DNA Methylation Dynamics in the Female Germline and Maternal-Effect Mutations That Disrupt Genomic Imprinting
Abstract
1. Introduction
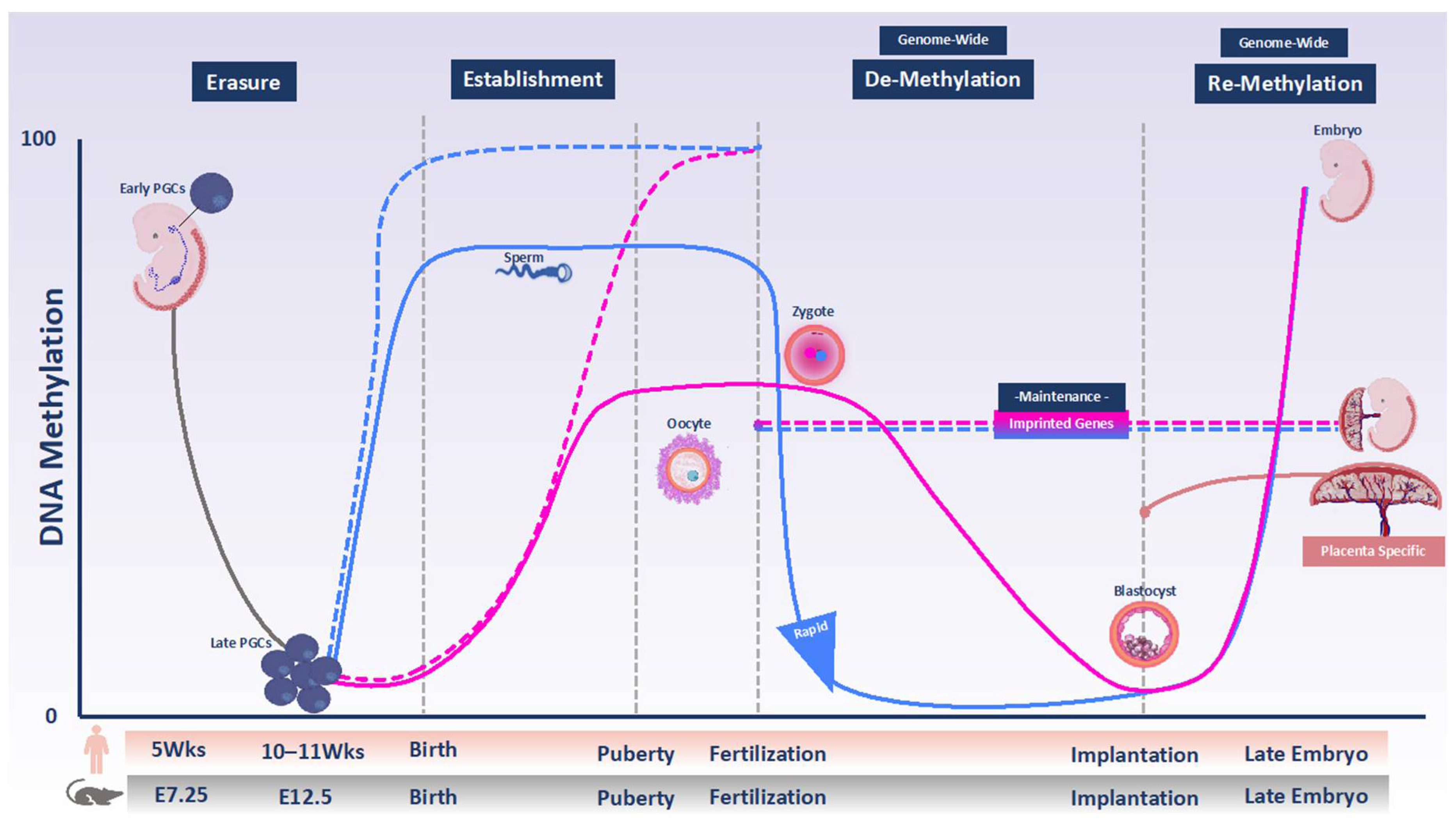
2. DNA Methylation Programming and Reprogramming in the Female Germline and Early Embryo
2.1. Primordial Germ Cells and Oocytes
2.2. Preimplantation Embryo
2.3. Post-Implantation Embryo
3. Methylation Patterning of Imprinted Genes
4. Global Loss of Imprinting Results in Hydatidiform Molar Pregnancies
5. Molar Pregnancies Indicate a Role for the Subcortical Maternal Complex in Ensuring Imprinting
6. Pathogenic Variants Identified within Human SCMC Genes
7. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Smith, Z.D.; Meissner, A. DNA Methylation: Roles in Mammalian Development. Nat. Rev. Genet. 2013, 14, 204–220. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.-M.; Lu, R.; Wang, P.; Yu, Y.; Chen, D.; Gao, L.; Liu, S.; Ji, D.; Rothbart, S.B.; Wang, Y.; et al. Structural Basis for DNMT3A-Mediated de Novo DNA Methylation. Nature 2018, 554, 387–391. [Google Scholar] [CrossRef]
- Petrussa, L.; Van de Velde, H.; De Rycke, M. Dynamic Regulation of DNA Methyltransferases in Human Oocytes and Preimplantation Embryos after Assisted Reproductive Technologies. Mol. Hum. Reprod. 2014, 20, 861–874. [Google Scholar] [CrossRef]
- Bostick, M.; Kim, J.K.; Estève, P.-O.; Clark, A.; Pradhan, S.; Jacobsen, S.E. UHRF1 Plays a Role in Maintaining DNA Methylation in Mammalian Cells. Science 2007, 317, 1760–1764. [Google Scholar] [CrossRef] [PubMed]
- Shirane, K.; Toh, H.; Kobayashi, H.; Miura, F.; Chiba, H.; Ito, T.; Kono, T.; Sasaki, H. Mouse Oocyte Methylomes at Base Resolution Reveal Genome-Wide Accumulation of Non-CpG Methylation and Role of DNA Methyltransferases. PLoS Genet. 2013, 9, e1003439. [Google Scholar] [CrossRef] [PubMed]
- Messerschmidt, D.M.; Knowles, B.B.; Solter, D. DNA Methylation Dynamics during Epigenetic Reprogramming in the Germline and Preimplantation Embryos. Genes Dev. 2014, 28, 812–828. [Google Scholar] [CrossRef]
- Hajkova, P.; Erhardt, S.; Lane, N.; Haaf, T.; El-Maarri, O.; Reik, W.; Walter, J.; Surani, M.A. Epigenetic Reprogramming in Mouse Primordial Germ Cells. Mech. Dev. 2002, 117, 15–23. [Google Scholar] [CrossRef]
- Henckel, A.; Chebli, K.; Kota, S.K.; Arnaud, P.; Feil, R. Transcription and histone methylation changes correlate with imprint acquisition in male germ cells. EMBO J. 2012, 31, 606–615. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Zhu, P.; Wu, X.; Li, X.; Wen, L.; Tang, F. Single-Cell Methylome Landscapes of Mouse Embryonic Stem Cells and Early Embryos Analyzed Using Reduced Representation Bisulfite Sequencing. Genome Res. 2013, 23, 2126–2135. [Google Scholar] [CrossRef] [PubMed]
- Stewart, K.R.; Veselovska, L.; Kelsey, G. Establishment and Functions of DNA Methylation in the Germline. Epigenomics 2016, 8, 1399–1413. [Google Scholar] [CrossRef] [PubMed]
- Smallwood, S.A.; Tomizawa, S.-I.; Krueger, F.; Ruf, N.; Carli, N.; Segonds-Pichon, A.; Sato, S.; Hata, K.; Andrews, S.R.; Kelsey, G. Dynamic CpG Island Methylation Landscape in Oocytes and Preimplantation Embryos. Nat. Genet. 2011, 43, 811–814. [Google Scholar] [CrossRef]
- Kobayashi, H.; Sakurai, T.; Imai, M.; Takahashi, N.; Fukuda, A.; Yayoi, O.; Sato, S.; Nakabayashi, K.; Hata, K.; Sotomaru, Y.; et al. Contribution of Intragenic DNA Methylation in Mouse Gametic DNA Methylomes to Establish Oocyte-Specific Heritable Marks. PLoS Genet. 2012, 8, e1002440. [Google Scholar] [CrossRef]
- Okae, H.; Chiba, H.; Hiura, H.; Hamada, H.; Sato, A.; Utsunomiya, T.; Kikuchi, H.; Yoshida, H.; Tanaka, A.; Suyama, M.; et al. Genome-Wide Analysis of DNA Methylation Dynamics during Early Human Development. PLoS Genet. 2014, 10, e1004868. [Google Scholar] [CrossRef]
- Veselovska, L.; Smallwood, S.A.; Saadeh, H.; Stewart, K.R.; Krueger, F.; Maupetit-Méhouas, S.; Arnaud, P.; Tomizawa, S.-I.; Andrews, S.; Kelsey, G. Deep Sequencing and de Novo Assembly of the Mouse Oocyte Transcriptome Define the Contribution of Transcription to the DNA Methylation Landscape. Genome Biol. 2015, 16, 209. [Google Scholar] [CrossRef]
- Santos, F.; Hendrich, B.; Reik, W.; Dean, W. Dynamic Reprogramming of DNA Methylation in the Early Mouse Embryo. Dev. Biol. 2002, 241, 172–182. [Google Scholar] [CrossRef] [PubMed]
- Gu, T.-P.; Guo, F.; Yang, H.; Wu, H.-P.; Xu, G.-F.; Liu, W.; Xie, Z.-G.; Shi, L.; He, X.; Jin, S.; et al. The Role of Tet3 DNA Dioxygenase in Epigenetic Reprogramming by Oocytes. Nature 2011, 477, 606–610. [Google Scholar] [CrossRef] [PubMed]
- Smallwood, S.A.; Kelsey, G. De Novo DNA Methylation: A Germ Cell Perspective. Trends Genet. 2012, 28, 33–42. [Google Scholar] [CrossRef] [PubMed]
- Amouroux, R.; Nashun, B.; Shirane, K.; Nakagawa, S.; Hill, P.W.; D’Souza, Z.; Nakayama, M.; Matsuda, M.; Turp, A.; Ndjetehe, E.; et al. De Novo DNA Methylation Drives 5hmC Accumulation in Mouse Zygotes. Nat. Cell Biol. 2016, 18, 225–233. [Google Scholar] [CrossRef]
- Zhu, P.; Guo, H.; Ren, Y.; Hou, Y.; Dong, J.; Li, R.; Lian, Y.; Fan, X.; Hu, B.; Gao, Y.; et al. Single-Cell DNA Methylome Sequencing of Human Preimplantation Embryos. Nat. Genet. 2018, 50, 12–19. [Google Scholar] [CrossRef]
- Barlow, D.P.; Bartolomei, M.S. Genomic Imprinting in Mammals. Cold Spring Harb. Perspect. Biol. 2014, 6, a018382. [Google Scholar] [CrossRef]
- Monk, D.; Mackay, D.J.G.; Eggermann, T.; Maher, E.R.; Riccio, A. Genomic Imprinting Disorders: Lessons on How Genome, Epigenome and Environment Interact. Nat. Rev. Genet. 2019, 20, 235–248. [Google Scholar] [CrossRef]
- Soellner, L.; Begemann, M.; Mackay, D.J.G.; Grønskov, K.; Tümer, Z.; Maher, E.R.; Temple, I.K.; Monk, D.; Riccio, A.; Linglart, A.; et al. Recent Advances in Imprinting Disorders. Clin. Genet. 2017, 91, 3–13. [Google Scholar] [CrossRef] [PubMed]
- Van den Veyver, I.B.; Al-Hussaini, T.K. Biparental Hydatidiform Moles: A Maternal Effect Mutation Affecting Imprinting in the Offspring. Hum. Reprod. Update 2006, 12, 233–242. [Google Scholar] [CrossRef]
- Demond, H.; Anvar, Z.; Jahromi, B.N.; Sparago, A.; Verma, A.; Davari, M.; Calzari, L.; Russo, S.; Jahromi, M.A.; Monk, D.; et al. A KHDC3L Mutation Resulting in Recurrent Hydatidiform Mole Causes Genome-Wide DNA Methylation Loss in Oocytes and Persistent Imprinting Defects Post-Fertilisation. Genome Med. 2019, 11, 84. [Google Scholar] [CrossRef] [PubMed]
