karyotype coding is defined by specific features: (1) the physical organization of the chromosome codes system information; (2) genomic topology provides context for individual genes; and (3) since different species display unique karyotypes or core genomes, karyotype coding is often species-specific. The key is that the order of gene and non-coding sequences along a chromosome represents a new ‘system inheritance,’ much like how the order of base pairs codes for ‘parts inheritance’ in mainstream ‘gene coding.’” [86].
There are many obstacles for most gene-based researchers to accept karyotype coding. The most significant one is the constraint of the somatic gene mutation theory, which is not compatible with genome-based theories. Another key factor is a lack of understanding of the multiple levels and types of bio-information. According to reductionist tradition, genetic information solely means gene-coded information, and the system information that organizes gene-level information is irrelevant. The Genome Architecture Theory classifies information management into information creation, preservation, modification, and usage [53,119,120]. Different types of coding have been linked to different types of inheritance, and karyotype coding is crucial for preserving combinational self-organization events (including chemical and physical events). Karyotype coding serves as a platform on which other organic codes can operate and accumulate, leading to increased biocomplexity and diversity [120,121,122,123].
Perhaps most importantly, new system information creation in biology is a process of self-creation. Under high-stress conditions likely to eliminate a system, the system’s cellular machinery will automatically switch into a mode that destroys the current genome and simultaneously forms new genomes using their own genomic materials. Changing the karyotype coding represents the only opportunity to pass on the information of life, at a level above individual cellular species. This capability of creating new from self-death is witnessed in rapid and massive drug-treatment-induced genome chaos, which produces drug resistance. As newly formed systems with new karyotype coding, these resistant individuals are no longer their original “selves.” Such a mechanism not only explains the basis for treatment-induced drug resistance [70,71,76,91], but also explains organismal macroevolution, especially when large numbers of new species emerge from massive extinction [8,124], (Heng et al., submitted). Of course, organismal evolution is more complex than cancer evolution. Somatic evolution selects stable genomes among many created unstable ones, and stable genomes are key to preserving system information. Interestingly, karyotype preservation is achieved in animals and plants through sex and the separation of the germline and somatic cells during development [8,63,125,126].
2.4. Two-Phased Evolutionary Model
A direct benefit of synthesizing all the above information (from redefining inheritance to illustrating genome chaos as a process for creating new system information) is to establish the model of two-phased cancer evolution. This model comprises a punctuated phase and a gradual stepwise phase. Within the punctuated phase, karyotype changes dominate, and genome-based macro-selection is the key driving force. Within the gradual phase, gene mutations and epigenetic alterations dominate, with microevolution as the key feature. This model can reconcile the contribution of genes as well as karyotypes. It is also useful for both cancer diagnosis and treatment [3,6,8,53,76,120]. As illustrated in Figure 1, the model of two-phased cancer evolution can acceptably integrate and unify stressors, various subtypes of genome chaos, system information creation by genome re-shuffling, the macroevolutionary selection of survivable new genomes, the clonal expansion of the “first cell”, and the involvement of immense gene mutations/epigenetic abnormalities that contribute to microevolution.
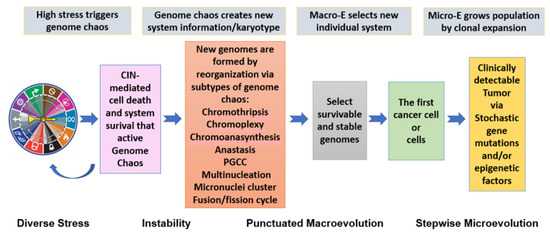
Figure 1.
New model of two-phased cancer evolution. Diverse stresses are represented by the hallmarks of cancer (modified from [90,127]). When stress is high enough to kill cells, it can trigger genome chaos. Genome chaos can manifest as many subtypes, both structural and numerical, of which only seven examples are listed here. Regardless of the individual subtypes, new system information is created. Evolution selects the first cancer cell or the first wave of cancer cells, which are then subject to microevolution to grow the cancer cell population. This process can be linked to large numbers of different gene mutations or pathways. For more information, please refer to [3,4,8,128].
3. Future Perspective
Clearly, the National Cancer Act has significantly altered the landscape of cancer research. It set in motion a chain of events, including government funding, infrastructure (the National Cancer Program), the promotion of bioindustry, and the National Cancer Institute (NCI)’s influence on the direction of national research, which has greatly promoted basic research. In addition to millions of manuscripts published and thousands of genes identified, a large body of molecular knowledge and experimental platforms/methodologies have been accumulated. Some impressive successes have also been achieved, such as imatinib, a drug for CML; an HPV vaccine for cervical cancer prevention; and the current development of immunotherapy. Interestingly, the data generated from the National Cancer Act have also unexpectedly sped up the effort to search for new frameworks beyond the gene, because the well-funded cancer field has accumulated so much molecular data that forcefully challenge many current biological theories [3,8].
Here are some further lessons learned in the past 50 years of the war on cancer.
First, the war metaphor has had unexpected consequences on researchers, physicians, and patients. The war mentality promotes a host of combative strategies against cancer. Treatments prioritize killing the enemy at all costs, using the most powerful weapons possible [76]. Combined with the rhetoric and ideas of searching for magic bullets and specific targets, the entire field has spent too much effort on molecular details in order to increase treatment certainty. Cancer is a complex adaptive system with evolution as its key mechanism; therefore, the current approach of understanding all possible molecular agents and systematically fighting them is both impossible and less useful.
