Time Course Analysis of Genome-Wide Identification of Mutations Induced by and Genes Expressed in Response to Carbon Ion Beam Irradiation in Rice (Oryza sativa L.)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and CIB Irradiation
2.2. Whole-Genome Mutation Analysis
2.3. RNA Extraction, Sequencing, and Gene Expression Profile Analysis
3. Results
3.1. Whole-Genome Resequencing of Irradiated Rice Seeds
3.2. Comprehensive Identification of Mutations Induced by CIB
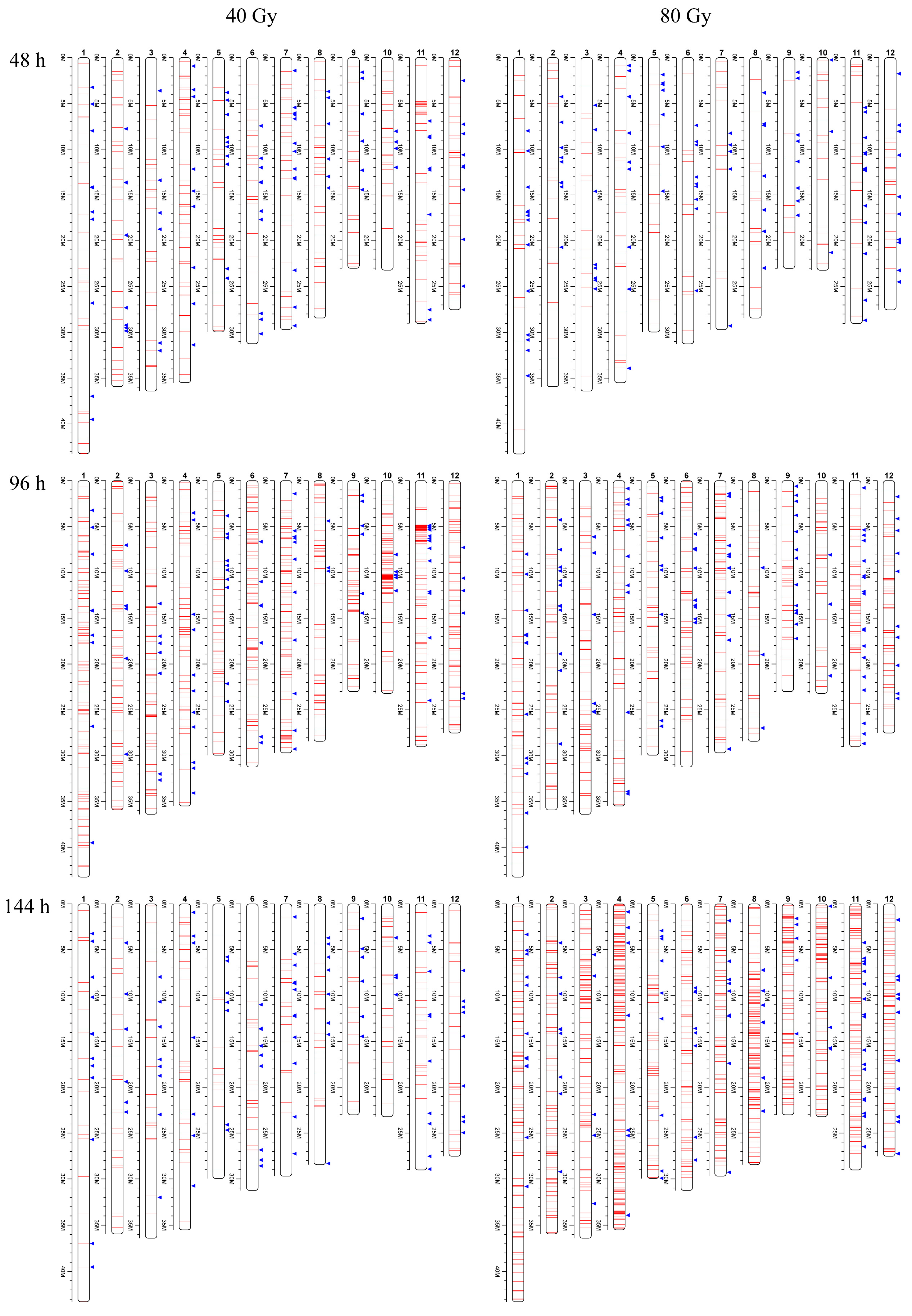
3.3. Mutation Frequency and Clustered DNA Damage Induced by CIB
3.4. Distribution and Rates of Mutations Induced by CIB Irradiation
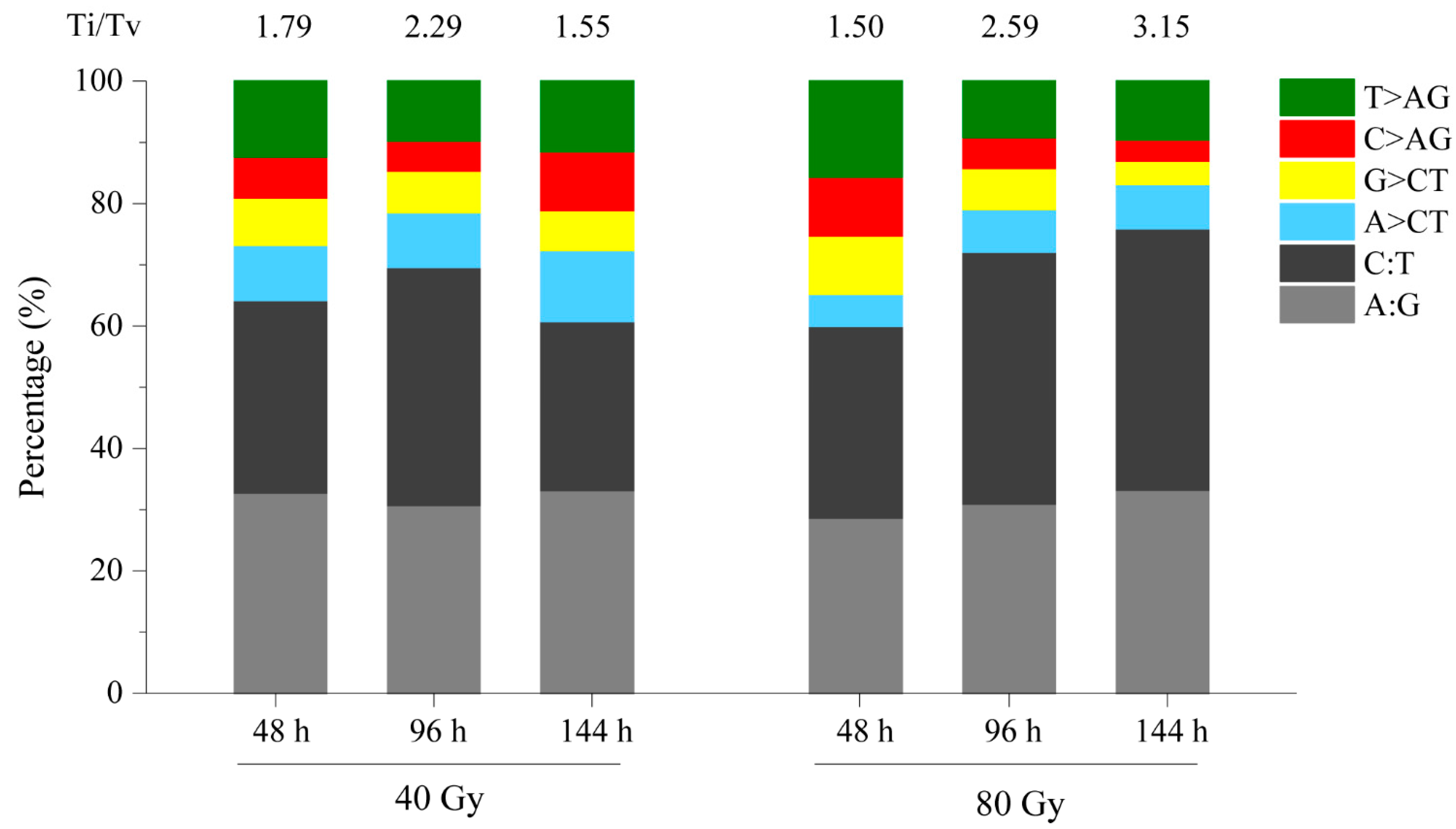
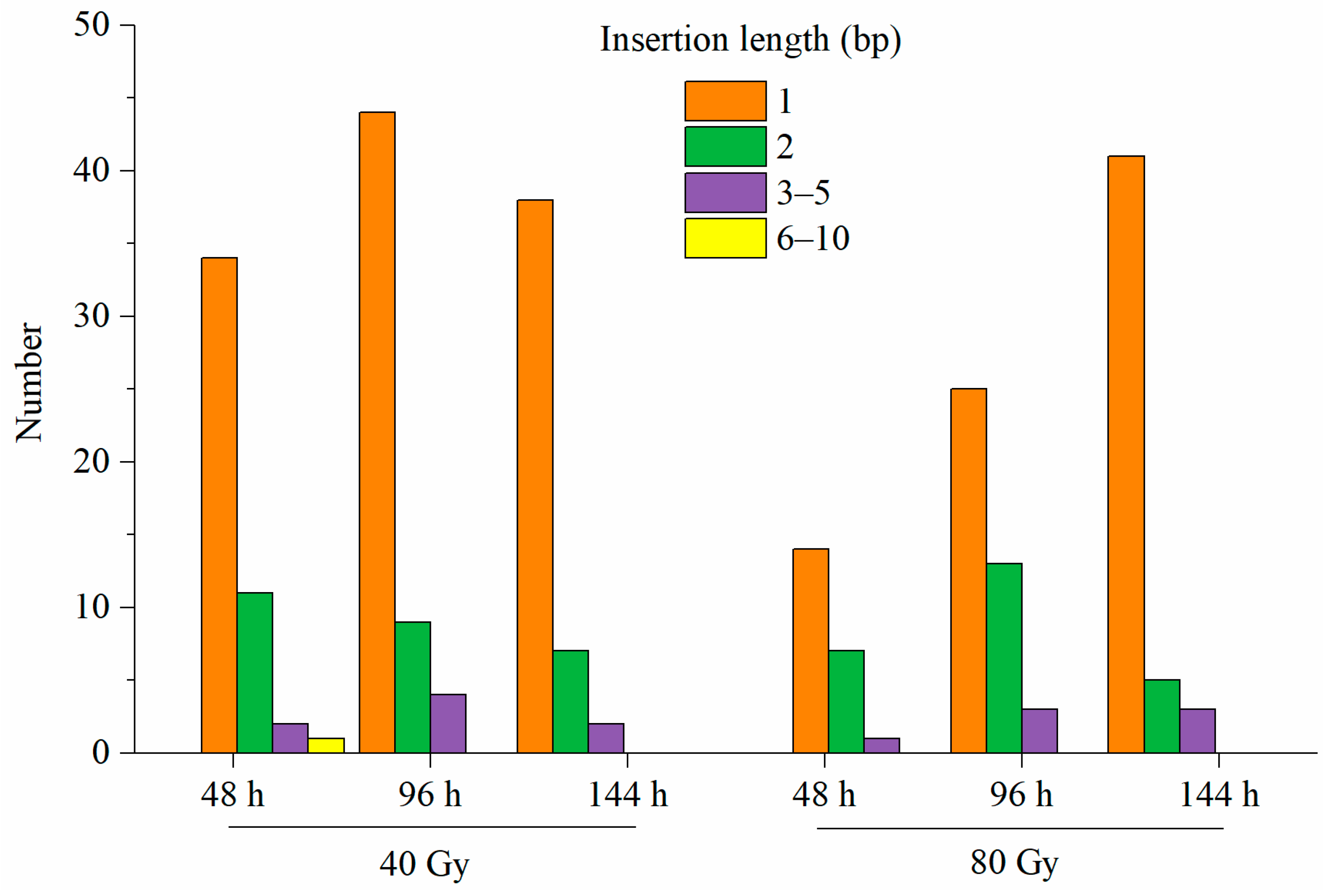
3.5. Characteristics of SBSs and InDels Induced by CIB Irradiation
3.6. Genome-Wide Expression Changes after CIB Irradiation
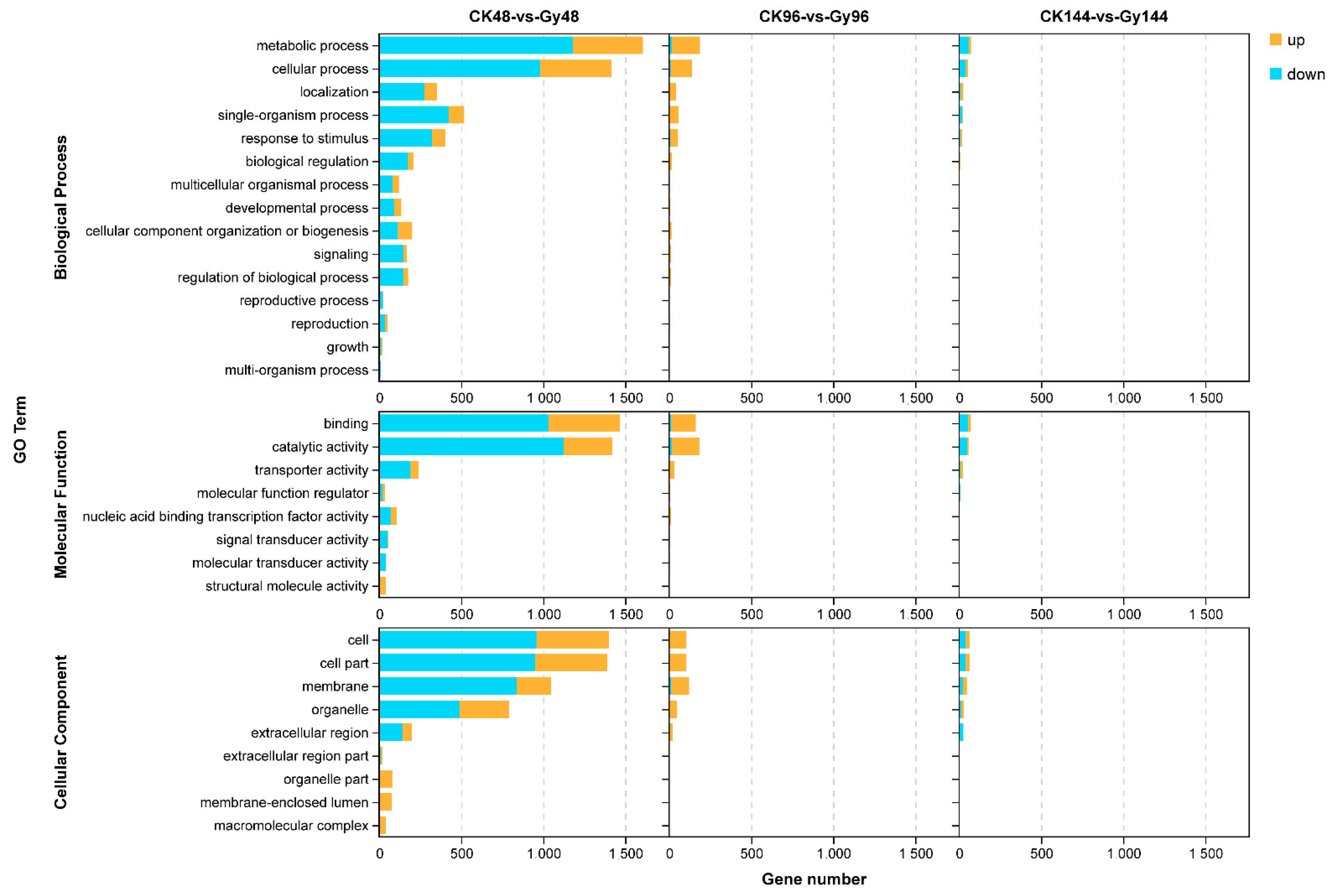
3.7. Time Course Enrichment Analysis of GO Terms and KEGG Pathways
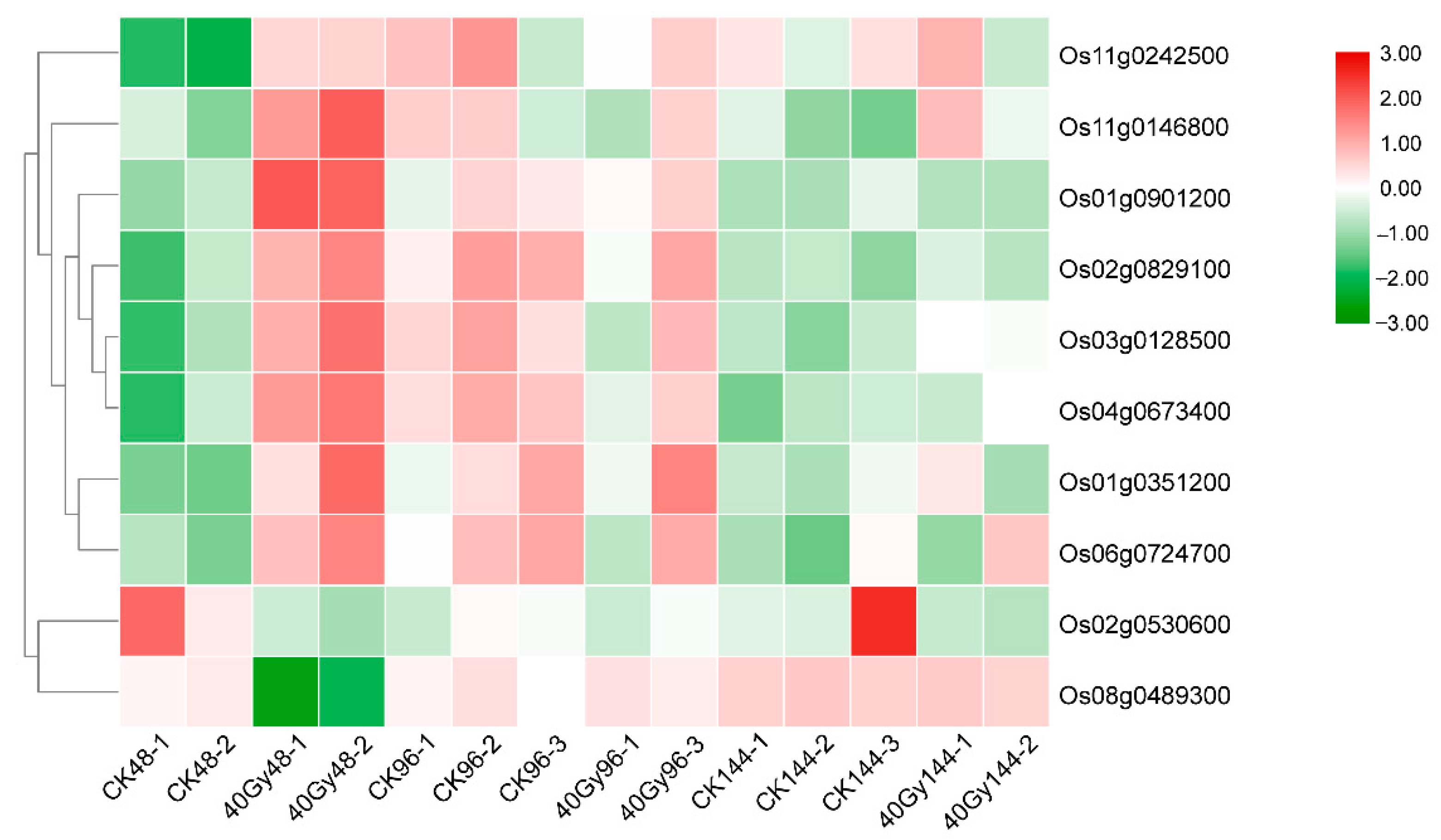
3.8. Variation in Transcript Levels of Damage/Repair-Related Genes Following CIB Irradiation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Saeb, A.T.M.; Al-Naqeb, D. The Impact of Evolutionary Driving Forces on Human Complex Diseases: A Population Genetics Approach. Scientifica 2016, 2016, 2079704. [Google Scholar] [CrossRef] [Green Version]
- Ossowski, S.; Schneeberger, K.; Lucas-Lledó, J.I.; Warthmann, N.; Clark, R.M.; Shaw, R.G.; Weigel, D.; Lynch, M. The Rate and Molecular Spectrum of Spontaneous Mutations in Arabidopsis thaliana. Science 2010, 327, 92–94. [Google Scholar] [CrossRef] [Green Version]
- Shirley, B.W.; Hanley, S.; Goodman, H.M. Effects of ionizing radiation on a plant genome: Analysis of two Arabidopsis transparent testa mutations. Plant Cell 1992, 4, 333–347. [Google Scholar] [CrossRef] [PubMed]
