Search for a Functional Genetic Variant Mimicking the Effect of SGLT2 Inhibitor Treatment
Abstract
:1. Introduction
2. Materials and Methods
2.1. UK Biobank Study Population
2.2. Genotyping and Imputation in the UK Biobank
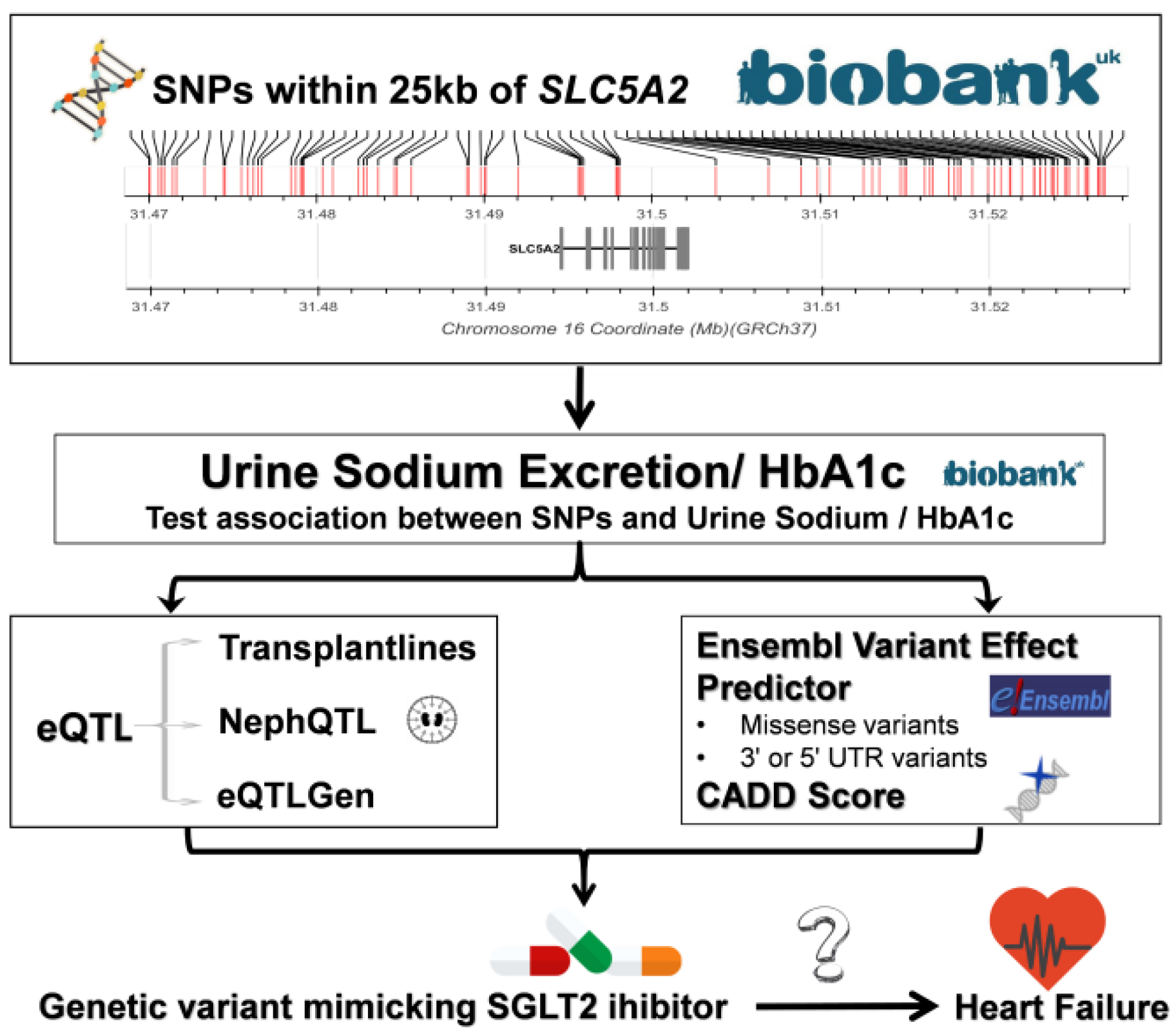
2.3. Candidate SNP Selection
2.4. SGLT2i Variants Based on Position
2.5. SLC5A2 eQTL Analyses
2.6. Functional Impact of SLC5A2 Variants
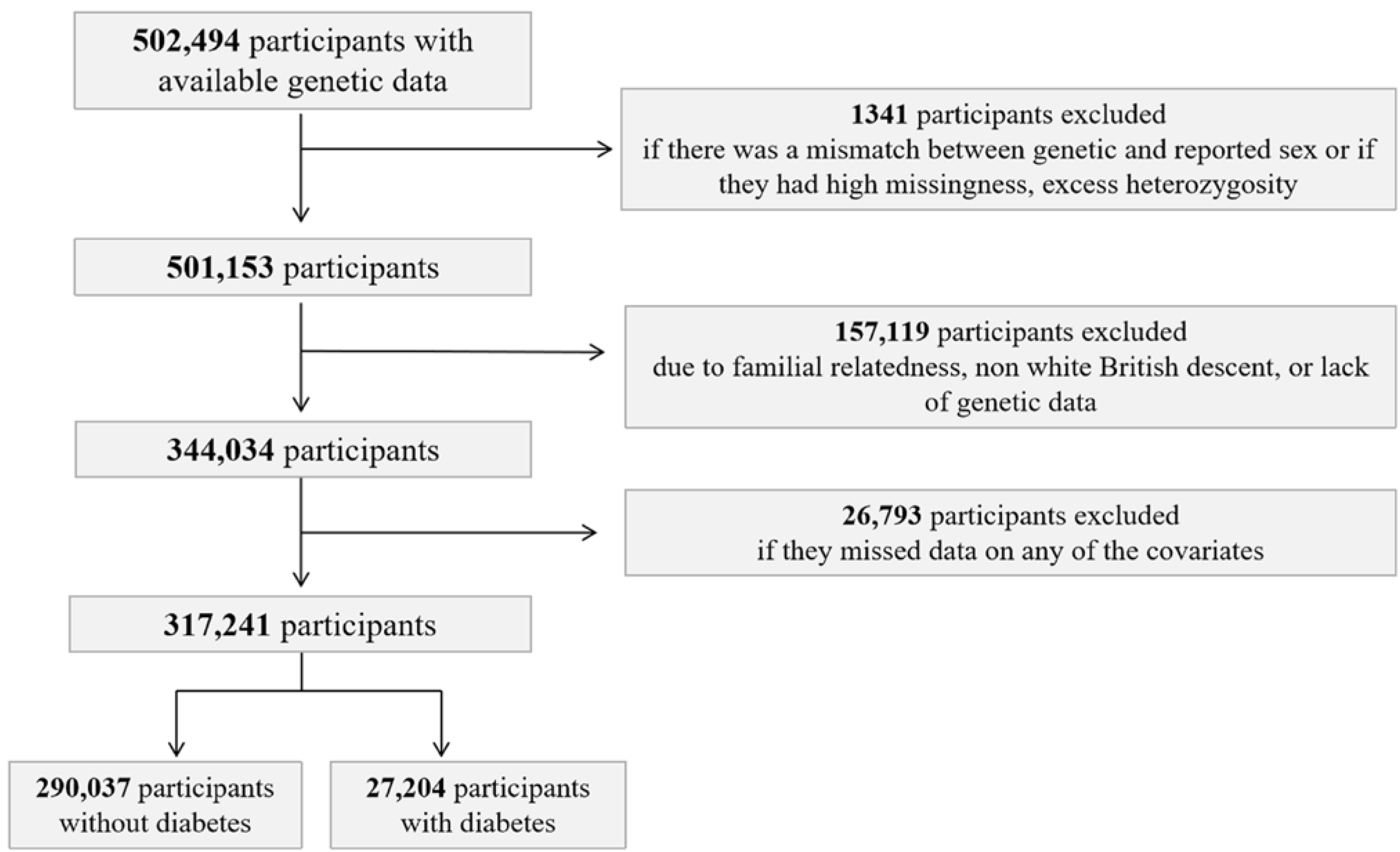
2.7. Exclusion Criteria
2.8. Statistical Analysis
3. Results
3.1. Population Characteristics
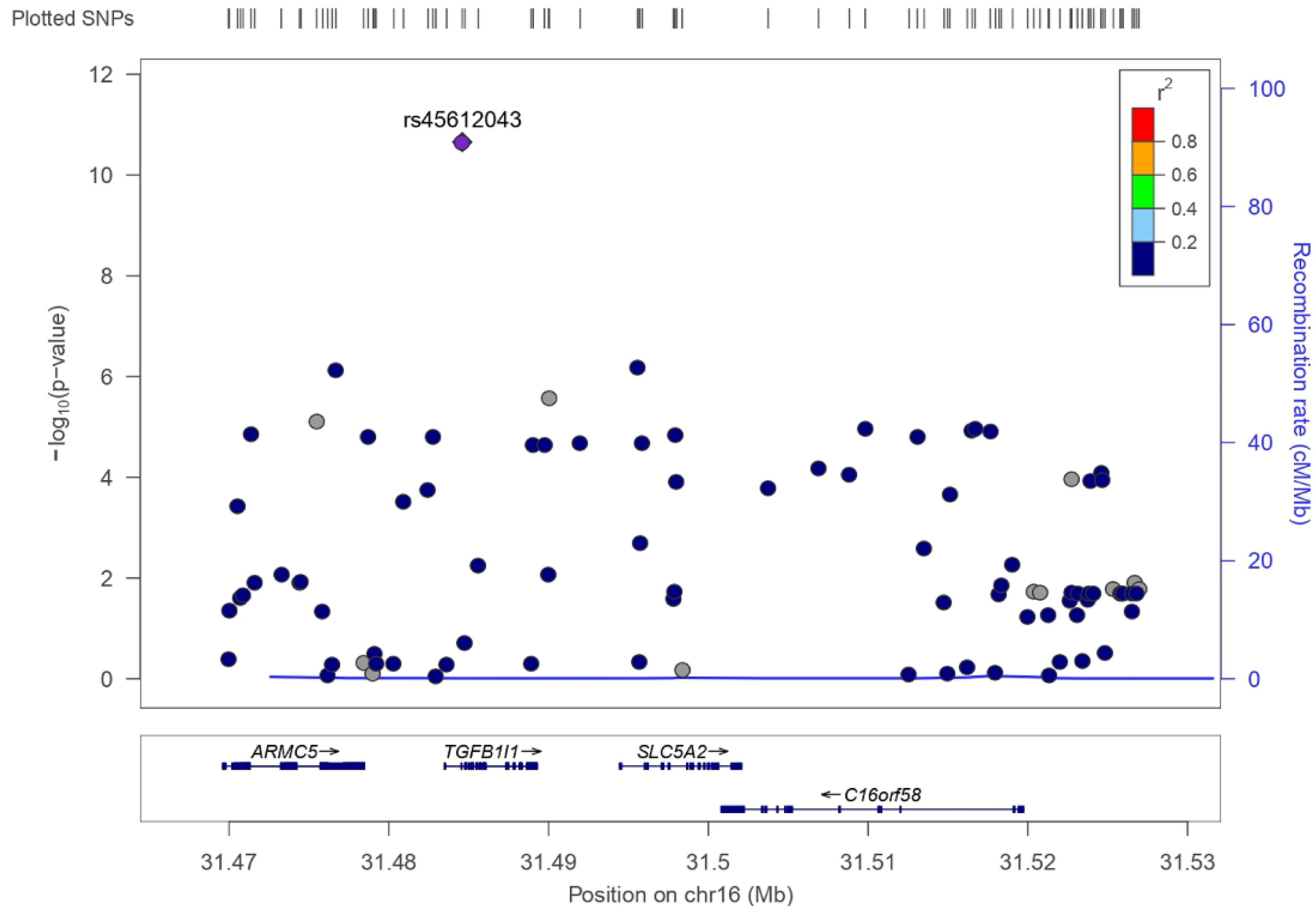
3.2. SLC5A2 Variants in UK Biobank
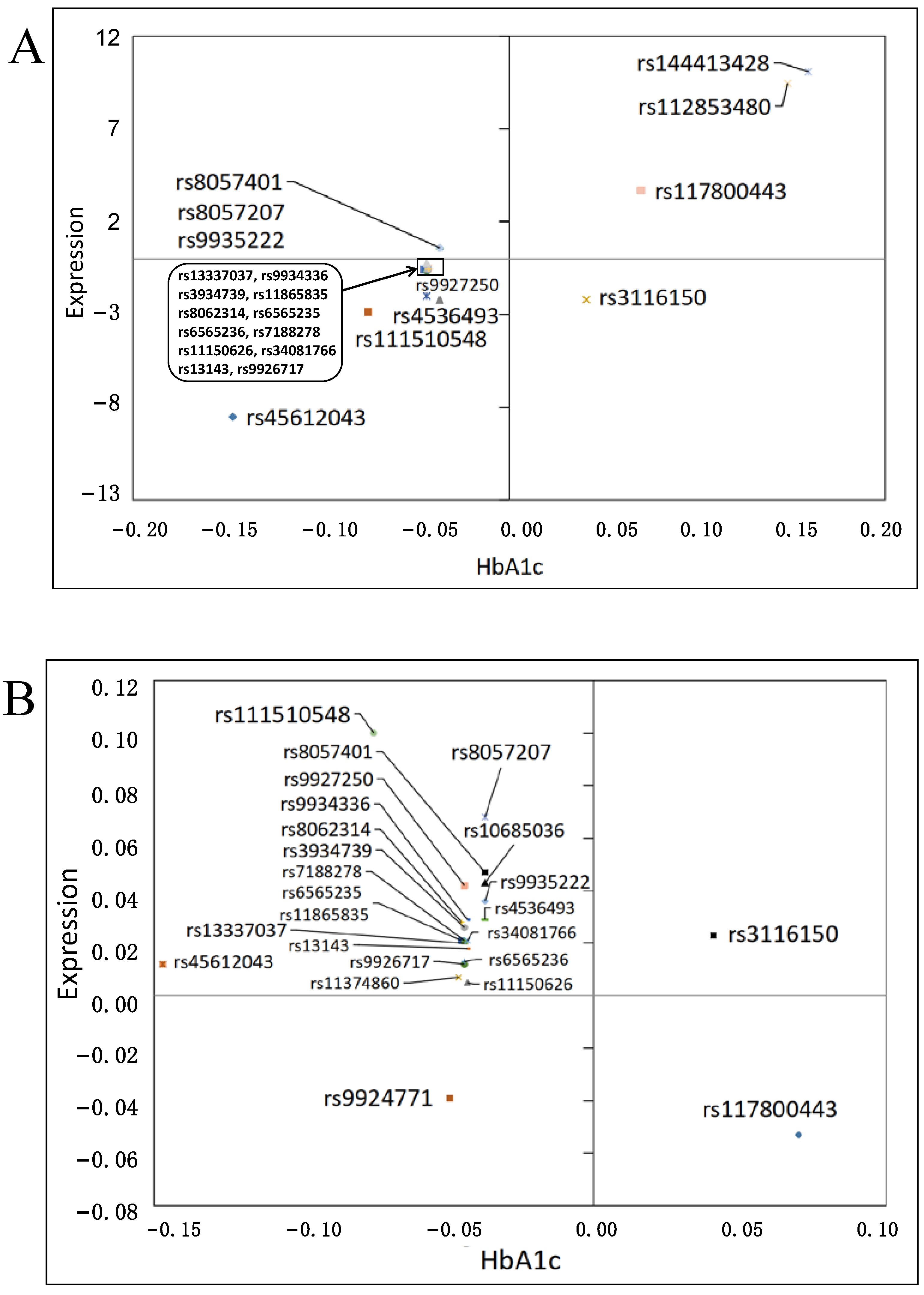
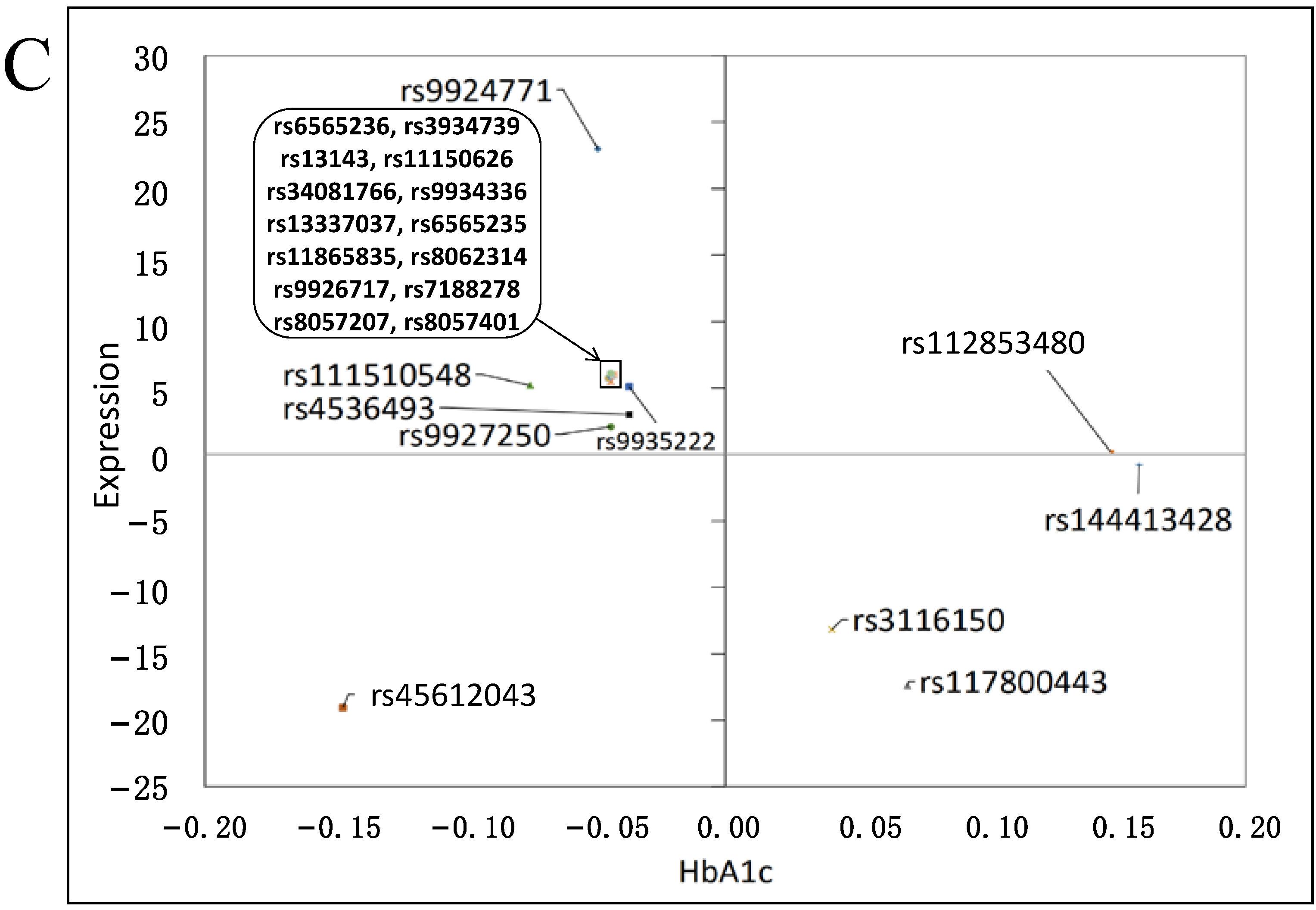
3.3. Effects on Gene Expression
3.4. Functional Impact
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cassis, P.; Locatelli, M.; Cerullo, D.; Corna, D.; Buelli, S.; Zanchi, C.; Villa, S.; Morigi, M.; Remuzzi, G.; Benigni, A.; et al. SGLT2 inhibitor dapagliflozin limits podocyte damage in proteinuric nondiabetic nephropathy. JCI Insight 2018, 3, e98720. [Google Scholar] [CrossRef] [Green Version]
- Coady, M.J.; El Tarazi, A.; Santer, R.; Bissonnette, P.; Sasseville, L.J.; Calado, J.; Lussier, Y.; Dumayne, C.; Bichet, D.G.; Lapointe, J.Y. MAP17 Is a Necessary Activator of Renal Na+/Glucose Cotransporter SGLT2. J. Am. Soc. Nephrol. 2017, 28, 85–93. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wells, R.G.; Mohandas, T.K.; Hediger, M.A. Localization of the Na+/glucose cotransporter gene SGLT2 to human chromosome 16 close to the centromere. Genomics 1993, 17, 787–789. [Google Scholar] [CrossRef] [PubMed]
- Ferrannini, E.; Baldi, S.; Frascerra, S.; Astiarraga, B.; Barsotti, E.; Clerico, A.; Muscelli, E. Renal Handling of Ketones in Response to Sodium-Glucose Cotransporter 2 Inhibition in Patients with Type 2 Diabetes. Diabetes Care 2017, 40, 771–776. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ni, L.; Yuan, C.; Chen, G.; Zhang, C.; Wu, X. SGLT2i: Beyond the glucose-lowering effect. Cardiovasc. Diabetol. 2020, 19, 98. [Google Scholar] [CrossRef]