Second, conducting basic science differs from finishing an engineering project, especially when the science is undergoing a framework change. For a defined engineering project, if there is a solid theoretical framework and implementable technologies, sufficient funding and labor can deliver the products, whether this is a nuclear bomb or moon landing. To cure an unknown, which requires both new theories and technical platforms, fulfilling a goal within a fixed amount of time will likely fail. The Human Genome Project (HGP) is a good example. Yes, we can sequence a human genome on time with improved technologies (an impressive engineering project), but we should not promise that we can decode the mystery of life and eliminate all diseases in our lifetime with said sequencing project (basic science).
During the past 50 years, both governments and research institutions have promised to cure cancer many times, to no avail [3]. From a promising consensus mentioned by the National Cancer Act of 1971: “There seems to be a consensus among cancer researchers that they are within striking distance of achieving the basic understanding of cancer cells which has eluded the most brilliant medical minds in the world” [1]; to President Bill Clinton’s statement in 2000, following the celebration of the success of the HGP: “It is now conceivable that our children and our children’s children will know the term cancer only as a constellation of stars…Genome science… will revolutionize the diagnosis, prevention and treatment of most, if not all, human diseases” [129]; to the NCI’s declaration that cancer will be cured by 2015; to President Obama’s promise in his 2016 State of the Union address: “For the loved ones we’ve all lost, for the family we can still save, let us make America the country that cures cancer once and for all” [130]; and to the proposed Moonshot to cure cancer in 2023 by a well-known cancer center [3,8], we witness one empty promise after another. Nuance is necessary here: optimism is good, but overstatements harm both scientists and patients.
Third, biology in general, and cancer research in particular, needs solid theories. We already have overwhelming data. Theoretical analyses and the re-examination of current concepts are urgently needed before we further push any data generation. For example, although single-cell data are interesting and potentially valuable, the system behavior of a cellular population is an emergent one, with contributions from both the average and outliers. Thus, we cannot study any disease by only using the averages of a population. Without the correct theoretical framework, without noticing the impact of outliers, we fail to see the big picture. Instead, we will only produce more data without biological insight, attributing the effects of outliers to the average.
As for the re-examination of basic theories in biology, both genomic and evolutionary theories should be prioritized. Specifically, how genes and karyotypes contribute to parts and system inheritance in cancer research, and the common pattern of cancer evolution, need more attention. It is important to separate gene mutations and chromosomal alterations in cancer research and diagnosis, because these alterations can be used as indices to monitor the different phases of cancer evolution. In addition, the multiple levels of genomic and non-genomic heterogeneity can be investigated using the concept of fuzzy inheritance [8]. Furthermore, heterogeneity, especially at the chromosomal level, can be used to monitor and/or predict general trends of cancer evolution.
The past 50 years in particular, with the Cancer Genome Project, have already produced sufficient data. What we need is to perform a systematic comparison to validate the model of two-phased cancer evolution and apply it to cancer diagnosis and treatment. For example, employing a moderate force of constraint in the microevolutionary phase could slow down cancer microevolution evolution without triggering genome chaos that would rapidly produce drug-resistant genomes. If successful, this approach can be integrated into adaptive therapy strategies [131].
Other theories, in the realms of information theory and complex system theory which originate in non-biological systems, need to be further developed to fit the biological context of cancer [120]. Currently, most cancer researchers still focus on gene-coded parts information and favor linear models to study cancer mechanisms. When applying information theory to study cancer, the first question should be: What types of information we should collect?; How much is minimal?; Additionally, when different types of information conflict, how do we prioritize them? When applying complex adaptive system theory in cancer research, researchers should know that the certainty illustrated in a linear model has very limited clinical value because the causative relationship they demonstrate is likely an illusion under simplistic experimental conditions.
One related issue is the misinformation perpetuated by the misguided use of preclinical assays. For example, even though the Nomenclature Committee on Cell Death has been warning the scientific community about the misuse of words and concepts that slow down progress in cancer research [132], most researchers simply ignore these caveats. Now it has become clear that the short-term benefit of induced cell death can paradoxically promote cancer, because dying cancer cells can emerge from the brink of death through genome chaos, including Anstasis (a cell recovery phenomenon that rescues cells from the brink of death) and PGCCs [8,71,76,77,91,117]. This can be explained as an example of why linear experimental models often cannot predict clinical realities when multiple types of evolution are involved.
In order to promote questioning of the current paradigm, as well as the creation of more accurate paradigms, our scientific culture needs to change. Institutionally, the NIH should promote theoretical study. Various organizations should actively promote much-needed debates to provide a competitive landscape for different ideas and approaches; for example, reviewers and grant agencies should ask authors to spell out the meaning of their presented data to see whether the data conflict with or support their initial rationale. Similarly, the NCI needs to discourage data generation without good ideas, because it is unwise to characterize all genes that can be linked to cancer—there are simply too many.
Moreover, we must increase education for scientists, physicians, and patients. In the future, it is essential to educate patients that cancer represents an evolutionary trade-off of cellular adaptation. To achieve various cellular functions (e.g., tissue repair, protection against bacterial infections), cells need to be able to change through cellular adaptation. However, such changes (genomic or non-genomic), although important for cellular function, can lead to various diseases, including cancer. Many genomic and environmental factors can contribute to cancer, and this becomes the main challenge for studying cancer genes for clinical use, because all these factors contribute to cancer but have very limited prediction power in the clinic. This evolutionary process is very complex and features high uncertainty; thus, the best method for individuals to prevent cancer is general lifestyle improvement. During treatment, paying attention to overall health status, as well as happiness and social support, should also be emphasized.
Additional education on complexity and uncertainty, as well as the limitations of molecular understandings of cancer, should also be extended to researchers and physicians. Some leaders in the field have already admitted this issue:
“We lack the conceptual paradigms and computational strategies for dealing with this complexity. Additionally, equally painful, we do not know how to integrate individual datasets, such as those deriving from cancer genome analyses, with other, equally important datasets, such as proteomics. This is most frustrating, since it is becoming increasingly apparent that a precise and truly useful understanding of the behavior of individual cancer cells and the tumors that they form will only come once we are able to integrate and then distill these data.” [2].
