Genome-Wide Association Study Using Whole-Genome Sequence Data for Fertility, Health Indicator, and Endoparasite Infection Traits in German Black Pied Cattle
Abstract
1. Introduction
2. Materials and Methods
2.1. Cow Traits
2.2. Genetic Architecture of Cow Traits
2.3. Whole-Genome Sequencing and Imputation of 50K Genotypes
2.4. Quality Control of Whole-Genome Sequence Genotypes
2.5. Genome-Wide Association Analyses
2.6. Candidate Gene Annotations and Pathway Analyses
3. Results
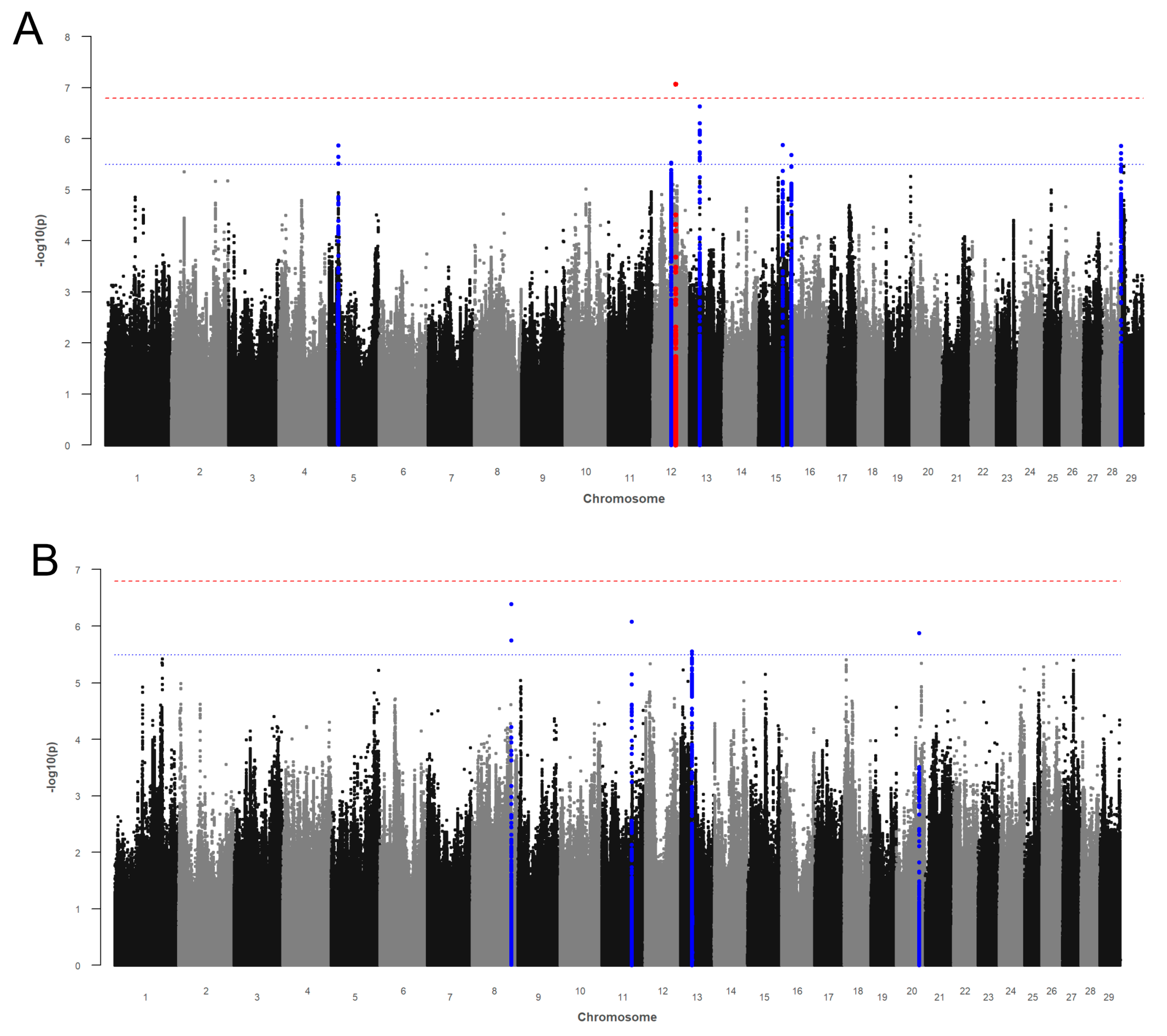
3.1. Female Fertility Traits
3.1.1. Calving-to-First Service Interval
3.1.2. Non-Return after Day 56
3.2. Health Indicator Traits
3.2.1. Somatic Cell Score
3.2.2. Fat-to-Protein Ratio
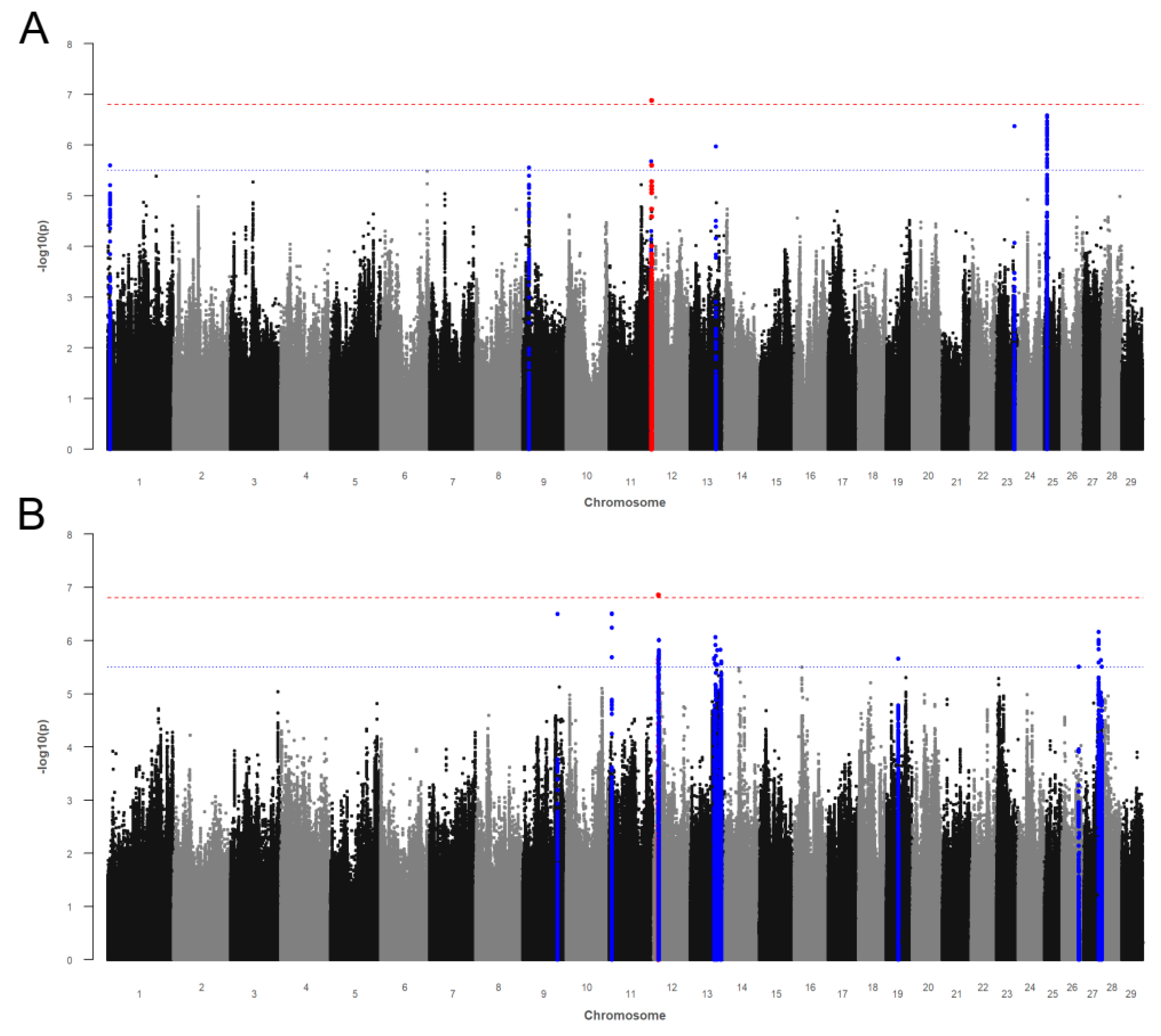
3.3. Endoparasite Infection Traits
3.3.1. Gastrointestinal Nematode Infections
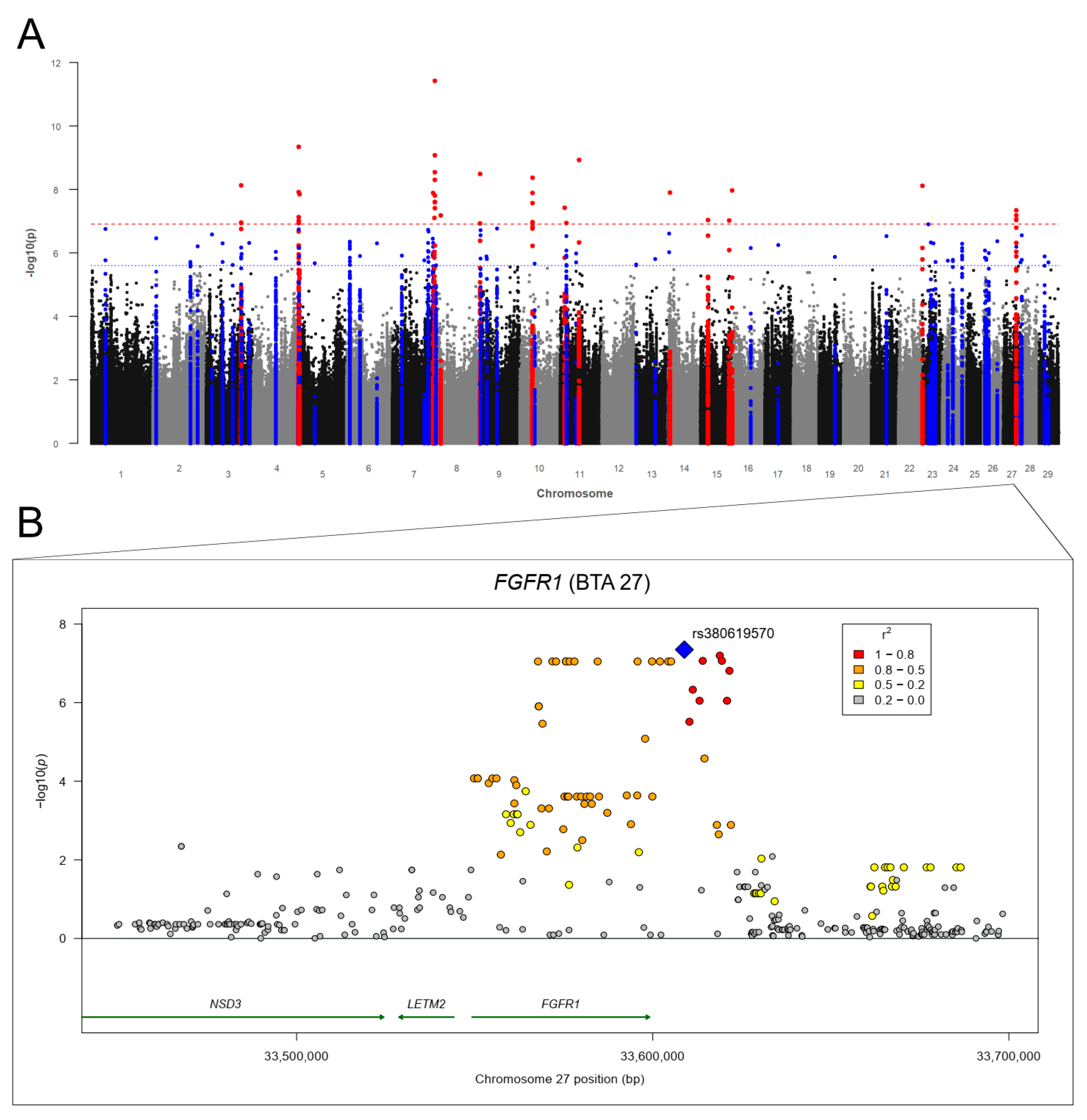
3.3.2. Liver Fluke (Fasciola hepatica) Infections
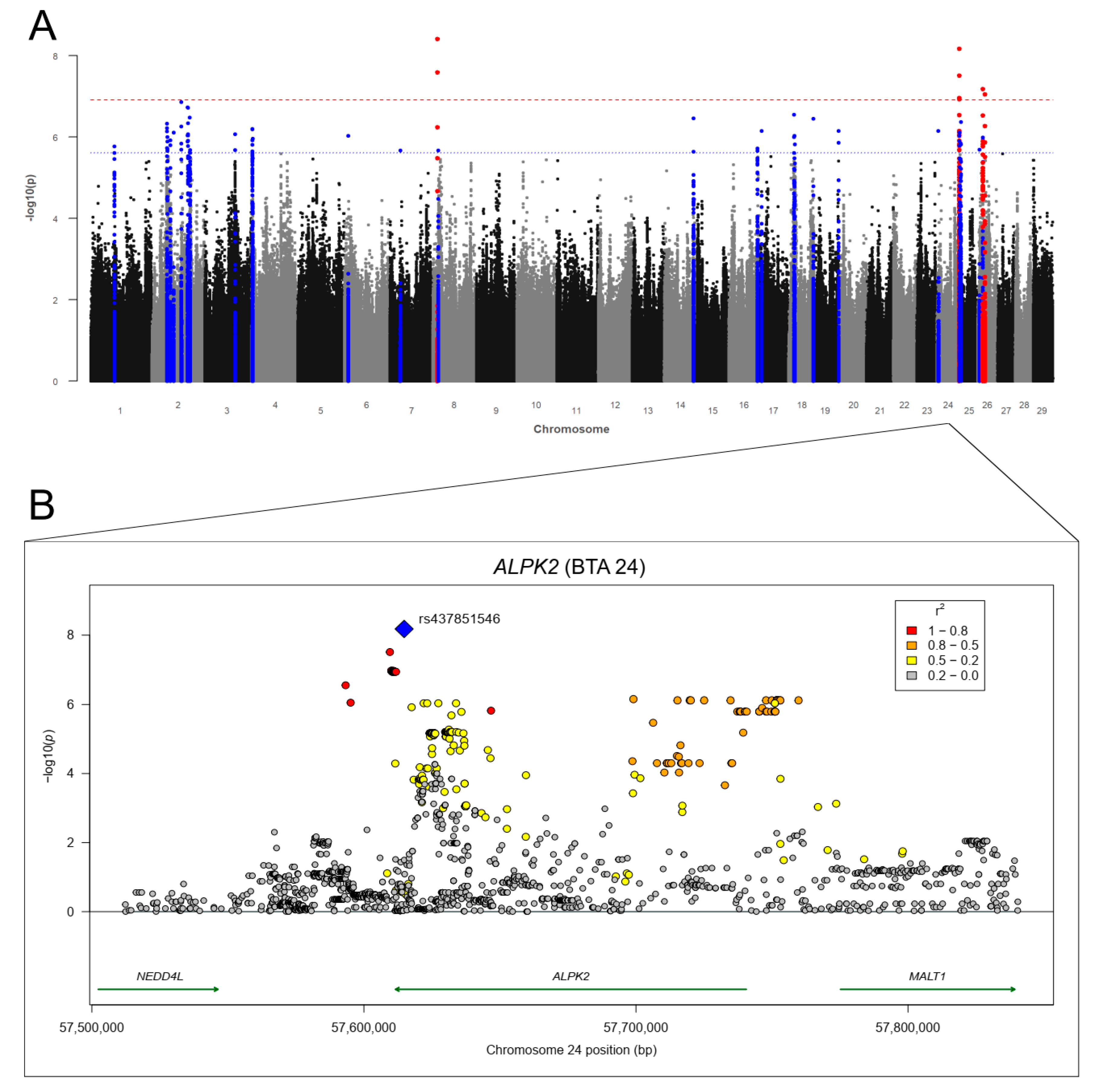
3.3.3. Bovine Lungworm (Dictyocaulus viviparus) Infections