- Zinman, B.; Wanner, C.; Lachin, J.M.; Fitchett, D.; Bluhmki, E.; Hantel, S.; Mattheus, M.; Devins, T.; Johansen, O.E.; Woerle, H.J.; et al. Empagliflozin, Cardiovascular Outcomes, and Mortality in Type 2 Diabetes. N. Engl. J. Med. 2015, 373, 2117–2128. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Xu, W.; Xu, Y.; Qian, Z. Iron Supplementation Improves Cardiovascular Outcomes in Patients with Heart Failure. Am. J. Med. 2019, 132, 955–963. [Google Scholar] [CrossRef]
- McMurray, J.J.V.; Solomon, S.D.; Inzucchi, S.E.; Køber, L.; Kosiborod, M.N.; Martinez, F.A.; Ponikowski, P.; Sabatine, M.S.; Anand, I.S.; Bělohlávek, J.; et al. Dapagliflozin in Patients with Heart Failure and Reduced Ejection Fraction. N. Engl. J. Med. 2019, 381, 1995–2008. [Google Scholar] [CrossRef] [Green Version]
- UK Biobank. UK Biobank: Protocol for a Large-Scale Prospective Epidemiological Resource; UK Biobank: Manchester, UK, 2007. [Google Scholar]
- Bycroft, C.; Freeman, C.; Petkova, D.; Band, G.; Elliott, L.T.; Sharp, K.; Motyer, A.; Vukcevic, D.; Delaneau, O.; O’Connell, J.; et al. Genome-wide genetic data on ~500,000 UK Biobank participants. bioRxiv 2017, 166298. [Google Scholar]
- Bycroft, C.; Freeman, C.; Petkova, D.; Band, G.; Elliott, L.T.; Sharp, K.; Motyer, A.; Vukcevic, D.; Delaneau, O.; O’Connell, J.; et al. The UK Biobank resource with deep phenotyping and genomic data. Nature 2018, 562, 203–209. [Google Scholar] [CrossRef] [Green Version]
- Weykamp, C. HbA1c: A review of analytical and clinical aspects. Ann. Lab Med. 2013, 33, 393–400. [Google Scholar] [CrossRef] [Green Version]
- Zanetti, D.; Bergman, H.; Burgess, S.; Assimes, T.L.; Bhalla, V.; Ingelsson, E. Urinary Albumin, Sodium, and Potassium and Cardiovascular Outcomes in the UK Biobank: Observational and Mendelian Randomization Analyses. Hypertension 2020, 75, 714–722. [Google Scholar] [CrossRef]
- Ahn, J.; Lee, J.H.; Lee, J.; Baek, J.Y.; Song, E.; Oh, H.S.; Kim, M.; Park, S.; Jeon, M.J.; Kim, T.Y.; et al. Association between urinary sodium levels and iodine status in Korea. Korean J. Internal Med. 2020, 35, 392–399. [Google Scholar] [CrossRef] [PubMed]
- Kawasaki, T.; Uezono, K.; Itoh, K.; Ueno, M. Prediction of 24-hour urinary creatinine excretion from age, body weight and height of an individual and its application. [Nihon koshu eisei zasshi] Jpn. J. Publ. Health 1991, 38, 567–574. [Google Scholar]