Clearly, any new conceptual paradigms must include genome- and information-based evolutionary theories. It is thus timely to introduce different theories of cancer to physicians, who can provide feedback based on clinical realities. It is crucial to understand that the same treatment can be good or bad for different individuals. Finally, the issue of overdiagnosis and overtreatment will also be considered, and the question of living with cancer rather than trying everything to kill all cancer cells should be addressed.
As we are writing, there are many news reports, official commemorations, and editorial comments from research journals celebrating the 50-year milestone of the war on cancer. Most are excited about the potential “golden age” of cancer treatment given our enormous body of molecular and genetic knowledge, where every tumor has a unique signature which is personalized targets [133,134]. We share the excitement, but caution that only the correct ideas and strategies will deliver.
Fifty years is a long time. We hope that the next 50 years will witness changes in attitudes towards cancer, with solid theories that will improve ideas and outcomes [3,8].
Author Contributions
J.H. and H.H.H. drafted the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
This manuscript is part of our series of publications on the subject of “the mechanisms of cancer and organismal evolution.” We appreciate Eric Heng for his discussions.
Conflicts of Interest
The authors declare no conflict of interest. Neither of the authors of this paper have any competing financial interests or non-financial interests.
References
- National Cancer Act of 1971. Available online: https://www.cancer.gov/about-nci/overview/history/national-cancer-act-1971 (accessed on 12 August 2021).
- Weinberg, R.A. Coming Full Circle—From Endless Complexity to Simplicity and Back Again. Cell 2014, 157, 267–271. [Google Scholar] [CrossRef] [Green Version]
- Heng, H.H. Debating Cancer: The Paradox in Cancer Research; World Scientific Publishing Co.: Singapore, 2015; ISBN 978-981-4520-84-3. [Google Scholar]
- Ye, C.J.; Sharpe, Z.; Heng, H.H. Origins and Consequences of Chromosomal Instability: From Cellular Adaptation to Genome Chaos-Mediated System Survival. Genes 2020, 11, 1162. [Google Scholar] [CrossRef]
- Heng, H.H. Cancer genome sequencing: The challenges ahead. Bioessays 2007, 29, 783–794. [Google Scholar] [CrossRef] [PubMed]
- Heng, H.H.; Stevens, J.B.; Liu, G.; Bremer, S.W.; Ye, K.J.; Reddy, P.-V.; Wu, G.S.; Wang, Y.A.; Tainsky, M.A.; Ye, C.J. Stochastic cancer progression driven by non-clonal chromosome aberrations. J. Cell. Physiol. 2006, 208, 461–472. [Google Scholar] [CrossRef] [PubMed]
- Heng, H.H. The genome-centric concept: Resynthesis of evolutionary theory. Bioessays 2009, 31, 512–525. [Google Scholar] [CrossRef] [PubMed]
- Heng, H.H. Genome Chaos: Rethinking Genetics, Evolution, and Molecular Medicine; Academic Press Elsevier: Cambridge, MA, USA, 2019; ISBN 978-012-8136-35-5. [Google Scholar]
- Mitelman, F. 50,000 Tumors, 40,000 Aberrations, and 300 Fusion Genes: How Much Remains? 50 Years of 46 Human Chromosomes: Progress in Cytogenetics; NCI, NIH: Bethesda, MD, USA, 2006. [Google Scholar]
- Park, S.Y.; Gönen, M.; Kim, H.J.; Michor, F.; Polyak, K. Cellular and genetic diversity in the progression of in situ human breast carcinomas to an invasive phenotype. J. Clin. Investig. 2010, 120, 636–644. [Google Scholar] [CrossRef] [Green Version]
- Jamal-Hanjani, M.; Wilson, G.A.; McGranahan, N.; Birkbak, N.J.; Watkins, T.B.K.; Veeriah, S.; Shafi, S.; Johnson, D.H.; Mitter, R.; Rosenthal, R.; et al. Tracking the Evolution of Non–Small-Cell Lung Cancer. N. Engl. J. Med. 2017, 376, 2109–2121. [Google Scholar] [CrossRef] [Green Version]
- Davoli, T.; Uno, H.; Wooten, E.C.; Elledge, S.J. Tumor aneuploidy correlates with markers of immune evasion and with re-duced response to immunotherapy. Science 2017, 355, eaaf8399. [Google Scholar] [CrossRef] [Green Version]
- Vargas-Rondón, N.; Villegas, V.E.; Rondón-Lagos, M. The role of chromosomalinstability in cancer and therapeutic responses. Cancers 2017, 10, 4. [Google Scholar] [CrossRef] [Green Version]
- Frias, S.; Ramos, S.; Salas, C.; Molina, B.; Sánchez, S.; Rivera-Luna, R. Nonclonal Chromosome Aberrations and Genome Chaos in Somatic and Germ Cells from Patients and Survivors of Hodgkin Lymphoma. Genes 2019, 10, 37. [Google Scholar] [CrossRef] [Green Version]
- Martincorena, I.; Campbell, P.J. Somatic mutation in cancer and normal cells. Science 2015, 349, 1483–1489. [Google Scholar] [CrossRef]