- Murdoch, S.; Djuric, U.; Mazhar, B.; Seoud, M.; Khan, R.; Kuick, R.; Bagga, R.; Kircheisen, R.; Ao, A.; Ratti, B.; et al. Mutations in NALP7 Cause Recurrent Hydatidiform Moles and Reproductive Wastage in Humans. Nat. Genet. 2006, 38, 300–302. [Google Scholar] [CrossRef] [PubMed]
- Parry, D.A.; Logan, C.V.; Hayward, B.E.; Shires, M.; Landolsi, H.; Diggle, C.; Carr, I.; Rittore, C.; Touitou, I.; Philibert, L.; et al. Mutations Causing Familial Biparental Hydatidiform Mole Implicate C6orf221 as a Possible Regulator of Genomic Imprinting in the Human Oocyte. Am. J. Hum. Genet. 2011, 89, 451–458. [Google Scholar] [CrossRef]
- Docherty, L.E.; Rezwan, F.I.; Poole, R.L.; Turner, C.L.S.; Kivuva, E.; Maher, E.R.; Smithson, S.F.; Hamilton-Shield, J.P.; Patalan, M.; Gizewska, M.; et al. Mutations in NLRP5 Are Associated with Reproductive Wastage and Multilocus Imprinting Disorders in Humans. Nat. Commun. 2015, 6, 8086. [Google Scholar] [CrossRef]
- Begemann, M.; Rezwan, F.I.; Beygo, J.; Docherty, L.E.; Kolarova, J.; Schroeder, C.; Buiting, K.; Chokkalingam, K.; Degenhardt, F.; Wakeling, E.L.; et al. Maternal Variants in NLRP and Other Maternal Effect Proteins Are Associated with Multilocus Imprinting Disturbance in Offspring. J. Med. Genet. 2018, 55, 497–504. [Google Scholar] [CrossRef]
- Tong, Z.B.; Gold, L.; Pfeifer, K.E.; Dorward, H.; Lee, E.; Bondy, C.A.; Dean, J.; Nelson, L.M. Mater, a Maternal Effect Gene Required for Early Embryonic Development in Mice. Nat. Genet. 2000, 26, 267–268. [Google Scholar] [CrossRef]
- Saitou, M.; Miyauchi, H. Gametogenesis from Pluripotent Stem Cells. Cell Stem Cell 2016, 18, 721–735. [Google Scholar] [CrossRef] [PubMed]
- Irie, N.; Weinberger, L.; Tang, W.W.C.; Kobayashi, T.; Viukov, S.; Manor, Y.S.; Dietmann, S.; Hanna, J.H.; Surani, M.A. SOX17 Is a Critical Specifier of Human Primordial Germ Cell Fate. Cell 2015, 160, 253–268. [Google Scholar] [CrossRef]
- Tang, W.W.C.; Dietmann, S.; Irie, N.; Leitch, H.G.; Floros, V.I.; Bradshaw, C.R.; Hackett, J.A.; Chinnery, P.F.; Surani, M.A. A Unique Gene Regulatory Network Resets the Human Germline Epigenome for Development. Cell 2015, 161, 1453–1467. [Google Scholar] [CrossRef] [PubMed]
- Guo, F.; Yan, L.; Guo, H.; Li, L.; Hu, B.; Zhao, Y.; Yong, J.; Hu, Y.; Wang, X.; Wei, Y.; et al. The Transcriptome and DNA Methylome Landscapes of Human Primordial Germ Cells. Cell 2015, 161, 1437–1452. [Google Scholar] [CrossRef] [PubMed]
- Yan, R.; Gu, C.; You, D.; Huang, Z.; Qian, J.; Yang, Q.; Cheng, X.; Zhang, L.; Wang, H.; Wang, P.; et al. Decoding Dynamic Epigenetic Landscapes in Human Oocytes Using Single-Cell Multi-Omics Sequencing. Cell Stem Cell 2021. [Google Scholar] [CrossRef] [PubMed]
- Bourc’his, D.; Xu, G.L.; Lin, C.S.; Bollman, B.; Bestor, T.H. Dnmt3L and the Establishment of Maternal Genomic Imprints. Science 2001, 294, 2536–2539. [Google Scholar] [CrossRef] [PubMed]
- Kaneda, M.; Hirasawa, R.; Chiba, H.; Okano, M.; Li, E.; Sasaki, H. Genetic Evidence for Dnmt3a-Dependent Imprinting during Oocyte Growth Obtained by Conditional Knockout with Zp3-Cre and Complete Exclusion of Dnmt3b by Chimera Formation. Genes Cells 2010, 15, 169–179. [Google Scholar] [CrossRef]
- Demond, H.; Kelsey, G. The Enigma of DNA Methylation in the Mammalian Oocyte. F1000Res 2020, 9, 146. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Zhu, P.; Yan, L.; Li, R.; Hu, B.; Lian, Y.; Yan, J.; Ren, X.; Lin, S.; Li, J.; et al. The DNA Methylation Landscape of Human Early Embryos. Nature 2014, 511, 606–610. [Google Scholar] [CrossRef]
- Brind’Amour, J.; Kobayashi, H.; Richard Albert, J.; Shirane, K.; Sakashita, A.; Kamio, A.; Bogutz, A.; Koike, T.; Karimi, M.M.; Lefebvre, L.; et al. LTR Retrotransposons Transcribed in Oocytes Drive Species-Specific and Heritable Changes in DNA Methylation. Nat. Commun. 2018, 9, 3331. [Google Scholar] [CrossRef]
- Gu, C.; Liu, S.; Wu, Q.; Zhang, L.; Guo, F. Integrative Single-Cell Analysis of Transcriptome, DNA Methylome and Chromatin Accessibility in Mouse Oocytes. Cell Res. 2019, 29, 110–123. [Google Scholar] [CrossRef] [PubMed]
- Gougeon, A. Dynamics of Follicular Growth in the Human: A Model from Preliminary Results. Hum. Reprod. 1986, 1, 81–87. [Google Scholar] [CrossRef] [PubMed]
- Hanna, C.W.; Demond, H.; Kelsey, G. Epigenetic Regulation in Development: Is the Mouse a Good Model for the Human? Hum. Reprod. Update 2018, 24, 556–576. [Google Scholar] [CrossRef] [PubMed]
- Seisenberger, S.; Peat, J.R.; Hore, T.A.; Santos, F.; Dean, W.; Reik, W. Reprogramming DNA Methylation in the Mammalian Life Cycle: Building and Breaking Epigenetic Barriers. Philos. Trans. R. Soc. B Biol. Sci. 2013, 368, 20110330. [Google Scholar] [CrossRef]
- Galan, A.; Diaz-Gimeno, P.; Poo, M.E.; Valbuena, D.; Sanchez, E.; Ruiz, V.; Dopazo, J.; Montaner, D.; Conesa, A.; Simon, C. Defining the Genomic Signature of Totipotency and Pluripotency during Early Human Development. PLoS ONE 2013, 8, e62135. [Google Scholar] [CrossRef]
- Yan, L.; Yang, M.; Guo, H.; Yang, L.; Wu, J.; Li, R.; Liu, P.; Lian, Y.; Zheng, X.; Yan, J.; et al. Single-Cell RNA-Seq Profiling of Human Preimplantation Embryos and Embryonic Stem Cells. Nat. Struct. Mol. Biol. 2013, 20, 1131–1139. [Google Scholar] [CrossRef] [PubMed]
- Dobson, A.T.; Raja, R.; Abeyta, M.J.; Taylor, T.; Shen, S.; Haqq, C.; Pera, R.A.R. The Unique Transcriptome through Day 3 of Human Preimplantation Development. Hum. Mol. Genet. 2004, 13, 1461–1470. [Google Scholar] [CrossRef] [PubMed]
- Hamada, H.; Okae, H.; Toh, H.; Chiba, H.; Hiura, H.; Shirane, K.; Sato, T.; Suyama, M.; Yaegashi, N.; Sasaki, H.; et al. Allele-Specific Methylome and Transcriptome Analysis Reveals Widespread Imprinting in the Human Placenta. Am. J. Hum. Genet. 2016, 99, 1045–1058. [Google Scholar] [CrossRef]
- Smith, Z.D.; Chan, M.M.; Humm, K.C.; Karnik, R.; Mekhoubad, S.; Regev, A.; Eggan, K.; Meissner, A. DNA Methylation Dynamics of the Human Preimplantation Embryo. Nature 2014, 511, 611–615. [Google Scholar] [CrossRef]
- Gerdes, P.; Richardson, S.R.; Mager, D.L.; Faulkner, G.J. Transposable Elements in the Mammalian Embryo: Pioneers Surviving through Stealth and Service. Genome Biol. 2016, 17, 100. [Google Scholar] [CrossRef]