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Daetwyler, H.D.; Capitan, A.; Pausch, H.; Stothard, P.; van Binsbergen, R.; Brondum, R.F.; Liao, X.; Djari, A.; Rodriguez, S.C.; Grohs, C.; et al. Whole-genome sequencing of 234 bulls facilitates mapping of monogenic and complex traits in cattle. Nat. Gen. 2014, 46, 858–867. [Google Scholar] [CrossRef] [PubMed]
- Das, A.; Panitz, F.; Gregersen, V.R.; Bendixen, C.; Holm, L.-E. Deep sequencing of Danish Holstein dairy cattle for variant detection and insight into potential loss-of-function variants in protein coding genes. BMC Genom. 2015, 16, 1043. [Google Scholar] [CrossRef] [PubMed]
- Schork, A.J.; Thompson, W.K.; Pham, P.; Torkamani, A.; Roddey, J.C.; Sullivan, P.F.; Kelso, J.R.; O’Donovan, M.C.; Furberg, H.; Schork, N.J.; et al. All SNPs are not created equal: Genome-wide association studies reveal a consistent pattern of enrichment among functionally annotated SNPs. PLoS Genet. 2013, 9, e1003449. [Google Scholar] [CrossRef]
- Van Binsbergen, R.; Bink, M.C.A.M.; Calus, M.P.L.; van Eeuwijk, F.A.; Hayes, B.J.; Hulsegge, I.; Veerkamp, R.F. Accuracy of imputation to whole-genome sequence data in Holstein Friesian cattle. Genet. Sel. Evol. 2014, 46, 41. [Google Scholar] [CrossRef]