- Morita, R.; Kusaba, M.; Iida, S.; Yamaguchi, H.; Nishio, T.; Nishimura, M. Molecular characterization of mutations induced by γ irradiation in rice. Genes Genet. Syst. 2009, 84, 361–370. [Google Scholar] [CrossRef] [Green Version]
- Belfield, E.J.; Gan, X.; Mithani, A.; Brown, C.; Jiang, C.; Franklin, K.; Alvey, E.; Wibowo, A.; Jung, M.; Bailey, K.; et al. Genome-wide analysis of mutations in mutant lineages selected following fast-neutron irradiation mutagenesis of Arabidopsis thaliana. Genome Res. 2012, 22, 1306–1315. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiao, X.-o.; Lin, W.; Li, K.; Feng, X.; Jin, H.; Zou, H. Genome-Wide Analysis of Artificial Mutations Induced by Ethyl Methanesulfonate in the Eggplant (Solanum melongena L.). Genes 2019, 10, 595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kazama, Y.; Ishii, K.; Hirano, T.; Wakana, T.; Yamada, M.; Ohbu, S.; Abe, T. Different mutational function of low- and high-linear energy transfer heavy-ion irradiation demonstrated by whole-genome resequencing of Arabidopsis mutants. Plant J. 2017, 92, 1020–1030. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tanaka, A.; Shikazono, N.; Hase, Y. Studies on Biological Effects of Ion Beams on Lethality, Molecular Nature of Mutation, Mutation Rate, and Spectrum of Mutation Phenotype for Mutation Breeding in Higher Plants. J. Radiat. Res. 2010, 51, 223–233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abe, T.; Kazama, Y.; Hirano, T. Ion Beam Breeding and Gene Discovery for Function Analyses Using Mutants. Nucl. Phys. News 2015, 25, 30–34. [Google Scholar] [CrossRef]
- Du, Y.; Luo, S.; Li, X.; Yang, J.; Cui, T.; Li, W.; Yu, L.; Feng, H.; Chen, Y.; Mu, J.; et al. Identification of Substitutions and Small Insertion-Deletions Induced by Carbon-Ion Beam Irradiation in Arabidopsis thaliana. Front. Plant Sci. 2017, 8. [Google Scholar] [CrossRef] [Green Version]