- Tan, K.; Zhang, Z.; Miao, K.; Yu, Y.; Sui, L.; Tian, J.; An, L. Dynamic Integrated Analysis of DNA Methylation and Gene Expression Profiles in in Vivo and in Vitro Fertilized Mouse Post-Implantation Extraembryonic and Placental Tissues. Mol. Hum. Reprod. 2016, 22, 485–498. [Google Scholar] [CrossRef]
- Zeng, Y.; Chen, T. DNA Methylation Reprogramming during Mammalian Development. Genes 2019, 10, 257. [Google Scholar] [CrossRef]
- Dahlet, T.; Argüeso Lleida, A.; Al Adhami, H.; Dumas, M.; Bender, A.; Ngondo, R.P.; Tanguy, M.; Vallet, J.; Auclair, G.; Bardet, A.F.; et al. Genome-Wide Analysis in the Mouse Embryo Reveals the Importance of DNA Methylation for Transcription Integrity. Nat. Commun. 2020, 11, 3153. [Google Scholar] [CrossRef] [PubMed]
- Christodoulou, N.; Weberling, A.; Strathdee, D.; Anderson, K.I.; Timpson, P.; Zernicka-Goetz, M. Morphogenesis of Extra-Embryonic Tissues Directs the Remodelling of the Mouse Embryo at Implantation. Nat. Commun. 2019, 10, 3557. [Google Scholar] [CrossRef] [PubMed]
- Hanna, C.W.; Peñaherrera, M.S.; Saadeh, H.; Andrews, S.; McFadden, D.E.; Kelsey, G.; Robinson, W.P. Pervasive Polymorphic Imprinted Methylation in the Human Placenta. Genome Res. 2016, 26, 756–767. [Google Scholar] [CrossRef] [PubMed]
- Camprubí, C.; Iglesias-Platas, I.; Martin-Trujillo, A.; Salvador-Alarcon, C.; Rodriguez, M.A.; Barredo, D.R.; Court, F.; Monk, D. Stability of Genomic Imprinting and Gestational-Age Dynamic Methylation in Complicated Pregnancies Conceived Following Assisted Reproductive Technologies. Biol. Reprod. 2013, 89, 50. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, D.I.; Blair, J.D.; Lott, P.; Yu, H.O.K.; Hong, D.; Crary, F.; Ashwood, P.; Walker, C.; Korf, I.; Robinson, W.P.; et al. The Human Placenta Methylome. Proc. Natl. Acad. Sci. USA 2013, 110, 6037–6042. [Google Scholar] [CrossRef]
- Peters, J. The Role of Genomic Imprinting in Biology and Disease: An Expanding View. Nat. Rev. Genet. 2014, 15, 517–530. [Google Scholar] [CrossRef] [PubMed]
- Bartolomei, M.S.; Ferguson-Smith, A.C. Mammalian Genomic Imprinting. Cold Spring Harb. Perspect. Biol. 2011, 3, a00259. [Google Scholar] [CrossRef]
- Inoue, A.; Jiang, L.; Lu, F.; Suzuki, T.; Zhang, Y. Maternal H3K27me3 Controls DNA Methylation-Independent Imprinting. Nature 2017, 547, 419–424. [Google Scholar] [CrossRef]
- Chen, Z.; Zhang, Y. Maternal H3K27me3-Dependent Autosomal and X Chromosome Imprinting. Nat. Rev. Genet. 2020, 21, 555–571. [Google Scholar] [CrossRef]
- Hanna, C.W. Placental Imprinting: Emerging Mechanisms and Functions. PLoS Genet. 2020, 16, e1008709. [Google Scholar] [CrossRef]
- Abramowitz, L.K.; Bartolomei, M.S. Genomic Imprinting: Recognition and Marking of Imprinted Loci. Curr. Opin. Genet. Dev. 2012, 22, 72–78. [Google Scholar] [CrossRef]
- Li, X.; Ito, M.; Zhou, F.; Youngson, N.; Zuo, X.; Leder, P.; Ferguson-Smith, A.C. A Maternal-Zygotic Effect Gene, Zfp57, Maintains Both Maternal and Paternal Imprints. Dev. Cell 2008, 15, 547–557. [Google Scholar] [CrossRef]
- Quenneville, S.; Verde, G.; Corsinotti, A.; Kapopoulou, A.; Jakobsson, J.; Offner, S.; Baglivo, I.; Pedone, P.V.; Grimaldi, G.; Riccio, A.; et al. In Embryonic Stem Cells, ZFP57/KAP1 Recognize a Methylated Hexanucleotide to Affect Chromatin and DNA Methylation of Imprinting Control Regions. Mol. Cell 2011, 44, 361–372. [Google Scholar] [CrossRef] [PubMed]
- Monteagudo-Sánchez, A.; Hernandez Mora, J.R.; Simon, C.; Burton, A.; Tenorio, J.; Lapunzina, P.; Clark, S.; Esteller, M.; Kelsey, G.; López-Siguero, J.P.; et al. The Role of ZFP57 and Additional KRAB-Zinc Finger Proteins in the Maintenance of Human Imprinted Methylation and Multi-Locus Imprinting Disturbances. Nucleic Acids Res. 2020, 48, 11394–11407. [Google Scholar] [CrossRef]
- Mackay, D.J.G.; Callaway, J.L.A.; Marks, S.M.; White, H.E.; Acerini, C.L.; Boonen, S.E.; Dayanikli, P.; Firth, H.V.; Goodship, J.A.; Haemers, A.P.; et al. Hypomethylation of Multiple Imprinted Loci in Individuals with Transient Neonatal Diabetes Is Associated with Mutations in ZFP57. Nat. Genet. 2008, 40, 949–951. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, N.; Coluccio, A.; Thorball, C.W.; Planet, E.; Shi, H.; Offner, S.; Turelli, P.; Imbeault, M.; Ferguson-Smith, A.C.; Trono, D. ZNF445 Is a Primary Regulator of Genomic Imprinting. Genes Dev. 2019, 33, 49–54. [Google Scholar] [CrossRef] [PubMed]
- Kagami, M.; Hara-Isono, K.; Matsubara, K.; Nakabayashi, K.; Narumi, S.; Fukami, M.; Ohkubo, Y.; Saitsu, H.; Takada, S.; Ogata, T. ZNF445: A homozygous truncating variant in a patient with Temple syndrome and multilocus imprinting disturbance. Clin. Epigenetics 2021, 13, 119. [Google Scholar] [CrossRef]
- Court, F.; Tayama, C.; Romanelli, V.; Martin-Trujillo, A.; Iglesias-Platas, I.; Okamura, K.; Sugahara, N.; Simón, C.; Moore, H.; Harness, J.V.; et al. Genome-Wide Parent-of-Origin DNA Methylation Analysis Reveals the Intricacies of Human Imprinting and Suggests a Germline Methylation-Independent Mechanism of Establishment. Genome Res. 2014, 24, 554–569. [Google Scholar] [CrossRef]
- Hanna, C.W.; Kelsey, G. The Specification of Imprints in Mammals. Heredity 2014, 113, 176–183. [Google Scholar] [CrossRef]
- Sanchez-Delgado, M.; Court, F.; Vidal, E.; Medrano, J.; Monteagudo-Sánchez, A.; Martin-Trujillo, A.; Tayama, C.; Iglesias-Platas, I.; Kondova, I.; Bontrop, R.; et al. Human Oocyte-Derived Methylation Differences Persist in the Placenta Revealing Widespread Transient Imprinting. PLoS Genet. 2016, 12, e1006427. [Google Scholar] [CrossRef] [PubMed]
- Yuen, R.K.; Jiang, R.; Peñaherrera, M.S.; McFadden, D.E.; Robinson, W.P. Genome-Wide Mapping of Imprinted Differentially Methylated Regions by DNA Methylation Profiling of Human Placentas from Triploidies. Epigenetics Chromatin 2011, 4, 10. [Google Scholar] [CrossRef]
- Barbaux, S.; Gascoin-Lachambre, G.; Buffat, C.; Monnier, P.; Mondon, F.; Tonanny, M.-B.; Pinard, A.; Auer, J.; Bessières, B.; Barlier, A.; et al. A Genome-Wide Approach Reveals Novel Imprinted Genes Expressed in the Human Placenta. Epigenetics 2012, 7, 1079–1090. [Google Scholar] [CrossRef] [PubMed]
- Hemberger, M.; Hanna, C.W.; Dean, W. Mechanisms of Early Placental Development in Mouse and Humans. Nat. Rev. Genet. 2020, 21, 27–43. [Google Scholar] [CrossRef] [PubMed]