- Yizhak, K.; Aguet, F.; Kim, J.; Hess, J.M.; Kübler, K.; Grimsby, J.; Frazer, R.; Zhang, H.; Haradhvala, N.J.; Rosebrock, D.; et al. RNA sequence analysis reveals macroscopic somatic clonal expansion across normal tissues. Science 2019, 364, eaaw0726. [Google Scholar] [CrossRef]
- de Magalhães, J.P. Every gene can (and possibly will) be associated with cancer. Trends Genet. 2021. [Google Scholar] [CrossRef]
- Boyle, E.A.; Li, Y.I.; Pritchard, J.K. An Expanded View of Complex Traits: From Polygenic to Omnigenic. Cell 2017, 169, 1177–1186. [Google Scholar] [CrossRef] [Green Version]
- McClellan, J.; King, M.-C. Genetic Heterogeneity in Human Disease. Cell 2010, 141, 210–217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rancati, G.; Pavelka, N.; Fleharty, B.; Noll, A.; Trimble, R.; Walton, K.; Perera, A.; Staehling-Hampton, K.; Seidel, C.W.; Li, R. Aneuploidy Underlies Rapid Adaptive Evolution of Yeast Cells Deprived of a Conserved Cytokinesis Motor. Cell 2008, 135, 879–893. [Google Scholar] [CrossRef] [Green Version]
- Iourov, I.Y.; Vorsanova, S.G.; Yurov, Y.B. Chromosomal mosaicism goes global. Mol. Cytogenet. 2008, 1, 26. [Google Scholar] [CrossRef] [Green Version]
- Iourov, I.Y.; Vorsanova, S.G.; Yurov, Y.B.; Kutsev, S.I. Ontogenetic and Pathogenetic Views on Somatic Chromosomal Mosaicism. Genes 2019, 10, 379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ye, C.J.; Chen, J.; Liu, G.; Heng, H.H. Somatic Genomic Mosaicism in Multiple Myeloma. Front. Genet. 2020, 11, 388. [Google Scholar] [CrossRef] [Green Version]
- Nurse, P. Biology must generate ideas as well as data. Nature 2021, 597, 305. [Google Scholar] [CrossRef] [PubMed]
- Duesberg, P.; Rausch, C.; Rasnick, D.; Hehlmann, R. Genetic instability of cancer cells is proportional to their degree of aneuploidy. Proc. Natl. Acad. Sci. USA 1998, 95, 13692–13697. [Google Scholar] [CrossRef] [Green Version]
- Duesberg, P.; Rasnick, D. Aneuploidy, the somatic mutation that makes cancer a species of its own. Cell Motil Cytoskelet. 2000, 47, 81–107. [Google Scholar] [CrossRef]
- Gibbs, W.W. Untangling the roots of cancer. Sci. Am. 2003, 289, 56–65. [Google Scholar] [CrossRef]
- Weaver, B.A.; Cleveland, D.W. Aneuploidy: Instigator and Inhibitor of Tumorigenesis. Cancer Res. 2007, 67, 10103–10105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pavelka, N.; Rancati, G.; Li, R. Dr Jekyll and Mr Hyde: Role of aneuploidy in cellular adaptation and cancer. Curr. Opin. Cell Biol. 2010, 22, 809–815. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Siegel, J.J.; Amon, A. New Insights into the Troubles of Aneuploidy. Annu. Rev. Cell Dev. Biol. 2012, 28, 189–214. [Google Scholar] [CrossRef] [Green Version]
- Ye, C.J.; Regan, S.; Liu, G.; Alemara, S.; Heng, H.H. Understanding aneuploidy in cancer through the lens of system inheritance, fuzzy inheritance and emergence of new genome systems. Mol. Cytogenet. 2018, 11, 31. [Google Scholar] [CrossRef]
- Soto, A.M.; Sonnenschein, C. The tissue organization field theory of cancer: A testable replacement for the somatic mutation theory. Bioessays 2011, 33, 332–340. [Google Scholar] [CrossRef] [Green Version]
- Baker, S.G. TOFT better explains experimental results in cancer research than SMT. Bioessays 2011, 33, 919–921. [Google Scholar] [CrossRef]
- Soto, A.M.; Sonnenschein, C. Paradoxes in Carcinogenesis: There Is Light at the End of That Tunnel! Disruptive Sci. Technol. 2013, 1, 154–156. [Google Scholar] [CrossRef] [Green Version]
- Huang, S.; Ernberg, I.; Kauffman, S. Cancer attractors: A systems view of tumors from a gene network dynamics and developmental perspective. Semin. Cell Dev. Biol. 2009, 20, 869–876. [Google Scholar] [CrossRef] [Green Version]
- Huang, S. Genetic and non-genetic instability in tumor progression: Link between the fitness landscape and the epigenetic landscape of cancer cells. Cancer Metastasis Rev. 2013, 32, 423–448. [Google Scholar] [CrossRef]
- Kulkarni, P.; Shiraishi, T.; Kulkarni, R.V. Cancer: Tilting at windmills? Mol. Cancer 2013, 12, 108. [Google Scholar] [CrossRef] [Green Version]
- Ao, P.; Galas, D.; Hood, L.; Yin, L.; Zhu, X.M. Towards predictive stochastic dynamical modeling of cancer genesis and progression. Interdiscip. Sci. Comput. Life Sci. 2010, 2, 140–144. [Google Scholar] [CrossRef] [Green Version]
- Noble, D. Cellular Darwinism: Regulatory networks, stochasticity, and selection in cancer development. Prog. Biophys. Mol. Biol. 2021, 165, 66–71. [Google Scholar] [CrossRef]