- Pausch, H.; MacLeod, I.M.; Fries, R.; Emmerling, R.; Bowman, P.J.; Daetwyler, H.D.; Goddard, M.E. Evaluation of the accuracy of imputed sequence variant genotypes and their utility for causal variant detection in cattle. Genet. Sel. Evol. 2017, 49, 24. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Lund, M.S.; Sahana, G.; Guldbrandtsen, B.; Sun, D.; Zhang, Q.; Su, G. Association analysis for udder health based on SNP-panel and sequence data in Danish Holsteins. Genet. Sel. Evol. 2015, 47, 50. [Google Scholar] [CrossRef][Green Version]
- Twomey, A.J.; Berry, D.P.; Evans, R.D.; Doherty, M.L.; Graham, D.A.; Purfield, D.C. Genome-wide association study of endo-parasite phenotypes using imputed whole-genome sequence data in dairy and beef cattle. Genet. Sel. Evol. 2019, 51, 15. [Google Scholar] [CrossRef] [PubMed]
- Hartwig, S.; Bennewitz, J. Züchterische Aspekte zur Konsolidierung und Weiterentwicklung lokaler Rinderpopulationen. Züchtungskunde 2014, 86, 19–24. [Google Scholar]
- Tijjani, A.; Utsunomiya, Y.T.; Ezekwe, A.G.; Nashiru, O.; Hannotee, O. Genome sequence analysis reveals selection signatures in endangered trypanotolerant West African Muturu cattle. Front. Genet. 2019, 10, 442. [Google Scholar] [CrossRef] [PubMed]
- Jaeger, M.P.H. Phenotypic, Quantitative Genetic and Genomic Characterization of the German Black and White Dual-Purpose Cattle Breed. Ph.D. Thesis, Justus-Liebig University Gießen, Giessen, Germany, 5 October 2018. [Google Scholar]
- Boettcher, P.J.; Hoffmann, I.; Baumung, R.; Drucker, A.G.; McManus, C.; Berg, P.; Stella, A.; Nilsen, L.B.; Moran, D.; Naves, M.; et al. Genetic ressources and genomics for adaptation of livestock to climate change. Front. Genet. 2015, 5, 461. [Google Scholar] [CrossRef]
- Mei, C.; Wang, H.; Zhu, W.; Wang, H.; Cheng, G.; Qu, K.; Guang, X.; Li, A.; Zhao, C.; Yang, W. Whole-genome sequencing of the endangered bovine species Gayal (Bos frontalis) provides new insights into its genetic features. Sci. Rep. 2016, 6, 19787. [Google Scholar] [CrossRef]