- Elbracht, M.; Mackay, D.; Begemann, M.; Kagan, K.O.; Eggermann, T. Disturbed Genomic Imprinting and Its Relevance for Human Reproduction: Causes and Clinical Consequences. Hum. Reprod. Update 2020, 26, 197–213. [Google Scholar] [CrossRef] [PubMed]
- Moein-Vaziri, N.; Fallahi, J.; Namavar-Jahromi, B.; Fardaei, M.; Momtahan, M.; Anvar, Z. Clinical and Genetic-Epignetic Aspects of Recurrent Hydatidiform Mole: A Review of Literature. Taiwan. J. Obstet. Gynecol. 2018, 57, 1–6. [Google Scholar] [CrossRef]
- Surani, M.A.; Barton, S.C.; Norris, M.L. Development of Reconstituted Mouse Eggs Suggests Imprinting of the Genome during Gametogenesis. Nature 1984, 308, 548–550. [Google Scholar] [CrossRef]
- McGrath, J.; Solter, D. Completion of Mouse Embryogenesis Requires Both the Maternal and Paternal Genomes. Cell 1984, 37, 179–183. [Google Scholar] [CrossRef]
- Li, Z.-K.; Wang, L.-Y.; Wang, L.-B.; Feng, G.-H.; Yuan, X.-W.; Liu, C.; Xu, K.; Li, Y.-H.; Wan, H.-F.; Zhang, Y.; et al. Generation of Bimaternal and Bipaternal Mice from Hypomethylated Haploid ESCs with Imprinting Region Deletions. Cell Stem Cell 2018, 23, 665–676.e4. [Google Scholar] [CrossRef]
- Qian, J.; Nguyen, N.M.P.; Rezaei, M.; Huang, B.; Tao, Y.; Zhang, X.; Cheng, Q.; Yang, H.; Asangla, A.; Majewski, J.; et al. Biallelic PADI6 Variants Linking Infertility, Miscarriages, and Hydatidiform Moles. Eur. J. Hum. Genet. 2018, 26, 1007–1013. [Google Scholar] [CrossRef]
- Judson, H.; Hayward, B.E.; Sheridan, E.; Bonthron, D.T. A Global Disorder of Imprinting in the Human Female Germ Line. Nature 2002, 416, 539–542. [Google Scholar] [CrossRef]
- El-Maarri, O.; Seoud, M.; Coullin, P.; Herbiniaux, U.; Oldenburg, J.; Rouleau, G.; Slim, R. Maternal Alleles Acquiring Paternal Methylation Patterns in Biparental Complete Hydatidiform Moles. Hum. Mol. Genet. 2003, 12, 1405–1413. [Google Scholar] [CrossRef][Green Version]
- Kou, Y.C.; Shao, L.; Peng, H.H.; Rosetta, R.; del Gaudio, D.; Wagner, A.F.; Al-Hussaini, T.K.; Van den Veyver, I.B. A Recurrent Intragenic Genomic Duplication, Other Novel Mutations in NLRP7 and Imprinting Defects in Recurrent Biparental Hydatidiform Moles. Mol. Hum. Reprod. 2008, 14, 33–40. [Google Scholar] [CrossRef][Green Version]
- Sanchez-Delgado, M.; Martin-Trujillo, A.; Tayama, C.; Vidal, E.; Esteller, M.; Iglesias-Platas, I.; Deo, N.; Barney, O.; Maclean, K.; Hata, K.; et al. Absence of Maternal Methylation in Biparental Hydatidiform Moles from Women with NLRP7 Maternal-Effect Mutations Reveals Widespread Placenta-Specific Imprinting. PLoS Genet. 2015, 11, e1005644. [Google Scholar] [CrossRef]
- Zhu, K.; Yan, L.; Zhang, X.; Lu, X.; Wang, T.; Yan, J.; Liu, X.; Qiao, J.; Li, L. Identification of a Human Subcortical Maternal Complex. Mol. Hum. Reprod. 2015, 21, 320–329. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Baibakov, B.; Dean, J. A Subcortical Maternal Complex Essential for Preimplantation Mouse Embryogenesis. Dev. Cell 2008, 15, 416–425. [Google Scholar] [CrossRef]
- Lu, X.; Gao, Z.; Qin, D.; Li, L. A Maternal Functional Module in the Mammalian Oocyte-To-Embryo Transition. Trends Mol. Med. 2017, 23, 1014–1023. [Google Scholar] [CrossRef] [PubMed]
- Bebbere, D.; Masala, L.; Albertini, D.F.; Ledda, S. The Subcortical Maternal Complex: Multiple Functions for One Biological Structure? J. Assist. Reprod. Genet. 2016, 33, 1431–1438. [Google Scholar] [CrossRef]
- Monk, D.; Sanchez-Delgado, M.; Fisher, R. NLRPs, the Subcortical Maternal Complex and Genomic Imprinting. Reproduction 2017, 154, R161–R170. [Google Scholar] [CrossRef] [PubMed]
- Akoury, E.; Zhang, L.; Ao, A.; Slim, R. NLRP7 and KHDC3L, the Two Maternal-Effect Proteins Responsible for Recurrent Hydatidiform Moles, Co-Localize to the Oocyte Cytoskeleton. Hum. Reprod. 2015, 30, 159–169. [Google Scholar] [CrossRef]
- Qian, J.; Cheng, Q.; Murdoch, S.; Xu, C.; Jin, F.; Chebaro, W.; Zhang, X.; Gao, H.; Zhu, Y.; Slim, R.; et al. The Genetics of Recurrent Hydatidiform Moles in China: Correlations between NLRP7 Mutations, Molar Genotypes and Reproductive Outcomes. Mol. Hum. Reprod. 2011, 17, 612–619. [Google Scholar] [CrossRef] [PubMed]
- Meyer, E.; Lim, D.; Pasha, S.; Tee, L.J.; Rahman, F.; Yates, J.R.W.; Woods, C.G.; Reik, W.; Maher, E.R. Germline Mutation in NLRP2 (NALP2) in a Familial Imprinting Disorder (Beckwith-Wiedemann Syndrome). PLoS Genet. 2009, 5, e1000423. [Google Scholar] [CrossRef]
- Messerschmidt, D.M. Should I Stay or Should I Go: Protection and Maintenance of DNA Methylation at Imprinted Genes. Epigenetics 2012, 7, 969–975. [Google Scholar] [CrossRef] [PubMed]
- Mahadevan, S.; Sathappan, V.; Utama, B.; Lorenzo, I.; Kaskar, K.; Van den Veyver, I.B. Maternally Expressed NLRP2 Links the Subcortical Maternal Complex (SCMC) to Fertility, Embryogenesis and Epigenetic Reprogramming. Sci. Rep. 2017, 7, 44667. [Google Scholar] [CrossRef]
- Tian, X.; Pascal, G.; Monget, P. Evolution and Functional Divergence of NLRP Genes in Mammalian Reproductive Systems. BMC Evol. Biol. 2009, 9, 202. [Google Scholar] [CrossRef]
- Zheng, P.; Dean, J. Role of Filia, a Maternal Effect Gene, in Maintaining Euploidy during Cleavage-Stage Mouse Embryogenesis. Proc. Natl. Acad. Sci. USA 2009, 106, 7473–7478. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.-J.; Yi, Z.; Gao, Z.; Qin, D.; Zhai, Y.; Chen, X.; Ou-Yang, Y.; Wang, Z.-B.; Zheng, P.; Zhu, M.-S.; et al. The Subcortical Maternal Complex Controls Symmetric Division of Mouse Zygotes by Regulating F-Actin Dynamics. Nat. Commun. 2014, 5, 4887. [Google Scholar] [CrossRef]
- Tashiro, F.; Kanai-Azuma, M.; Miyazaki, S.; Kato, M.; Tanaka, T.; Toyoda, S.; Yamato, E.; Kawakami, H.; Miyazaki, T.; Miyazaki, J.-I. Maternal-Effect Gene Ces5/Ooep/Moep19/Floped Is Essential for Oocyte Cytoplasmic Lattice Formation and Embryonic Development at the Maternal-Zygotic Stage Transition. Genes Cells 2010, 15, 813–828. [Google Scholar] [CrossRef]
- Kaneda, M.; Okano, M.; Hata, K.; Sado, T.; Tsujimoto, N.; Li, E.; Sasaki, H. Essential Role for de Novo DNA Methyltransferase Dnmt3a in Paternal and Maternal Imprinting. Nature 2004, 429, 900–903. [Google Scholar] [CrossRef] [PubMed]
- Alazami, A.M.; Awad, S.M.; Coskun, S.; Al-Hassan, S.; Hijazi, H.; Abdulwahab, F.M.; Poizat, C.; Alkuraya, F.S. TLE6 Mutation Causes the Earliest Known Human Embryonic Lethality. Genome Biol. 2015, 16, 240. [Google Scholar] [CrossRef]