- Shirasawa, K.; Hirakawa, H.; Nunome, T.; Tabata, S.; Isobe, S. Genome-wide survey of artificial mutations induced by ethyl methanesulfonate and γ rays in tomato. Plant Biotechnol. J. 2016, 14, 51–60. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Zheng, Y.-C.; Cui, H.-R.; Fu, H.-W.; Shu, Q.-Y.; Huang, J.-Z. Frequency and type of inheritable mutations induced by γ rays in rice as revealed by whole genome sequencing. J. Zhejiang Univ. Sci. B 2016, 17, 905–915. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hirano, T.; Kazama, Y.; Ishii, K.; Ohbu, S.; Shirakawa, Y.; Abe, T. Comprehensive identification of mutations induced by heavy-ion beam irradiation in Arabidopsis thaliana. Plant J. 2015, 82, 93–104. [Google Scholar] [CrossRef] [PubMed]
- Ichida, H.; Morita, R.; Shirakawa, Y.; Hayashi, Y.; Abe, T. Targeted exome sequencing of unselected heavy-ion beam-irradiated populations reveals less-biased mutation characteristics in the rice genome. Plant J. 2019, 98, 301–314. [Google Scholar] [CrossRef]
- Yang, G.; Luo, W.; Zhang, J.; Yan, X.; Du, Y.; Zhou, L.; Li, W.; Wang, H.; Chen, Z.; Guo, T. Genome-Wide Comparisons of Mutations Induced by Carbon-Ion Beam and γ-Rays Irradiation in Rice via Resequencing Multiple Mutants. Front. Plant Sci. 2019, 10. [Google Scholar] [CrossRef] [Green Version]
- Li, F.; Shimizu, A.; Nishio, T.; Tsutsumi, N.; Kato, H. Comparison and Characterization of Mutations Induced by γ-Ray and Carbon-Ion Irradiation in Rice (Oryza sativa L.) Using Whole-Genome Resequencing. G3 Genes Genomes Genet. 2019, 9, 3743–3751. [Google Scholar] [CrossRef] [Green Version]
- Mannuss, A.; Trapp, O.; Puchta, H. Gene regulation in response to DNA damage. Biochim. Biophys. Acta Gene Regul. Mech. 2012, 1819, 154–165. [Google Scholar] [CrossRef] [PubMed]