- Davies, P.C.; Lineweaver, C.H. Cancer tumors as Metazoa 1.0: Tapping genes of ancient ancestors. Phys. Biol. 2011, 8, 015001. [Google Scholar] [CrossRef]
- Wilkins, A.S. The enemy within: An epigenetic role of retrotransposons in cancer initiation. Bioessays 2010, 32, 856–865. [Google Scholar] [CrossRef] [PubMed]
- Gluckman, P.D.; Beedle, A.S.; Hanson, M.A.; Low, F.M. Human Growth: Evolutionary and Life History Perspectives. Nestle Nutr. Inst. Workshop Ser. 2013, 71, 89–102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horne, S.D.; Chowdhury, S.K.; Heng, H.H. Stress, genomic adaptation, and the evolutionary trade-off. Front Genet. 2014, 5, 92. [Google Scholar] [CrossRef] [Green Version]
- Feinberg, A.P.; Ohlsson, R.; Henikoff, S. The epigenetic progenitor origin of human cancer. Nat. Rev. Genet. 2006, 7, 21–33. [Google Scholar] [CrossRef] [PubMed]
- Jaffe, L.F. A Calcium-Based Theory of Carcinogenesis. Adv. Cancer Res. 2005, 94, 231–263. [Google Scholar] [CrossRef]
- Levin, M. Bioelectrical approaches to cancer as a problem of the scaling of the cellular self. Prog. Biophys. Mol. Biol. 2021, 165, 102–113. [Google Scholar] [CrossRef]
- Ewald, P.W. An evolutionary perspective on parasitism as a cause of cancer. Adv. Parasitol. 2009, 68, 21–43. [Google Scholar] [CrossRef]
- Warburg, O. On Respiratory Impairment in Cancer Cells. Science 1956, 124, 269–270. [Google Scholar] [CrossRef] [PubMed]
- A Loeb, L.; Springgate, C.F.; Battula, N. Errors in DNA replication as a basis of malignant changes. Cancer Res. 1974, 34, 2311–2321. [Google Scholar]
- Heppner, G.H. Tumor heterogeneity. Cancer Res. 1984, 44, 2259–2265. [Google Scholar] [PubMed]
- Heppner, G.H.; Miller, B.E. Therapeutic implications of tumor heterogeneity. Semin. Oncol. 1989, 16, 91–105. [Google Scholar] [PubMed]
- Raza, A. The First Cell: And the Human Costs of Pursuing Cancer to the Last. Basic Books; Hachette Book Group, Inc.: New York, NY, USA, 2019. [Google Scholar]
- Heng, J.; Heng, H.H. Two-phased evolution: Genome chaos-mediated information creation and maintenance. Prog. Biophys. Mol. Biol. 2021, 165, 29–42. [Google Scholar] [CrossRef]
- Heng, H.H.; Liu, G.; Stevens, J.B.; Abdallah, B.Y.; Horne, S.D.; Ye, K.J.; Bremer, S.W.; Chowdhury, S.K.; Ye, C.J. Karyotype heterogeneity and unclassified chromosomal abnormalities. Cytogenet. Genome Res. 2013, 139, 144–157. [Google Scholar] [CrossRef]
- Heng, H.; Chen, W.Y.; Wang, Y.C. Effects of pingyanymycin on chromosomes: A possible structural basis for chromosome aberration. Mutat. Res. Mol. Mech. Mutagen. 1988, 199, 199–205. [Google Scholar] [CrossRef]
- Heng, H.H. Chapter 12: Bio-complexity challenging reductionism. In Handbook on System and Complexity in Health; Sturmberg, J., Martin, C., Eds.; Springer: Berlin/Heidelberg, Germany, 2013; pp. 193–208. [Google Scholar]
- Tez, M.; Tez, S. Cancer is the chaotic search for adaptation to previously unknown environments. Theor. Biol. Forum 2016, 109, 149–154. [Google Scholar]
- Wu, S.; Turner, K.M.; Nguyen, N.-P.; Raviram, R.; Erb, M.; Santini, J.; Luebeck, J.; Rajkumar, U.; Diao, Y.; Li, B.; et al. Circular ecDNA promotes accessible chromatin and high oncogene expression. Nature 2019, 575, 699–703. [Google Scholar] [CrossRef]
- Raghuram, G.V.; Chaudhary, S.; Johari, S.; Mittra, I. Illegitimate and Repeated Genomic Integration of Cell-Free Chromatin in the Aetiology of Somatic Mosaicism, Ageing, Chronic Diseases and Cancer. Genes 2019, 10, 407. [Google Scholar] [CrossRef] [Green Version]
- Huxley, J. Cancer Biology: Comparative and Genetic. Biol. Rev. 1956, 31, 474–514. [Google Scholar] [CrossRef]
- van Valen, L.; Mairorana, V.C. Hela, a new microbial species. Evol Theor. 1991, 10, 71–74. [Google Scholar]
- Ye, C.J.; Liu, G.; Bremer, S.W.; Heng, H.H.Q. The dynamics of cancer chromosomes and genomes. Cytogenet. Genome Res. 2007, 118, 237–246. [Google Scholar] [CrossRef]
- Heng, H.H. Elimination of altered karyotypes by sexual reproduction preserves species identity. Genome 2007, 50, 517–524. [Google Scholar] [CrossRef] [PubMed]
- Vincent, M.D. The Animal within: Carcinogenesis and the Clonal Evolution of Cancer Cells Are Speciation Events Sensu Stricto. Evolution 2010, 64, 1173–1183. [Google Scholar] [CrossRef] [PubMed]
- Bloomfield, M.; Duesberg, P. Inherent variability of cancer-specific aneuploidy generates metastases. Mol. Cytogenet. 2016, 9, 90. [Google Scholar] [CrossRef] [Green Version]
- Paul, D. Cancer as a form of life: Musings of the cancer and evolution symposium. Prog. Biophys. Mol. Biol. 2021, 165, 120–139. [Google Scholar] [CrossRef]