- Xu, Y.; Shi, Y.; Fu, J.; Yu, M.; Feng, R.; Sang, Q.; Liang, B.; Chen, B.; Qu, R.; Li, B.; et al. Mutations in PADI6 Cause Female Infertility Characterized by Early Embryonic Arrest. Am. J. Hum. Genet. 2016, 99, 744–752. [Google Scholar] [CrossRef]
- Maddirevula, S.; Coskun, S.; Alhassan, S.; Elnour, A.; Alsaif, H.S.; Ibrahim, N.; Abdulwahab, F.; Arold, S.T.; Alkuraya, F.S. Female Infertility Caused by Mutations in the Oocyte-Specific Translational Repressor PATL2. Am. J. Hum. Genet. 2017, 101, 603–608. [Google Scholar] [CrossRef]
- Wang, X.; Song, D.; Mykytenko, D.; Kuang, Y.; Lv, Q.; Li, B.; Chen, B.; Mao, X.; Xu, Y.; Zukin, V.; et al. Novel Mutations in Genes Encoding Subcortical Maternal Complex Proteins May Cause Human Embryonic Developmental Arrest. Reprod. Biomed. Online 2018, 36, 698–704. [Google Scholar] [CrossRef]
- Mu, J.; Wang, W.; Chen, B.; Wu, L.; Li, B.; Mao, X.; Zhang, Z.; Fu, J.; Kuang, Y.; Sun, X.; et al. Mutations in NLRP2 and NLRP5 Cause Female Infertility Characterised by Early Embryonic Arrest. J. Med. Genet. 2019, 56, 471–480. [Google Scholar] [CrossRef]
- Deveault, C.; Qian, J.H.; Chebaro, W.; Ao, A.; Gilbert, L.; Mehio, A.; Khan, R.; Tan, S.L.; Wischmeijer, A.; Coullin, P.; et al. NLRP7 Mutations in Women with Diploid Androgenetic and Triploid Moles: A Proposed Mechanism for Mole Formation. Hum. Mol. Genet. 2009, 18, 888–897. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hayward, B.E.; De Vos, M.; Talati, N.; Abdollahi, M.R.; Taylor, G.R.; Meyer, E.; Williams, D.; Maher, E.R.; Setna, F.; Nazir, K.; et al. Genetic and Epigenetic Analysis of Recurrent Hydatidiform Mole. Hum. Mutat. 2009, 30, E629–E639. [Google Scholar] [CrossRef]
- Xu, Y.; Qian, Y.; Liu, Y.; Wang, Q.; Wang, R.; Zhou, Y.; Zhang, C.; Pang, Z.; Ye, H.; Xue, S.; et al. A Novel Homozygous Variant in NLRP5 Is Associate with Human Early Embryonic Arrest in a Consanguineous Chinese Family. Clin. Genet. 2020, 98, 69–73. [Google Scholar] [CrossRef]
- Begemann, M.; Spengler, S.; Kanber, D.; Haake, A.; Baudis, M.; Leisten, I.; Binder, G.; Markus, S.; Rupprecht, T.; Segerer, H.; et al. Silver-Russell Patients Showing a Broad Range of ICR1 and ICR2 Hypomethylation in Different Tissues. Clin. Genet. 2011, 80, 83–88. [Google Scholar] [CrossRef]
- Bens, S.; Kolarova, J.; Beygo, J.; Buiting, K.; Caliebe, A.; Eggermann, T.; Gillessen-Kaesbach, G.; Prawitt, D.; Thiele-Schmitz, S.; Begemann, M.; et al. Phenotypic Spectrum and Extent of DNA Methylation Defects Associated with Multilocus Imprinting Disturbances. Epigenomics 2016, 8, 801–816. [Google Scholar] [CrossRef]
- Cubellis, M.V.; Pignata, L.; Verma, A.; Sparago, A.; Del Prete, R.; Monticelli, M.; Calzari, L.; Antona, V.; Melis, D.; Tenconi, R.; et al. Loss-of-Function Maternal-Effect Mutations of PADI6 Are Associated with Familial and Sporadic Beckwith-Wiedemann Syndrome with Multi-Locus Imprinting Disturbance. Clin. Epigenetics 2020, 12, 139. [Google Scholar] [CrossRef]
| Gene | Family | hg19 Position | GenBank | cDNA Mutation | Protein Mutation | Mutation Effect | gnomAD_Exomeall MAF | gnomAD_Genomeall MAF | SIFT | Polyphen | Inheritance | Domain/Exon | Pregnancy Outcomes | Country | Ref |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NALP7 | MoLb1 | Chr19: 55452298 | NM_206828 | IVS3+1G>A | 2 splicing isoforms: -inclusion of the first 4 bp of intron 3 between exons 3 and 4, addition of two aa followed by a stop codon -exclusion of exon 3 | Splicing mutation (Splice donor) | 0.00E + 00 | 0.00E + 00 | NA | NA | Autosomal recessive (Homozygous) | Intron 3 | Recurrent hydatidiform moles | Lebanon | [25] |
| MoPa61 | Chr19: 55445856 | IVS7+1G>A | inclusion of the entire intron 7 | Splicing mutation | 5.17E-05 | NA | NA | NA | Autosomal recessive (Homozygous) | Intron 7 | Complete hydatidiform mole, spontaneous abortion (7–20 weeks) | Pakistan | |||
| MoGe2 | Chr19: 55449464 | 2077C>T | p.Arg693Trp | Missense mutation | 2.74E-04 | 6.69E-04 | NA | NA | Autosomal recessive (Homozygous) | Exon 5 | Complete hydatidiform mole | Germany | |||
| MoIn68 | Chr19: 55449463 | 2078G>C | p.Arg693Pro | Missense mutation | 4.77E-05 | NA | tolerated (0.07) | benign (0.056) | Autosomal recessive (Homozygous) | Exon 5 | Complete hydatidiform mole | India | |||
| MoIn69-2 | Chr19: 55441939 | c.2738A>G | p.Asn913Ser | Missense mutation | 1.35E-04 | 7.33E-04 | deleterious (0) | probably_damaging (0.991) | Autosomal recessive (Compound heterozygous) | Exon 5 | Complete hydatidiform mole and invasive mole | India | |||
| Chr19: 55449463 | c.2078G>C | p.Arg693Pro | Missense mutation | 4.77E-05 | NA | tolerated (0.07) | benign (0.056) | Exon 9 | |||||||
| NLRP7 | Family 6 | Chr19: 55447768 | NM_001127255.1 | c.2161C>T | p.Arg721Trp | Missense mutations | 5.97E-05 | NA | NA | NA | Compound Heterozygous | Exon 7 | BWS–MLID | Germany | [28] |
| Chr19: 55445006 | c. 2573T>C | p. Ile858Thr | 7.16E-05 | 6.37E-05 | deleterious (0) | benign (0.351) | Exon 8 | ||||||||
| Family 7 | Chr19: 55451438 | c.749T>G | p.Phe250Cys | Missense mutations | 4.57E-04 | 4.14E-04 | deleterious (0) | possibly_ damaging (0.88) | Compound Heterozygous (Mother) | NACHT domain | BWS and TNDM | ||||
| Chr19: 55451083 | c. 1104T>G | p.Ile368Met | 4.84E-04 | 5.49E-04 | NA | NA | Heterozygous in Proband | Exon 4 | |||||||
| Family 8 | Chr19: 55447773 | NM_206828.2 | c.2156C>T | p.Ala719Val | Missense mutation | 1.05E-03 | 1.05E-03 | deleterious (0.01) | probably_damaging (0.963) | Heterozygous (Mother and Proband) | Exon 6 | SRS | UK | [28] | |
| China | [91] | ||||||||||||||
| Italy | [105] | ||||||||||||||
| Patient 1 and 2 | Chr19: 55449463 | NM_001127255.1 | c. 2078G>C | p.Arg693Pro | Missense mutation | 4.77E-05 | NA | tolerated (0.07) | benign (0.056) | Autosomal recessive | Exon 5 | Complete hydatidiform moles | UK | [84] | |
| Patient 3 | Chr19: 55449184_55454887 del | c.-39-1769_2129+ 228del | Deletion of exons 2-5 | NA | NA | NA | NA | Autosomal recessive | 5′UTR | ||||||
| Patient 4 | Chr19: 55449523 | c.2018C>G | p.Ser673Ter | Nonsense mutation | 3.98E-06 | NA | NA | NA | Compound Heterozygous | Exon 5 | |||||
| Chr19: 55447768 | c.2161C>T | p.Arg721Trp | Missense mutation | 5.97E-05 | NA | NA | NA | Exon 6 | |||||||