- Missirian, V.; Conklin, P.A.; Culligan, K.M.; Huefner, N.D.; Britt, A.B. High atomic weight, high-energy radiation (HZE) induces transcriptional responses shared with conventional stresses in addition to a core “DSB” response specific to clastogenic treatments. Front. Plant Sci. 2014, 5. [Google Scholar] [CrossRef] [Green Version]
- Ochoa, M.; Tierra, W.; Tupuna-Yerovi, D.S.; Guanoluisa, D.; Otero, X.L.; Ruales, J. Assessment of cadmium and lead contamination in rice farming soils and rice (Oryza sativa L.) from Guayas province in Ecuador. Environ. Pollut. 2020, 260, 114050. [Google Scholar] [CrossRef]
- Hada, M.; Georgakilas, A.G. Formation of Clustered DNA Damage after High-LET Irradiation: A Review. J. Radiat. Res. 2008, 49, 203–210. [Google Scholar] [CrossRef]
- Tokuyama, Y.; Furusawa, Y.; Ide, H.; Yasui, A.; Terato, H. Role of isolated and clustered DNA damage and the post-irradiating repair process in the effects of heavy ion beam irradiation. J. Radiat. Res. 2015, 56, 446–455. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, Q.; Wang, J.; Qi, J.; Peng, D.; Guan, B.; Zhang, J.; Li, Z.; Zhang, H.; Li, T.; Shi, Y.; et al. Increased chromosomal instability characterizes metastatic renal cell carcinoma. Transl. Oncol. 2021, 14, 100929. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Xie, L.; Yao, Z.; Zhou, Y.; Zhou, W.; Wang, J.; Sun, Y.; Gong, C. Caragana korshinskii phenylalanine ammonialyase is up-regulated in the phenylpropanoid biosynthesis pathway in response to drought stress. Biotechnol. Biotechnol. Equip. 2019, 33, 842–854. [Google Scholar] [CrossRef] [Green Version]
- Odahara, M.; Kishita, Y.; Sekine, Y. MSH1 maintains organelle genome stability and genetically interacts with RECA and RECG in the moss Physcomitrella patens. Plant J. 2017, 91, 455–465. [Google Scholar] [CrossRef] [Green Version]