- Kawasaki, T.; Itoh, K.; Uezono, K.; Sasaki, H. A simple method for estimating 24 h urinary sodium and potassium excretion from second morning voiding urine specimen in adults. Clin. Exp. Pharmacol. Physiol. 1993, 20, 7–14. [Google Scholar] [CrossRef] [PubMed]
- UK Biobank. UK Biobank Biomarker Enhancement Project-Companion Document to Accompany HbA1c Biomarker Data; UK Biobank: Manchester, UK, 2018. [Google Scholar]
- Eisenga, M.F.; Gomes-Neto, A.W.; van Londen, M.; Ziengs, A.L.; Douwes, R.M.; Stam, S.P.; Oste, M.C.J.; Knobbe, T.J.; Hessels, N.R.; Buunk, A.M.; et al. Rationale and design of TransplantLines: A prospective cohort study and biobank of solid organ transplant recipients. BMJ Open 2018, 8, e024502. [Google Scholar] [CrossRef] [PubMed]
- Gillies, C.E.; Putler, R.; Menon, R.; Otto, E.; Yasutake, K.; Nair, V.; Hoover, P.; Lieb, D.; Li, S.; Eddy, S.; et al. An eQTL Landscape of Kidney Tissue in Human Nephrotic Syndrome. Am. J. Hum. Genet 2018, 103, 232–244. [Google Scholar] [CrossRef] [Green Version]
- Võsa, U.; Claringbould, A.; Westra, H.-J.; Bonder, M.J.; Deelen, P.; Zeng, B.; Kirsten, H.; Saha, A.; Kreuzhuber, R.; Kasela, S.; et al. Unraveling the polygenic architecture of complex traits using blood eQTL metaanalysis. bioRxiv 2018, 447367. [Google Scholar]
- Butler, J.; Hamo, C.E.; Filippatos, G.; Pocock, S.J.; Bernstein, R.A.; Brueckmann, M.; Cheung, A.K.; George, J.T.; Green, J.B.; Januzzi, J.L.; et al. The potential role and rationale for treatment of heart failure with sodium-glucose co-transporter 2 inhibitors. Eur. J. Heart Fail. 2017, 19, 1390–1400. [Google Scholar] [CrossRef] [Green Version]
- Morris, A.P.; Le, T.H.; Wu, H.; Akbarov, A.; van der Most, P.J.; Hemani, G.; Smith, G.D.; Mahajan, A.; Gaulton, K.J.; Nadkarni, G.N.; et al. Trans-ethnic kidney function association study reveals putative causal genes and effects on kidney-specific disease aetiologies. Nat. Commun. 2019, 10, 29. [Google Scholar] [CrossRef] [Green Version]
- Das, S.; Forer, L.; Schonherr, S.; Sidore, C.; Locke, A.E.; Kwong, A.; Vrieze, S.I.; Chew, E.Y.; Levy, S.; McGue, M.; et al. Next-generation genotype imputation service and methods. Nat. Gen. 2016, 48, 1284–1287. [Google Scholar] [CrossRef] [Green Version]