| Family E | Chr19: 55451235_55451248 | NM_206828.3 | c.939_952 dup 14 | p.Tyr318Cys fsTer7 | Frameshift mutation | 2.39E-05 | 1.27E-04 | NA | NA | Compound Heterozygous | Exon 4 | Familial biparental hydatidiform mole | UK | [106] | |
| Chr19: 55449511 | c.2030delT | p.Leu677Pro fsTer6 | Mutations | NA | NA | NA | NA | Exon 5 | |||||||
| Family N | Chr19: 55449523 | c.2018C>G | p.Ser673Ter | Nonsense mutation | 3.98E-06 | NA | NA | NA | Autosomal recessive | Exon 5 | Pakistan | ||||
| Family J | chr19: 55452305 | c.346A>T | p.Lys116Ter | Nonsense mutation | NA | NA | NA | NA | Autosomal recessive | Exon 3 | Pakistan | ||||
| Family K | Chr19: 55449463 | c.2078G>C | p.Arg693Pro | Missense mutation | 4.77E-05 | NA | tolerated (0.07) | benign (0.056) | Autosomal recessive | Exon 5 | Pakistan | ||||
| Family L | Chr19: 55451049 | c.1138G>C | p.Gly380Arg | Missense mutation | 6.66E-04 | NA | NA | NA | Heterozygous | Exon 4 | Pakistan | ||||
| Singleton 1 | chr19: 55445994 | c.2334G>A | p.Trp778Ter | Nonsense mutation | NA | NA | tolerated (0.16) | probably_damaging (0.95) | Autosomal recessive | Exon 7 | Molar pregnancy | Pakistan | |||
| Singleton 2 | Chr19: 55450731 | c.1456dupG | p.Glu486Gly fsTer42 | Frameshift mutation | NA | NA | NA | NA | Autosomal recessive | Exon 4 | Punjabi | ||||
| Singleton 4 | Chr19: 55450994 | c.1193T>G | p.Leu398Arg | Missense mutation | 3.48E-05 | NA | tolerated (0.16) | probably_damaging (0.95) | Autosomal recessive | Exon 4 | Pakistan | ||||
| Singleton 5 | Chr19: 55449463 | c.2078G>C | p.Arg693Pro | Missense mutation | 4.77E-05 | NA | tolerated (0.07) | benign (0.056) | Autosomal recessive | Exon 5 | Pakistan | ||||
| Singleton 6 | Chr19: 55452802 | c.277+1G>C | Splicing mutation | NA | NA | NA | NA | Autosomal recessive | Intron 2 | Pakistan | |||||
| Singleton 7 | Chr19: 55450994 | c.1193T>G | p.Leu398Arg | Missense mutation | 3.48E-05 | NA | tolerated (0.16) | probably_damaging (0.95) | Autosomal recessive | Exon 4 | Pakistan | ||||
| MoCh76 | Chr19: 55452356 | NM_206828.2 | c.295G>T | p.Glu99Ter | Nonsense mutation | NA | NA | NA | NA | Compound Heterozygous | Exon 3 | BiCHM | China | [91] | |
| Chr19: 55449571 | c. 1970A>T | p. Asp657Val | Missense mutation | NA | NA | NA | NA | Exon 5 | |||||||
| Ch29 | Chr19: 55447764 | c.2165A>G | p.Asp722Gly | Missense mutation | 3.98E-06 | NA | deleterious (0.05) | possibly_ damaging (0.574) | Autosomal recessive | Exon 6 | BiCHM | ||||
| Ch77 | Chr19: 55450893 | c.1294C>T | p.Arg432Ter | Nonsense mutations | 3.61E-05 | 3.19E-05 | NA | NA | Compound Heterozygous | Exon 4 | CHM | ||||
| Chr19: 55445108 | c.2471+1G>A | p.Leu825Ter | NA | NA | NA | NA | Exon 7 | ||||||||
| Ch101 | Chr19: 55449440 | c. 2101C>T | p.Arg701Cys | Missense mutation | 1.99E-05 | NA | tolerated (1) | benign (0.018) | Compound Heterozygous | Exon 5 | BiCHM | ||||
| Chr19: 55449463 | c.2078G>A | p.Arg693Gln | 7.95E-06 | 3.19E-05 | tolerated (0.07) | benign (0.056) | |||||||||
| MoCh195 | Chr19: 32436314_55448111 del1218 | c.2130-312_2300+ 1737del1218 | NA | NA | NA | NA | Exon 6 | CHM | |||||||
| MoCh200 | Chr19: 55450487_55450562 del76 | c.1625_1700 del76 | p.Met542Thr fsTer2 | Frameshift mutation | NA | NA | NA | NA | Compound Heterozygous | Exon 4 | HM | ||||
| Chr19: 55445108 | c. 2471+1G>A | p.Leu825Ter | Nonsense mutation | NA | NA | NA | NA | Exon 7 | |||||||
| MoCh293 | Chr19: 55450893 | c.1294C>T | p.Arg432Ter | Nonsense mutation | 3.61E-05 | 3.19E-05 | NA | NA | Compound Heterozygous | Exon 4 | HM | ||||
| Chr19: 55447773 | c.2156C>T | p.Ala719Val | Missense mutation | 1.05E-03 | 1.05E-03 | deleterious (0.01) | probably_damaging (0.963) | Exon 6 | |||||||
| MoCh73 | Chr19: 55451050 | c.1137G>C | p.Lys379Asn | Missense mutation | 5.01E-03 | 6.08E-03 | NA | NA | Heterozygous | Exon 4 | CHM | ||||
| MoCh71 | Chr19: 55452829 | c.251G>A | p.Cys84Tyr | Missense mutation | 4.53E-04 | 3.20E-04 | tolerated (0.05) | benign (0.079) | Heterozygous | Exon 2 | AnCHM | ||||
| MoCh193 | Chr19: 55451050 | c.1137G>C | p.Lys379Asn | Missense mutation | 5.01E-03 | 6.08E-03 | NA | NA | Heterozygous | Exon 4 | HM | ||||
| MoCh190 | Chr19: 55445860 | c.2468T>A | p.Leu823Ter | Nonsense mutation | NA | NA | NA | NA | Heterozygous | Exon 7 | AnCHM | ||||
| MoCh71 | Chr19: 55452829 | NM_001127255.1 | c.251G>A | p.Cys84Tyr | Missense Mutation | 4.53E-04 | 3.20E-04 | tolerated (0.05) | benign (0.079) | Heterozygous | Exon 2 | CHM, PHM (with no family history of moles) | China | [105] | |
| MoIt96 | Chr19: 55451720 | c.467G>A | p.Arg156Gln | Missense mutation | 7.25E-03 | 8.09E-03 | tolerated (0.23) | benign (0.125) | Heterozygous | Exon 4 | HM (with no family history of moles) | Italia | |||
| MoCh73 | Chr19: 55451050 | c.1137G>C | p.Lys379Asn | Missense mutation | 5.01E-03 | 6.08E-03 | NA | NA | Exon 4 | China | |||||
| MoCa57 | Chr19: 55450991 | c.1196G>A | p.Cys399Tyr | Missense mutations | 4.72E-04 | 2.87E-04 | deleterious (0) | probably_damaging (1) | Compound Heterozygous | Exon 4 | CHM/IM (with no family history of moles) | Morocco and Algeria | |||
| Chr19: 55450727 | c.1460G>A | p.Gly487Glu | 5.31E-02 | 1.29E-01 | tolerated (0.12) | benign (0.094) | Exon 4 | ||||||||
| MoCa88 | Chr19: 55450655 | c.1532A>G | p.Lys511Arg | Missense mutation | 1.33E-02 | 2.91E-02 | deleterious (0.01) | possibly_ damaging (0.701) | Heterozygous | Exon 4 | Recurrent spontaneous abortions, 2 twins Hashimoto disease (with no family history of moles) | Morocco and UK | |||
| Ch101 | Chr19: 55449440 | c.2101C>T | p.Arg701Cys | Missense mutations | 1.99E-05 | NA | tolerated (1) | benign (0.018) | Compound Heterozygous | Exon 5 | CHM (with no family history of moles) | China | |||
| Chr19: 55449463 | c.2078G>A | p.Arg693Gln | 7.95E-06 | 3.19E-05 | tolerated (0.07) | benign (0.056) | |||||||||
| MoCa94 | Chr19: 55447773 | c.2156C>T | p.Ala719Val | Missense mutation | 1.05E-03 | 1.05E-03 | deleterious (0.01) | probably_damaging (0.963) | Heterozygous | Exon 6 | PHM (with no family history of moles) | Italy | |||
| Ch29 | Chr19: 55447764 | c.2165A>G | p.Asp722Gly | Missense mutation | 3.98E-06 | NA | deleterious (0.05) | possibly_ damaging (0.574) | Autosomal recessive (Homozygous) | Exon 6 | PHM, BiCHM, CHM (with no family history of moles) | China | |||