- Rowan, B.A.; Oldenburg, D.J.; Bendich, A.J. RecA maintains the integrity of chloroplast DNA molecules in Arabidopsis. J. Exp. Bot. 2010, 61, 2575–2588. [Google Scholar] [CrossRef] [PubMed]
- Manova, V.; Gruszka, D. DNA damage and repair in plants—From models to crops. Front. Plant Sci. 2015, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gu, Z.; Pan, W.; Chen, W.; Lian, Q.; Wu, Q.; Lv, Z.; Cheng, X.; Ge, X. New perspectives on the plant PARP family: Arabidopsis PARP3 is inactive, and PARP1 exhibits predominant poly (ADP-ribose) polymerase activity in response to DNA damage. BMC Plant Biol. 2019, 19, 364. [Google Scholar] [CrossRef] [Green Version]
- Wynn, E.; Purfeerst, E.; Christensen, A. Mitochondrial DNA Repair in an Arabidopsis thaliana Uracil N-Glycosylase Mutant. Plants 2020, 9, 261. [Google Scholar] [CrossRef] [Green Version]
- Boesch, P.; Ibrahim, N.; Paulus, F.; Cosset, A.; Tarasenko, V.; Dietrich, A. Plant mitochondria possess a short-patch base excision DNA repair pathway. Nucleic Acids Res. 2009, 37, 5690–5700. [Google Scholar] [CrossRef] [Green Version]
- Elo, A.; Lyznik, A.; Gonzalez, D.O.; Kachman, S.D.; Mackenzie, S.A. Nuclear Genes That Encode Mitochondrial Proteins for DNA and RNA Metabolism Are Clustered in the Arabidopsis Genome. Plant Cell 2003, 15, 1619–1631. [Google Scholar] [CrossRef] [Green Version]
- Feng, W.; Hale, C.J.; Over, R.S.; Cokus, S.J.; Jacobsen, S.E.; Michaels, S.D. Large-scale heterochromatin remodeling linked to overreplication-associated DNA damage. Proc. Natl. Acad. Sci. USA 2017, 114, 406–411. [Google Scholar] [CrossRef] [Green Version]
- Mahapatra, K.; Roy, S. An insight into the mechanism of DNA damage response in plants- role of SUPPRESSOR OF γ RESPONSE 1: An overview. Mutat. Res. Mol. Mech. Mutagen. 2020, 819–820, 111689. [Google Scholar] [CrossRef]