- McCarthy, S.; Das, S.; Kretzschmar, W.; Delaneau, O.; Wood, A.R.; Teumer, A.; Kang, H.M.; Fuchsberger, C.; Danecek, P.; Sharp, K.; et al. A reference panel of 64,976 haplotypes for genotype imputation. Nat. Genet. 2016, 48, 1279–1283. [Google Scholar] [CrossRef] [Green Version]
- Wozniak, M.B.; Le Calvez-Kelm, F.; Abedi-Ardekani, B.; Byrnes, G.; Durand, G.; Carreira, C.; Michelon, J.; Janout, V.; Holcatova, I.; Foretova, L.; et al. Integrative genome-wide gene expression profiling of clear cell renal cell carcinoma in Czech Republic and in the United States. PLoS ONE 2013, 8, e57886. [Google Scholar] [CrossRef] [Green Version]
- Wain, L.V.; Vaez, A.; Jansen, R.; Joehanes, R.; van der Most, P.J.; Erzurumluoglu, A.M.; O’Reilly, P.F.; Cabrera, C.P.; Warren, H.R.; Rose, L.M.; et al. Novel Blood Pressure Locus and Gene Discovery Using Genome-Wide Association Study and Expression Data Sets From Blood and the Kidney. Hypertension 2017, 70, e4–e9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghezzi, C.; Loo, D.D.F.; Wright, E.M. Physiology of renal glucose handling via SGLT1, SGLT2 and GLUT2. Diabetologia 2018, 61, 2087–2097. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cohen, C.D.; Frach, K.; Schlondorff, D.; Kretzler, M. Quantitative gene expression analysis in renal biopsies: A novel protocol for a high-throughput multicenter application. Kidney Int. 2002, 61, 133–140. [Google Scholar] [CrossRef] [Green Version]
- Yates, A.D.; Achuthan, P.; Akanni, W.; Allen, J.; Allen, J.; Alvarez-Jarreta, J.; Amode, M.R.; Armean, I.M.; Azov, A.G.; Bennett, R.; et al. Ensembl 2020. Nucl. Acids Res. 2019, 48, D682–D688. [Google Scholar] [CrossRef] [PubMed]
- Hunt, S.E.; McLaren, W.; Gil, L.; Thormann, A.; Schuilenburg, H.; Sheppard, D.; Parton, A.; Armean, I.M.; Trevanion, S.J.; Flicek, P.; et al. Ensembl variation resources. Database 2018, 2018, bay119. [Google Scholar] [CrossRef] [PubMed]
- Amendola, L.M.; Dorschner, M.O.; Robertson, P.D.; Salama, J.S.; Hart, R.; Shirts, B.H.; Murray, M.L.; Tokita, M.J.; Gallego, C.J.; Kim, D.S.; et al. Actionable exomic incidental findings in 6503 participants: Challenges of variant classification. Genom. Res. 2015, 25, 305–315. [Google Scholar] [CrossRef] [Green Version]