| MoUs99 | Chr19: 55447681 | c.2248C>G | p.Leu750Val | Missense mutation | 5.29E-04 | 9.56E-05 | NA | NA | Exon 5 | PHM, CHM, HM (Familial recurrent HMs) | Mexico | ||||
| Ch77 | Chr19: 55450893 | c.1294C>T | p.Arg432Ter | Nonsense mutations | 3.61E-05 | 3.19E-05 | NA | NA | Compound Heterozygous | Exon 4 | CHM | China | |||
| Chr19: 55445108 | c.2471+1 G>A | p.Leu825Ter | NA | NA | NA | NA | Intron 7 | ||||||||
| MoFr101 | Chr19: 55439063 | c.2891T>C | p.Leu964Pro | Missense mutation | NA | NA | deleterious (0) | probably_damaging (1) | Autosomal recessive (Homozygous) | Exon 10 | PHM | France | |||
| NLRP5 | Family 1 | Chr19: 56544020 | NM_153447.4 | c.2320T>C | p.Cys774Arg | Missense mutation | 4.02E-06 | NA | deleterious (0) | probably_damaging (0.997) | Compound Heterozygous (mother and 2 probands) | LRR domain | Proband 1 with SRS-MLID (heterozygous c.2320T > C) | UK | [27] |
| Chr19: 56539263 | c.1664G>T | p.Gly555Val | Missense mutation | NA | NA | deleterious (0) | probably_damaging (0.944) | NACHT domain | Proband 2 with BWS-MLID (heterozygous c.1664G > T) | ||||||
| Family 2 | Chr19: 56544053 | c.2353C>T | p.Gln785Ter | Nonsense mutation | 8.43E-05 | NA | NA | NA | Compound Heterozygous in the mother and proband 1. c.2840T > C not inherited by either affected offspring | LRR domain | Proband 1 with BWS–MLID. Proband 2 with a clinically non-specific autism and obesity–MLID | UK | |||
| Chr19: 56552341 | c.2840T>C | p.Leu947Pro | Missense mutation | 2.61E-04 | 2.23E-04 | deleterious (0) | probably_damaging (0.996) | ||||||||
| Family 3 | Chr19: 56515174 | c.155T>C | p.Met52Thr | Missense mutation | 8.02E-06 | NA | tolerated (0.07) | benign (0.007) | Compound Heterozygous | DAPIN domain (N-terminal effector) | Proband with BWS–MLID | UK | |||
| Chr19: 56515245 | c.226G>C | p.Glu76Gln | 8.02E-06 | NA | deleterious (0) | probably_damaging (0.999) | |||||||||
| Family 4 | Chr19: 56538755 | c.1156_1158 dupCCT | p.386dupPro | Missense mutation | 4.03E-06 | NA | NA | NA | Heterozygous in the mother but not inherited in either twin | NACHT domain | Proband (one of discordant monozygotic pair) was SRS–MLID | Germany | |||
| Family 5 | Chr19: 56539298 | c.1699A>G | p.Met567Val | Missense mutation | 4.42E-05 | NA | tolerated (0.11) | benign (0.017) | NACHT domain | MLID, presenting with atypical | UK | ||||
| clinical features of BWS and Prader–Willi syndrome | |||||||||||||||
| Family 6 | Chr19: 56515311 | c.292C>T | p.Gln98Ter | Nonsense mutation | NA | NA | NA | NA | Compound Heterozygous | Pyrin | Recurrent early embryonic arrest | China | [104] | ||
| Chr19: 56539680 | c.2081C>T | p.Thr694Ile | Missense mutation | NA | NA | deleterious (0) | probably_damaging (0.973) | LRR | |||||||
| Family 7 | Chr19: 56538465 | c.866G>A | p.Gly289Glu | Missense mutation | NA | NA | deleterious (0) | probably_damaging (1) | Compound Heterozygous | NACHT | |||||
| Chr19: 56569626 | c.3320C>T | p.Thr1107Ile | Missense mutation | NA | 3.19E-05 | deleterious (0) | probably_damaging (0.993) | LRR | |||||||
| Family 1 | Chr19: 56538660 | c.1061C>T | p.Pro354Leu | Missense mutation | 1.21E-05 | 3.19E-05 | deleterious (0.03) | probably_damaging (0.999) | Autosomal recessive | NACHT | Recurrent early embryonic arrest | China | [107] | ||
| NLRP2 | Family 1 | Chr19: 55494543 | NM_017852.4 | c.1479_1480 delAG | p.Arg493Ser fsTer32 | Frameshift mutation | 7.56E-05 | NA | NA | NA | Autosomal recessive (Homozygous Mother), Heterozygous in both probands | LRR domain | MLID | Germany | [28] |
| Family | Autosomal recessive consanguineous family | Proband with BWS–MLID | Pakistan | [92] | |||||||||||
| Family 2 | Chr19: 55497553 | c.2237delA | p.Asn746Thr fsTer4 | Frameshift mutation | 3.98E-06 | NA | NA | NA | Heterozygous mother and proband | Exon 8 | Proband with SRS | Germany | Family previously reported in [108] and [109] | ||
| Family 3 | Chr19: 55505788 | c.2860_2861 delTG | p.Cys954Gln fsTer18 | Frameshift mutation | NA | NA | NA | NA | Heterozygous mother | Exon 11/LRR domain | Proband 47, XXY, Symmetrical growth restriction and developmental delay | Germany | [28] | ||
| Family 4 | Chr19: 55485901 | c.314C>T | p.Pro105Leu | Missense mutation | 2.79E-05 | NA | tolerated (0.15) | possibly_ damaging (0.604) | Heterozygous mother | Exon 3 | TNDM | [28] | |||
| Family 5 | Chr19: 55494951 | c.1885T>C | p.Ser629Pro | Missense mutations | 1.01E-03 | 1.12E-03 | deleterious (0) | probably_damaging (0.959) | Compound Heterozygous (Mother and Proband) | Exon 6 | SRS | UK | [28] | ||
| Chr19: 55501424 | c. 2401G>A | p. Ala801Thr | 9.17E-03 | 1.27E-02 | tolerated (0.51) | benign (0.097) | Exon 9 | ||||||||
| Family 1 | Chr19: 55495027 | NM_017852.5 | c.1961C>A | p.Ser654Ter | Nonsense mutation | NA | NA | NA | NA | Autosomal recessive | Exon 6 | MLID | China | [104] | |
| Family 2 | Chr19: 55493839 | c.773T>C | p.Phe258Ser | Missense mutation | 3.98E-06 | NA | deleterious (0) | probably_damaging (0.993) | Compound Heterozygous | NACHT | |||||
| Chr19: 55497571 | c.2254C>T | p.Arg752Ter | Nonsense mutation | 3.98E-06 | NA | NA | NA | Exon 9 | |||||||
| Family 3 | Chr19: 55493591 | c.525G>C | p.Trp175Cys | Missense mutation | NA | NA | tolerated (0.06) | probably_damaging (0.979) | Compound Heterozygous | Exon 6 | |||||
| Chr19: 55501876 | c.2544A>T | p.Glu848Asp | Missense mutation | NA | NA | deleterious (0.01) | probably_damaging (0.994) | LRR | |||||||
| Family 4 | Chr19: 55493728 | c.662C>T | p.Thr221Met | Missense mutation | 8.85E-02 | 9.05E-02 | deleterious (0.04) | probably_damaging (0.989) | Compound Heterozygous | NACHT | |||||
| Chr19: 55494913 | c.1847A>T | p.Glu616Val | Missense mutation | 7.96E-06 | NA | deleterious (0.04) | benign (0.405) | Exon8 | |||||||
| Family 5 | Chr19: 55493728 | c.662C>T | p.Thr221Met | Missense mutation | 8.85E-02 | 9.05E-02 | deleterious (0.04) | probably_damaging (0.989) | Compound Heterozygous | NACHT | |||||
| Chr19: 55494534 | c.1469C>T | p.Arg490Cys | Missense mutation | 1.28E-04 | 3.20E-05 | deleterious (0.01) | benign (0.03) | Exon7 | |||||||
| KHDC3L | Family L | Chr6: 74072455 | NM_001017361.3 | c.3G>T | p.Met1Ile next available downstream ATG codon lies at residue 14 | Loss of start codon | 3.98E-06 | NA | deleterious (0) | probably_damaging (0.916) | Autosomal recessive (consanguineous family) | Exon1 | Familial Biparental Hydatidiform Mole | Pakistan | [26] |