- Baselet, B.; Belmans, N.; Coninx, E.; Lowe, D.; Janssen, A.; Michaux, A.; Tabury, K.; Raj, K.; Quintens, R.; Benotmane, M.A.; et al. Functional Gene Analysis Reveals Cell Cycle Changes and Inflammation in Endothelial Cells Irradiated with a Single X-ray Dose. Front. Pharmacol. 2017, 8. [Google Scholar] [CrossRef] [Green Version]
- Georgakilas, A.G.; O’Neill, P.; Stewart, R.D. Induction and Repair of Clustered DNA Lesions: What Do We Know So Far? Radiat. Res. 2013, 180, 100–109. [Google Scholar] [CrossRef]
- Ishii, K.; Kazama, Y.; Morita, R.; Hirano, T.; Ikeda, T.; Usuda, S.; Hayashi, Y.; Ohbu, S.; Motoyama, R.; Nagamura, Y.; et al. Linear Energy Transfer-Dependent Change in Rice Gene Expression Profile after Heavy-Ion Beam Irradiation. PLoS ONE 2016, 11, e0160061. [Google Scholar] [CrossRef]
- Jia, Q.; Dulk-Ras, A.D.; Shen, H.; Hooykaas, P.J.J.; de Pater, S. Poly(ADP-ribose)polymerases are involved in microhomology mediated back-up non-homologous end joining in Arabidopsis thaliana. Plant. Mol. Biol. 2013, 82, 339–351. [Google Scholar] [CrossRef]
- Song, J.; Keppler, B.D.; Wise, R.R.; Bent, A.F. PARP2 Is the Predominant Poly(ADP-Ribose) Polymerase in Arabidopsis DNA Damage and Immune Responses. PLoS Genet. 2015, 11, e1005200. [Google Scholar] [CrossRef] [PubMed]




| Sample | Clean Data (bp) | HQ Clean Data (bp) | Total Reads | Total Unmapped Reads | Total Mapped Reads | Average Depth |
|---|---|---|---|---|---|---|
| CK-40Gy | 1.2 × 1010 | 10,699,457,320 | 71,555,678 | 9,994,707 (13.97%) | 61,560,971 (86.03%) | 32.16× |
| 40Gy-48h | 6.539 × 1010 | 60,338,857,661 | 403,481,808 | 102,161,779 (25.32%) | 301,320,029 (74.68%) | 175.20× |
| 40Gy-96h | 5.475 × 1010 | 51,836,646,543 | 346,316,164 | 116,046,170 (33.51%) | 230,269,994 (66.49%) | 146.69× |