- Pruim, R.J.; Welch, R.P.; Sanna, S.; Teslovich, T.M.; Chines, P.S.; Gliedt, T.P.; Boehnke, M.; Abecasis, G.R.; Willer, C.J. LocusZoom: Regional visualization of genome-wide association scan results. Bioinformatics 2010, 26, 2336–2337. [Google Scholar] [CrossRef] [Green Version]
- Verma, S.; McMurray, J.J.V. SGLT2 inhibitors and mechanisms of cardiovascular benefit: A state-of-the-art review. Diabetologia 2018, 61, 2108–2117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosano, G.; Quek, D.; Martínez, F. Sodium-Glucose Co-transporter 2 Inhibitors in Heart Failure: Recent Data and Implications for Practice. Card. Fail. Rev. 2020, 6, e31. [Google Scholar] [CrossRef] [PubMed]
- Packer, M.; Anker, S.D.; Butler, J.; Filippatos, G.; Pocock, S.J.; Carson, P.; Januzzi, J.; Verma, S.; Tsutsui, H.; Brueckmann, M.; et al. Cardiovascular and Renal Outcomes with Empagliflozin in Heart Failure. N. Engl. J. Med. 2020, 383, 1413–1424. [Google Scholar] [CrossRef] [PubMed]
- Lopaschuk, G.D.; Verma, S. Mechanisms of Cardiovascular Benefits of Sodium Glucose Co-Transporter 2 (SGLT2) Inhibitors: A State-of-the-Art Review. JACC. Basic Transl. Sci. 2020, 5, 632–644. [Google Scholar] [CrossRef] [PubMed]
- Scheen, A.J. Reduction in HbA1c with SGLT2 inhibitors vs. DPP-4 inhibitors as add-ons to metformin monotherapy according to baseline HbA1c: A systematic review of randomized controlled trials. Diabetes Metabol. 2020, 46, 186–196. [Google Scholar] [CrossRef]
- Cowie, M.R.; Fisher, M. SGLT2 inhibitors: Mechanisms of cardiovascular benefit beyond glycaemic control. Nat. Rev. Cardiol. 2020, 17, 761–772. [Google Scholar] [CrossRef]
- Buse, J.B.; Wexler, D.J.; Tsapas, A.; Rossing, P.; Mingrone, G.; Mathieu, C.; D’Alessio, D.A.; Davies, M.J. 2019 update to: Management of hyperglycaemia in type 2 diabetes, 2018. A consensus report by the American Diabetes Association (ADA) and the European Association for the Study of Diabetes (EASD). Diabetologia 2020, 63, 221–228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Monobe, K.; Noso, S.; Babaya, N.; Hiromine, Y.; Taketomo, Y.; Niwano, F.; Yoshida, S.; Yasutake, S.; Minohara, T.; Kawabata, Y.; et al. Clinical and genetic determinants of urinary glucose excretion in patients with diabetes mellitus. J. Diabetes Investig. 2020. [Google Scholar] [CrossRef]