- Walen, K.H. Mitosis is not the only distributor of mutated cells: Non-mitotic endopolyploid cells produce reproductive genome-reduced cells. Cell Biol. Int. 2010, 34, 867–872. [Google Scholar] [CrossRef]
- Erenpreisa, J.; Kalejs, M.; Cragg, M. Mitotic catastrophe and endomitosis in tumour cells: An evolutionary key to a molecular solution. Cell Biol. Int. 2005, 29, 1012–1018. [Google Scholar] [CrossRef]
- Erenpreisa, J.; Salmina, K.; Anatskaya, O.; Cragg, M.S. Paradoxes of cancer: Survival at the brink. Semin. Cancer Biol. 2020. [Google Scholar] [CrossRef]
- Heng, H.H.; Stevens, J.B.; Lawrenson, L.; Liu, G.; Ye, K.J.; Bremer, S.W.; Ye, C.J. Patterns of genome dynamics and cancer evolution. Cell. Oncol. Off. J. Int. Soc. Cell. Oncol. 2008, 30, 513–514. [Google Scholar] [CrossRef]
- Zhang, S.; Mercado-Uribe, I.; Xing, Z.; Sun, B.; Kuang, J.; Liu, J. Generation of cancer stem-like cells through the formation of polyploid giant cancer cells. Oncogene 2013, 33, 116–128. [Google Scholar] [CrossRef]
- Niu, N.; Mercado-Uribe, I.; Liu, J. Dedifferentiation into blastomere-like cancer stem cells via formation of polyploid giant cancer cells. Oncogene 2017, 36, 4887–4900. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Niu, N.; Zhang, J.; Qi, L.; Shen, W.; Donkena, K.V.; Feng, Z.; Liu, J. Polyploid Giant Cancer Cells (PGCCs): The Evil Roots of Cancer. Curr. Cancer Drug Targets 2019, 19, 360–367. [Google Scholar] [CrossRef] [PubMed]
- Liu, J. The dualistic origin of human tumors. Semin. Cancer Biol. 2018, 53, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Liu, J. The “life code”: A theory that unifies the human life cycle and the origin of human tumors. Semin. Cancer Biol. 2020, 60, 380–397. [Google Scholar] [CrossRef]
- Ye, J.C.; Horne, S.; Zhang, J.Z.; Jackson, L.; Heng, H.H. Therapy Induced Genome Chaos: A Novel Mechanism of Rapid Cancer Drug Resistance. Front. Cell Dev. Biol. 2021, 9, 676344. [Google Scholar] [CrossRef]
- Zaitceva, V.; Kopeina, G.S.; Zhivotovsky, B. Anastasis: Return Journey from Cell Death. Cancers 2021, 13, 3671. [Google Scholar] [CrossRef]
- Stevens, J.B.; Liu, G.; Bremer, S.W.; Ye, K.J.; Xu, W.; Xu, J.; Sun, Y.; Wu, G.S.; Savasan, S.; Krawetz, S.A.; et al. Mitotic Cell Death by Chromosome Fragmentation. Cancer Res. 2007, 67, 7686–7694. [Google Scholar] [CrossRef] [Green Version]
- Pienta, K.; Hammarlund, E.; Austin, R.; Axelrod, R.; Brown, J.; Amend, S. Cancer cells employ an evolutionarily conserved polyploidization program to resist therapy. Semin. Cancer Biol. 2020. [Google Scholar] [CrossRef]
- Navin, N.; Kendall, J.; Troge, J.; Andrews, P.; Rodgers, L.; McIndoo, J.; Cook, K.; Stepansky, A.; Levy, D.; Esposito, D.; et al. Tumour evolution inferred by single-cell sequencing. Nature 2011, 472, 90–94. [Google Scholar] [CrossRef] [Green Version]
- Sottoriva, A.; Kang, H.; Ma, Z.; Graham, T.A.; Salomon, M.P.; Zhao, J.; Marjoram, P.; Siegmund, K.; Press, M.F.; Shibata, D.; et al. A Big Bang model of human colorectal tumor growth. Nat. Genet. 2015, 47, 209–216. [Google Scholar] [CrossRef] [PubMed]
- Shapiro, J.A. What can evolutionary biology learn from cancer biology? Prog. Biophys. Mol. Biol. 2021, 165, 19–28. [Google Scholar] [CrossRef] [PubMed]
- Shapiro, J.; Noble, D. The value of treating cancer as an evolutionary disease. Prog. Biophys. Mol. Biol. 2021, 165, 1–2. [Google Scholar] [CrossRef] [PubMed]
- Furst, R. The Importance of Henry H. Heng’s Genome Architecture Theory. Prog. Biophys. Mol. Biol. 2021, 165, 153–156. [Google Scholar] [CrossRef]
- Vendramin, R.; Litchfield, K.; Swanton, C. Cancer evolution: Darwin and beyond. EMBO J. 2021, 40, e108389. [Google Scholar] [CrossRef] [PubMed]
- Ye, C.J.; Stilgenbauer, L.; Moy, A.; Liu, G.; Heng, H.H. What Is Karyotype Coding and Why Is Genomic Topology Important for Cancer and Evolution? Front. Genet. 2019, 10, 1082. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boveri, T. Concerning the Origin of Malignant Tumours by Theodor Boveri. Translated and annotated by Henry Harris. J. Cell Sci. 2008, 121 (Suppl. 1), 1–84. [Google Scholar] [CrossRef]
- Heng, H.H.; Liu, G.; Stevens, J.B.; Bremer, S.W.; Ye, K.J.; Abdallah, B.Y.; Horne, S.D.; Ye, C.J. Decoding the genome beyond sequencing: The new phase of genomic research. Genomics 2011, 98, 242–252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ye, C.J.; Stevens, J.B.; Liu, G.; Bremer, S.W.; Jaiswal, A.S.; Ye, K.J.; Lin, M.-F.; Lawrenson, L.; Lancaster, W.D.; Kurkinen, M.; et al. Genome based cell population heterogeneity promotes tumorigenicity: The evolutionary mechanism of cancer. J. Cell. Physiol. 2008, 219, 288–300. [Google Scholar] [CrossRef] [Green Version]