| Family T | Chr6: 74072970 | c.322_325 delGACT | p.Asp108Ile fsTer30 | Frameshift mutation | 2.39E-05 | NA | NA | NA | Exon 2 | Complete Hydatidiform Mole | Tunisia | ||||
| Family W | Chr6: 74072453 | c.1A>G | p.Met1Val | Missense mutation | NA | NA | deleterious (0) | probably_damaging (0.916) | Compound Heterozygous | Exon 1 | Complete Hydatidiform Mole | Asia | |||
| Chr6: 74072969 | c.322_325 delGACT | p.Asp108Ile fsTer30 | Frameshift mutation | 2.39E-05 | NA | NA | NA | Exon 2 | |||||||
| Patient D | Chr6: 74072453 | c.1A>G | p.Met1Val | Start codon loss | NA | NA | deleterious (0) | probably_damaging (0.916) | Autosomal recessive | BiCHM | Iran | [24] | |||
| TLE6 | Family 1 | Chr19: 2993572 | NM_001143986.2 | c.1529C>A | p.Ser510Tyr | Missense mutation | NA | NA | deleterious (0) | probably_damaging (0.912) | Homozygous in 2 probands | WD40 domain repeats (Cterminal) | Early embryonic Arrest (1,2 and 4 cell stage) | Saudi Arabia | [100] |
| Family 2 | Homozygous in consanguineous family | ||||||||||||||
| PADI6 | Family 1 | Chr1: 17720537 | NM_207421.4 | c.1141C>T | p.Gln381Ter | Nonsense mutation | NA | NA | NA | NA | Homozygous in consanguineous family | PAD domain | Early Embryonic Arrest (arrested at the 2- to 4-cell stage) | China | [101] |
| Family 2 | Chr1: 17727858 | c.2009_2010 del | p.Glu670Gly fsTer48 | Frameshift mutation | NA | NA | NA | NA | Compound Heterozygous | PAD domain | Early Embryonic Arrest (arrested at the 1- to 2-cell stage) | ||||
| Chr1: 17708541 | c.633T>A | p.His211Gln | Missense mutation | 3.21E-05 | NA | deleterious (0.02) | probably_damaging (0.936) | PAD middle domain | |||||||
| Family 3 | Chr1: 17722159 | c.1618G>A | p.Gly540Arg | Missense mutation | 4.08E-06 | NA | tolerated (0.05) | benign (0.159) | Compound Heterozygous | PAD domain | Early Embryonic Arrest (arrested between the 2- and 5-cell stages) | ||||
| Chr1: 17718616 | c.970C>T | p.Gln324Ter | Nonsense mutation | NA | NA | NA | NA | ||||||||
| Family | Chr1: 17725285 | c.1793A>G | p.Asn598Ser | Missense mutation | NA | NA | tolerated (0.05) | probably_damaging (0.911) | Compound Heterozygous | PAD domain | Recurrent hydatidiform moles (RHM) | China | [80] | ||
| Chr1: 17727894 | c.2045 G>A | p. Arg682Gln | Missense mutation | 8.03E-06 | NA | deleterious (0) | probably_damaging (0.992) | ||||||||
| Family 1 | Chr1: 17718714 | c.1067G>A | p.Trp356Ter | Nonsense mutation | NA | NA | NA | NA | Probands mother is Compound Heterozygous | PAD domain (Exon 10) | Beckwith-Wiedemann syndrome with multi-locus imprinting disturbance | Italy | [110] | ||
| Chr1: 17727743 | c.1894C>G | p.Pro632Ala | Missense mutation | 4.01E-06 | NA | deleterious (0) | probably_damaging (1) | PAD domain (Exon 17) | |||||||
| Family 2 | Chr1: 17721538 | c.1429A>G | p.Met477Val | Missense mutation | 4.01E-06 | NA | tolerated (0.48) | possibly_ damaging (0.452) | Proband’s mother is Compound Heterozygous | PAD domain (Exon 13) | |||||
| Chr1: 17727929 | c.2080C>T | p.Pro694Ser | 8.05E-06 | NA | deleterious (0) | probably_damaging (1) | PAD domain (Exon 17) | ||||||||
| Family 3 | Chr1: 17727855 | c.2006delC | p.Thr669Lys fsTer85 | Frameshift deletion | NA | NA | NA | NA | Heterozygous | PAD domain (Exon 17) | |||||
| PADI6 (hg38) | Family 9 | Chr1: 17388820 | NM_207421.3 | c.902G>A | p.Arg301Gln | Missense mutations | NA | NA | deleterious (0) | probably_damaging (1) | Compound Heterozygous (Mother) Proband not tested | Exon 8 | SRS | [28] | |
| Chr1: 17394415 | c.1298C>T | p.Pro433Leu | NA | 2.63E-05 | deleterious (0) | probably_damaging (1) | Exon 11 | ||||||||
| Family 10 | Chr1: 17394024 | c.1124T>C | p.Leu375Ser | Missense mutations | NA | NA | deleterious (0.01) | probably_damaging (0.915) | Compound Heterozygous (Mother) | Exon 10 | BWS–MLID | ||||
| Chr1: 17397091 | c.1639G>A | p.Asp547Asn | NA | 5.06E-04 | tolerated (1) | benign (0.005) | Heterozygous in Proband | Exon 14 | |||||||
| Family 11 | Chr1: 17392197 | c.1046A>G | p.Asp349Gly | Missense mutation | NA | NA | tolerated (0.37) | probably_damaging (0.953) | Heterozygous (Mother) | Exon 9 | SRS | Germany | |||
| Family 12 | Chr1: 17379985 | c.433A>G | p.Lys145Glu | Missense mutation | NA | 6.57E-06 | deleterious (0.02) | possibly_damaging (0.612) | Heterozygous (Mother) | Exon 4 | SRS | Germany | |||
| OOEP (hg38) | Family 13 | Chr6: 73369684 | NM_001080507.2 | c.109C>T | p.Arg37Trp | Missense mutation | NA | 3.29E-05 | deleterious (0.04) | benign (0.135) | Autosomal recessive (Homozygous Mother), Heterozygous proband | Exon 1 | TNDM | ||
| UHRF1 (hg38) | Family 14 | Chr19: 4930782 | NM_013282.4 | c.514G>A | p.Val172Met | Missense mutation | NA | NA | deleterious (0) | probably_damaging (0.952) | Heterozygous (Mother and Proband) | Exon 3 | SRS | ||
| ZAR1 (hg38) | Family 15 | Chr4: 48492438 | NM_175619.2 | c.130G>T | p.Glu44Cys | Missense mutation | NA | NA | deleterious (0.01) | possibly_damaging (0.748) | Heterozygous (Mother and Proband) | Exon 1 | mild macroglossia, and high birth weight, but no other features of BWS |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Anvar, Z.; Chakchouk, I.; Demond, H.; Sharif, M.; Kelsey, G.; Van den Veyver, I.B. DNA Methylation Dynamics in the Female Germline and Maternal-Effect Mutations That Disrupt Genomic Imprinting. Genes 2021, 12, 1214. https://doi.org/10.3390/genes12081214
Anvar Z, Chakchouk I, Demond H, Sharif M, Kelsey G, Van den Veyver IB. DNA Methylation Dynamics in the Female Germline and Maternal-Effect Mutations That Disrupt Genomic Imprinting. Genes. 2021; 12(8):1214. https://doi.org/10.3390/genes12081214
Chicago/Turabian StyleAnvar, Zahra, Imen Chakchouk, Hannah Demond, Momal Sharif, Gavin Kelsey, and Ignatia B. Van den Veyver. 2021. "DNA Methylation Dynamics in the Female Germline and Maternal-Effect Mutations That Disrupt Genomic Imprinting" Genes 12, no. 8: 1214. https://doi.org/10.3390/genes12081214
APA StyleAnvar, Z., Chakchouk, I., Demond, H., Sharif, M., Kelsey, G., & Van den Veyver, I. B. (2021). DNA Methylation Dynamics in the Female Germline and Maternal-Effect Mutations That Disrupt Genomic Imprinting. Genes, 12(8), 1214. https://doi.org/10.3390/genes12081214