- Enigk, U.; Breitfeld, J.; Schleinitz, D.; Dietrich, K.; Halbritter, J.; Fischer-Rosinsky, A.; Enigk, B.; Muller, I.; Spranger, J.; Pfeiffer, A.; et al. Role of genetic variation in the human sodium-glucose cotransporter 2 gene (SGLT2) in glucose homeostasis. Pharmacogenomics 2011, 12, 1119–1126. [Google Scholar] [CrossRef]
- Drexel, H.; Leiherer, A.; Saely, C.H.; Brandtner, E.M.; Geiger, K.; Vonbank, A.; Fraunberger, P.; Muendlein, A. Are SGLT2 polymorphisms linked to diabetes mellitus and cardiovascular disease? Prospective study and meta-analysis. Biosci. Rep. 2019, 39. [Google Scholar] [CrossRef] [Green Version]
- Zimdahl, H.; Haupt, A.; Brendel, M.; Bour, L.; Machicao, F.; Salsali, A.; Broedl, U.C.; Woerle, H.J.; Haring, H.U.; Staiger, H. Influence of common polymorphisms in the SLC5A2 gene on metabolic traits in subjects at increased risk of diabetes and on response to empagliflozin treatment in patients with diabetes. Pharmacogenet. Genom. 2017, 27, 135–142. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.A.; McMahon, G.; Karhunen, V.; Wade, K.H.; Corbin, L.J.; Hughes, D.A.; Smith, G.D.; Lawlor, D.A.; Jarvelin, M.R.; Timpson, N.J. Common variation at 16p11.2 is associated with glycosuria in pregnancy: Findings from a genome-wide association study in European women. Hum. Mol. Genet. 2020, 29, 2098–2106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, S.; Wu, X.; Pei, Y.; Wang, W.; Zheng, K.; Qiu, E.; Zhang, X. PTHR1 May Be Involved in Progression of Osteosarcoma by Regulating miR-124-3p-AR-Tgfb1i1, miR-27a-3p-PPARG-Abca1, and miR-103/590-3p-AXIN2 Axes. DNA Cell Biol. 2019, 38, 1323–1337. [Google Scholar] [CrossRef] [PubMed]
- Elliott, P.; Muller, D.C.; Schneider-Luftman, D.; Pazoki, R.; Evangelou, E.; Dehghan, A.; Neal, B.; Tzoulaki, I. Estimated 24-Hour Urinary Sodium Excretion and Incident Cardiovascular Disease and Mortality Among 398 628 Individuals in UK Biobank. Hypertension 2020, 76, 683–691. [Google Scholar] [CrossRef]
- Law, P.J.; Timofeeva, M.; Fernandez-Rozadilla, C.; Broderick, P.; Studd, J.; Fernandez-Tajes, J.; Farrington, S.; Svinti, V.; Palles, C.; Orlando, G.; et al. Association analyses identify 31 new risk loci for colorectal cancer susceptibility. Nat. Commun. 2019, 10, 2154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Characteristics | Mean (SD)/n (%) |