| 40Gy-144h | 5.209 × 1010 | 51,735,401,305 | 345,632,662 | 68,450,817 (19.80%) | 277,181,845 (80.20%) | 139.55× |
| CK80Gy | 1.281 × 1010 | 12,669,338,558 | 84,647,388 | 5,273,573 (6.23%) | 79,373,815 (93.77%) | 34.32× |
| 80Gy48h | 4.043 × 1010 | 39,926,015,703 | 266,742,060 | 32,349,666 (12.13%) | 234,392,394 (87.87%) | 108.33× |
| 80Gy96h | 4.081 × 1010 | 40,346,411,461 | 269,666,882 | 19,379,819 (7.19%) | 250,287,063 (92.81%) | 109.34× |
| 80Gy144h | 4.807 × 1010 | 47,692,554,563 | 318,715,200 | 24,827,815 (7.79%) | 293,887,385 (92.21%) | 128.79× |
| Treatment | Time | Total | Number of Mutations | Proportion of Each Mutation (%) | ||||
|---|---|---|---|---|---|---|---|---|
| SBSs | Insertions | Deletions | SBSs | Insertions | Deletions | |||
| 40 Gy | 48 h | 447 | 299 | 94 | 54 | 66.89% | 21.03% | 12.08% |
| 96 h | 1304 | 1133 | 114 | 57 | 86.89% | 8.74% | 4.37% | |
| 144 h | 333 | 199 | 87 | 47 | 59.76% | 26.13% | 14.11% | |
| 80 Gy | 48 h | 202 | 115 | 57 | 30 | 56.93% | 28.22% | 14.85% |
| 96 h | 828 | 717 | 70 | 41 | 86.59% | 8.45% | 4.95% | |
| 144 h | 1307 | 1183 | 75 | 49 | 90.51% | 5.74% | 3.75% | |
| Os ID | Gene Description | AT ID | GO Biological Processes |
|---|---|---|---|
| Os01g0351200 | Poly synthetase 2-A | AT4G02390 | DNA ADP-ribosylation; double-stranded break repair via non-homologous end joining; protein poly-ADP-ribosylation |
| Os01g0901200 | RecA protein | AT2G19490 | DNA repair |
| Os02g0829100 | Poly synthetase 3 | AT5G22470 | Double-stranded break repair; protein poly-ADP-ribosylation |
| Os03g0128500 | POLD2-Putative DNA polymerase delta complex subunit | AT2G42120 | DNA strand elongation involving DNA replication |
| Os04g0673400 | Uracil-DNA glycosylase | AT3G18630 | Base-excision repair; AP site formation via deaminated base removal |