- Heng, H.H.; Bremer, S.W.; Stevens, J.B.; Horne, S.D.; Liu, G.; Abdallah, B.Y.; Ye, K.J.; Ye, C.J. Chromosomal instability (CIN): What it is and why it is crucial to cancer evolution. Cancer Metastasis Rev. 2013, 32, 325–340. [Google Scholar] [CrossRef]
- Liu, G.; Stevens, J.B.; Horne, S.D.; Abdallah, B.Y.; Ye, K.J.; Bremer, S.W.; Ye, C.J.; Chen, D.J.; Heng, H.H. Genome chaos: Survival strategy during crisis. Cell Cycle 2014, 13, 528–537. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shapiro, J. How Chaotic Is Genome Chaos? Cancers 2021, 13, 1358. [Google Scholar] [CrossRef] [PubMed]
- Pellestor, F.; Gatinois, V. Chromoanagenesis: A piece of the macroevolution scenario. Mol. Cytogenet. 2020, 13, 3. [Google Scholar] [CrossRef] [PubMed]
- Pellestor, F.; Gaillard, J.; Schneider, A.; Puechberty, J.; Gatinois, V. Chromoanagenesis, the mechanisms of a genomic chaos. Semin. Cell Dev. Biol. 2021. [Google Scholar] [CrossRef]
- Stephens, P.J.; Greenman, C.D.; Fu, B.; Yang, F.; Bignell, G.R.; Mudie, L.J.; Pleasance, E.D.; Lau, K.W.; Beare, D.; Stebbings, L.A.; et al. Massive Genomic Rearrangement Acquired in a Single Catastrophic Event during Cancer Development. Cell 2011, 144, 27–40. [Google Scholar] [CrossRef]
- Horne, S.D.; Heng, H.H. Genome Chaos, Chromothripsis and Cancer Evolution. J. Cancer Stud. Ther. 2014, 1, 1–6. [Google Scholar]
- Heng, H.H. Chapter 5: Cancer Genomic Landscape, Ecology and Evolution of Cancer; Ujvari, B., Roche, B., Thomas, F., Eds.; Elseiver: Amsterdam, The Netherlands, 2017; pp. 69–86. [Google Scholar]
- Baca, S.C.; Prandi, D.; Lawrence, M.S.; Mosquera, J.M.; Romanel, A.; Drier, Y.; Park, K.; Kitabayashi, N.; Macdonald, T.Y.; Ghandi, M.; et al. Punctuated Evolution of Prostate Cancer Genomes. Cell 2013, 153, 666–677. [Google Scholar] [CrossRef] [Green Version]
- Crasta, K.; Ganem, N.; Dagher, R.; Lantermann, A.B.; Ivanova, E.V.; Pan, Y.; Nezi, L.; Protopopov, A.; Chowdhury, D.; Pellman, D. DNA breaks and chromosome pulverization from errors in mitosis. Nature 2012, 482, 53–58. [Google Scholar] [CrossRef]
- Forment, J.V.; Kaidi, A.; Jackson, S.P. Chromothripsis and cancer: Causes and consequences of chromosome shattering. Nat. Rev. Cancer 2012, 12, 663–670. [Google Scholar] [CrossRef] [PubMed]
- Holland, A.J.; Cleveland, D.W. Chromoanagenesis and cancer: Mechanisms and consequences of localized, complex chromosomal rearrangements. Nat. Med. 2012, 18, 1630–1638. [Google Scholar] [CrossRef] [Green Version]
- Inaki, K.; Liu, E.T. Structural mutations in cancer: Mechanistic and functional insights. Trends Genet. 2012, 28, 550–559. [Google Scholar] [CrossRef] [PubMed]
- Jones, M.J.; Jallepalli, P.V. Chromothripsis: Chromosomes in crisis. Dev. Cell. 2012, 23, 908–917. [Google Scholar] [CrossRef] [Green Version]
- Liu, P.; Erez, A.; Nagamani, S.C.S.; Dhar, S.U.; Kołodziejska, K.E.; Dharmadhikari, A.V.; Cooper, M.L.; Wiszniewska, J.; Zhang, F.; Withers, M.A.; et al. Chromosome catastrophes involve replication mechanisms generating complex genomic rearrangements. Cell 2001, 146, 889–890. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malhotra, A.; Lindberg, M.; Faust, G.G.; Leibowitz, M.L.; Clark, R.A.; Layer, R.M.; Quinlan, A.R.; Hall, I.M. Breakpoint profiling of 64 cancer genomes reveals numerous complex rear-rangements spawned by homology-independent mechanisms. Genome Res. 2013, 23, 762–776. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Righolt, C.; Mai, S. Shattered and stitched chromosomes-chromothripsis and chromoanasynthesis-manifestations of a new chromosome crisis? Genes Chromosomes Cancer 2012, 51, 975–981. [Google Scholar] [CrossRef] [PubMed]
- Setlur, S.R.; Lee, C. Tumor Archaeology Reveals that Mutations Love Company. Cell 2012, 149, 959–961. [Google Scholar] [CrossRef] [Green Version]
- Tubio, J.M.; Estivill, X. Cancer: When catastrophe strikes a cell. Nature 2011, 470, 476–477. [Google Scholar] [CrossRef] [PubMed]
- Heng, H.H. Karyotypic Chaos, A Form of Non-Clonal Chromosome Aberrations, Plays A Key Role for Cancer Progression and Drug Resistance; FASEB: Nuclear Structure and Cancer; Vermont Academy: Saxtons River, VT, USA, 2009. [Google Scholar]
- Heng, H.H.; Regan, S.M.; Liu, G.; Ye, C.J. Why it is crucial to analyze non clonal chromosome aberrations or NCCAs? Mol. Cytogenet. 2016, 9, 15. [Google Scholar] [CrossRef] [Green Version]