|---|---|
| Age (years) | 57.39 (7.99) |
| Female | 169,688 (53.49%) |
| BMI (kg/m2) | 27.39 (4.73) |
| Diastolic blood pressure (mm Hg) | 82.15 (8.54) * |
| Systolic blood pressure (mm Hg) | 133.75 (17.95) * |
| Resting heart rate | 69.32 (11.24) ** |
| HbA1c (mmol/mol) | 35.93 (6.45) |
| LDL (mmol/L) | 3.57 (0.87) # |
| HDL (mmol/L) | 1.45 (0.38) ## |
| Urinary sodium/creatinine ratio (mg/g) | 2145.87 (1198.64) |
| Predicted 24 h urinary sodium excretion (mg/day) | 1894.40 (563.79) |
| Heart failure | 9850 (3.1%) |
| Hypertension | 117,196 (36.97%) * |
| Diabetes | 27,204 (8.58%) |
| SNP | CHR | EFAL | NEFAL | MAF (Minor Allele) | β | SE | p |
|---|---|---|---|---|---|---|---|
| rs45612043 | 16 | C | A | 0.043 (C) | −0.147 | 0.022 | 2.22 × 10−11 |
| rs9924771 | 16 | G | A | 0.348 (A) | −0.049 | 0.010 | 6.53 × 10−7 |
| rs111510548 | 16 | C | T | 0.098 (C) | −0.075 | 0.015 | 7.58 × 10−7 |
| rs1251169601 | 16 | C | CAAAAAAAAAAAA | 0.410 (C) | 0.046 | 0.010 | 2.72 × 10−6 |
| rs11374860 | 16 | TG | T | 0.279 (TG) | −0.046 | 0.010 | 7.80 × 10−6 |
| rs11865835 | 16 | C | T | 0.290 (C) | −0.045 | 0.010 | 1.09 × 10−5 |
| rs8062314 | 16 | A | C | 0.290 (A) | −0.045 | 0.010 | 1.11 × 10−5 |
| rs6565235 | 16 | T | C | 0.290 (T) | −0.045 | 0.010 | 1.20 × 10−5 |
| rs6565236 | 16 | T | A | 0.278 (T) | −0.044 | 0.010 | 1.23 × 10−5 |
| rs9926717 | 16 | G | A | 0.292 (G) | −0.044 | 0.010 | 1.40 × 10−5 |
| rs144413428 | 16 | A | G | 0.015 (A) | 0.159 | 0.037 | 1.47 × 10−5 |
| rs13337037 | 16 | A | G | 0.275 (A) | −0.044 | 0.010 | 1.56 × 10−5 |
| rs7188278 | 16 | T | C | 0.290 (T) | −0.044 | 0.010 | 1.56 × 10−5 |
| rs3934739 | 16 | T | C | 0.277 (T) | −0.044 | 0.010 | 1.56 × 10−5 |
| rs11150626 | 16 | C | T | 0.278 (C) | −0.043 | 0.010 | 2.06 × 10−5 |
| rs9934336 | 16 | A | G | 0.277 (A) | −0.043 | 0.010 | 2.11 × 10−5 |
| rs34081766 | 16 | A | C | 0.277 (A) | −0.043 | 0.010 | 2.27 × 10−5 |
| rs13143 | 16 | T | C | 0.278 (T) | −0.043 | 0.010 | 2.27 × 10−5 |
| rs9927250 | 16 | G | A | 0.220 (A) | −0.044 | 0.011 | 6.61 × 10−5 |