| Os06g0724700 | Phosphatidylinositol kinase and FAT domain-containing protein | AT5G40820 | DNA repair, double-stranded break repair via non-homologous end joining, response to γ radiation |
| Os11g0146800 | OsDMC1B | ||
| Os11g0242500 | Cyclin-dependent kinase | ||
| Os02g0530600 | Polysynthetase 3 | AT5G22470 | Double-stranded break repair, protein poly-ADP-ribosylation |
| Os08g0489300 | Methyladenine glycosylase | AT1G13635 | Base-excision repair |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, J.; Peng, Z.; Liu, Q.; Yang, G.; Zhou, L.; Li, W.; Wang, H.; Chen, Z.; Guo, T. Time Course Analysis of Genome-Wide Identification of Mutations Induced by and Genes Expressed in Response to Carbon Ion Beam Irradiation in Rice (Oryza sativa L.). Genes 2021, 12, 1391. https://doi.org/10.3390/genes12091391
Zhang J, Peng Z, Liu Q, Yang G, Zhou L, Li W, Wang H, Chen Z, Guo T. Time Course Analysis of Genome-Wide Identification of Mutations Induced by and Genes Expressed in Response to Carbon Ion Beam Irradiation in Rice (Oryza sativa L.). Genes. 2021; 12(9):1391. https://doi.org/10.3390/genes12091391
Chicago/Turabian StyleZhang, Jian, Ziai Peng, Qiling Liu, Guili Yang, Libin Zhou, Wenjian Li, Hui Wang, Zhiqiang Chen, and Tao Guo. 2021. "Time Course Analysis of Genome-Wide Identification of Mutations Induced by and Genes Expressed in Response to Carbon Ion Beam Irradiation in Rice (Oryza sativa L.)" Genes 12, no. 9: 1391. https://doi.org/10.3390/genes12091391
APA StyleZhang, J., Peng, Z., Liu, Q., Yang, G., Zhou, L., Li, W., Wang, H., Chen, Z., & Guo, T. (2021). Time Course Analysis of Genome-Wide Identification of Mutations Induced by and Genes Expressed in Response to Carbon Ion Beam Irradiation in Rice (Oryza sativa L.). Genes, 12(9), 1391. https://doi.org/10.3390/genes12091391