- Duesberg, P. Chromosomal Chaos and Cancer. Sci. Am. 2007, 296, 52–59. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.Z.; Spektor, A.; Cornils, H.; Francis, J.M.; Jackson, E.K.; Liu, S.; Meyerson, M.; Pellman, D. Chromothripsis from DNA damage in micronuclei. Nature 2015, 522, 179–184. [Google Scholar] [CrossRef] [Green Version]
- Maciejowski, J.; Li, Y.; Bosco, N.; Campbell, P.J.; de Lange, T. Chromothripsis and Kataegis Induced by Telomere Crisis. Cell 2015, 163, 1641–1654. [Google Scholar] [CrossRef] [Green Version]
- Heng, H.H.; Stevens, J.B.; Bremer, S.W.; Liu, G.; Abdallah, B.Y.; Ye, C.J. Evolutionary Mechanisms and Diversity in Cancer. Adv. Cancer Res. 2011, 112, 217–253. [Google Scholar] [CrossRef] [PubMed]
- Morishita, M.; Muramatsu, T.; Suto, Y.; Hirai, M.; Konishi, T.; Hayashi, S.; Shigemizu, D.; Tsunoda, T.; Moriyama, K.; Inazawa, J. Chromothripsis-like chromosomal rearrangements induced by ionizing radiation using proton microbeam irradiation system. Oncotarget 2016, 7, 10182–10192. [Google Scholar] [CrossRef]
- Mardin, B.R.; Drainas, A.P.; Waszak, S.M.; Weischenfeldt, J.; Isokane, M.; Stütz, A.M.; Raeder, B.; Efthymiopoulos, T.; Buccitelli, C.; Se-gura-Wang, M.; et al. A cell-based model system links chromothripsis with hyperploidy. Mol. Syst. Biol. 2015, 11, 828. [Google Scholar] [CrossRef] [PubMed]
- Ye, C.J.; Liu, G.; Heng, H.H. Experimental Induction of Genome Chaos. Methods Mol. Biol. 2018, 337–352. [Google Scholar] [CrossRef]
- Ye, C.J.; Sharpe, Z.; Alemara, S.; Mackenzie, S.; Liu, G.; Abdallah, B.; Horne, S.; Regan, S.; Heng, H.H. Micronuclei and Genome Chaos: Changing the System Inheritance. Genes 2019, 10, 366. [Google Scholar] [CrossRef] [Green Version]
- Heng, J.; Heng, H.H. Genome chaos: Creating new genomic information essential for cancer macroevolution. Semin. Cancer Biol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Heng, J.; Heng, H.H. Karyotype coding: The creation and maintenance of system information for complexity and biodiversity. Biosystems 2021, 208, 104476. [Google Scholar] [CrossRef]
- Barbieri, M. What is code biology? Biosystems 2018, 164, 1–10. [Google Scholar] [CrossRef]
- Barbieri, M. Overview of the third special issue in code biology. Biosystems 2021, 210, 104553. [Google Scholar] [CrossRef]
- Barbieri, M. A new theory of development: The generation of complexity in ontogenesis. Philos. Trans. R. Soc. A Math. Phys. Eng. Sci. 2016, 374, 20150148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crkvenjakov, R.; Heng, H.H. Further illusions: On key evolutionary mechanisms that could never fit with Modern Synthesis. Prog. Biophys. Mol. Biol. 2021. [Google Scholar] [CrossRef] [PubMed]
- Wilkins, A.S.; Holliday, R. The Evolution of Meiosis from Mitosis. Genetics 2009, 181, 3–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gorelick, R.; Heng, H.H. Sex Reduces Genetic Variation: A Multidisciplinary Review. Evolution 2010, 65, 1088–1098. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. The hallmarks of cancer. Cell 2000, 100, 57–70. [Google Scholar] [CrossRef] [Green Version]
- Heng, E.; Moy, A.; Liu, G.; Heng, H.H.; Zhang, K. ER Stress and Micronuclei Cluster: Stress Response Contributes to Genome Chaos in Cancer. Front. Cell Dev. Biol. 2021, 9, 673188. [Google Scholar] [CrossRef]
- The White House Office of the Press Secretary, 2000. Available online: https://www.genome.gov/10001356/june-2000-white-house-event (accessed on 1 December 2021).
- Transcript: President Obama’s Final State of the Union Address. Available online: https://www.npr.org/2016/01/12/462831088/president-obama-state-of-the-union-transcript (accessed on 8 December 2021).
- Gatenby, R.A.; Silva, A.S.; Gillies, R.J.; Frieden, B.R. Adaptive therapy. Cancer Res. 2009, 69, 4894–4903. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kroemer, G.; Galluzzi, L.; Vandenabeele, P.; Abrams, J.; Alnemri, E.S.; Baehrecke, E.H.; Blagosklonny, M.V.; El-Deiry, W.S.; Golstein, P.; Green, D.R.; et al. Classification of cell death: Recommendations of the Nomenclature Committee on Cell Death 2009. Cell Death Differ. 2009, 16, 3–11. [Google Scholar] [CrossRef] [PubMed]
- The Cancer Genome Atlas Research Network. Comprehensive, Integrative Genomic Analysis of Diffuse Lower-Grade Gliomas. N. Engl. J. Med. 2015, 372, 2481–2498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Available online: https://www.cityofhope.org/breakthroughs/the-war-on-cancer-at-50-where-are-we (accessed on 12 August 2021).
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).