| rs8057207 | 16 | T | C | 0.357 (T) | −0.037 | 0.010 | 8.26 × 10−5 |
| rs112853480 | 16 | C | T | 0.016 (C) | 0.148 | 0.038 | 8.71 × 10−5 |
| rs10685036 | 16 | TTA | T | 0.359 (TTA) | −0.037 | 0.010 | 1.10 × 10−4 |
| rs8057401 | 16 | T | C | 0.356 (T) | −0.037 | 0.010 | 1.14 × 10−4 |
| rs9935222 | 16 | A | C | 0.354 (A) | −0.037 | 0.010 | 1.19 × 10−4 |
| rs3116150 | 16 | A | G | 0.235 (A) | 0.041 | 0.011 | 1.24 × 10−4 |
| rs4536493 | 16 | G | A | 0.312 (A) | −0.037 | 0.010 | 1.65 × 10−4 |
| rs117800443 | 16 | A | G | 0.066 (A) | 0.070 | 0.019 | 1.80 × 10−4 |
| SNP | CHR | Locus | Allele | Consequence | Symbol | CADD Score |
|---|---|---|---|---|---|---|
| rs9926717 | 16 | 31,471,378 | G | 3′ prime UTR variant, NMD transcript variant | ARMC5 | 8.528 |
| rs111510548 | 16 | 31,476,695 | C | 3′ prime UTR variant | ARMC5 | 4.538 |
| rs45612043 | 16 | 31,484,598 | C | 5′ prime UTR variant | TGFB1I1 | 12.790 * |
| rs13143 | 16 | 31,489,033 | T | 3′ prime UTR variant, NMD transcript variant | TGFB1I1 | 6.091 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, S.; Said, M.A.; Groot, H.E.; van der Most, P.J.; Thio, C.H.L.; van de Vegte, Y.J.; Verweij, N.; Snieder, H.; van der Harst, P. Search for a Functional Genetic Variant Mimicking the Effect of SGLT2 Inhibitor Treatment. Genes 2021, 12, 1174. https://doi.org/10.3390/genes12081174
Wang S, Said MA, Groot HE, van der Most PJ, Thio CHL, van de Vegte YJ, Verweij N, Snieder H, van der Harst P. Search for a Functional Genetic Variant Mimicking the Effect of SGLT2 Inhibitor Treatment. Genes. 2021; 12(8):1174. https://doi.org/10.3390/genes12081174
Chicago/Turabian StyleWang, Siqi, M. Abdullah Said, Hilde E. Groot, Peter J. van der Most, Chris H. L. Thio, Yordi J. van de Vegte, Niek Verweij, Harold Snieder, and Pim van der Harst. 2021. "Search for a Functional Genetic Variant Mimicking the Effect of SGLT2 Inhibitor Treatment" Genes 12, no. 8: 1174. https://doi.org/10.3390/genes12081174
APA StyleWang, S., Said, M. A., Groot, H. E., van der Most, P. J., Thio, C. H. L., van de Vegte, Y. J., Verweij, N., Snieder, H., & van der Harst, P. (2021). Search for a Functional Genetic Variant Mimicking the Effect of SGLT2 Inhibitor Treatment. Genes, 12(8), 1174. https://doi.org/10.3390/genes12081174