- Doublet, A.-C.; Restoux, G.; Fritz, S.; Balberini, L.; Fayolle, G.; Hozé, C.; Laloe, D.; Croiseau, P. Intensified use of reproductive technologies and reduced dimensions of breeding schemes put genetic diversity at risk in dairy cattle breeds. Animals 2020, 10, 1903. [Google Scholar] [CrossRef]
- TGRDEU. Zentrale Dokumentation Tiergenetischer Ressourcen in Deutschland, Bundesanstalt für Landwirtschaft und Ernährung. 2019. Available online: https://tgrdeu.genres.de/ (accessed on 24 May 2021).
- Mügge, B.; Lutz, W.E.; Südbeck, H.; Zelfel, S. Deutsche Holsteins: Die Geschichte einer Zucht; Verlag Eugen Ulmer: Stuttgart, Germany, 1999. [Google Scholar]
- Grothe, P.O. Holstein-Friesian: Eine Rasse Geht um die Welt; Landwirtschaftsverlag: Münster, Germany, 1993. [Google Scholar]
- May, K.; Brügemann, K.; König, S.; Strube, C. Patent gastrointestinal nematode infections in organically and conventionally pastured dairy cows and their impact on individual milk fertility parameters. Vet. Parasitol. 2017, 245, 119–127. [Google Scholar] [CrossRef]
- Biedermann, G. Zuchtplanung für die Erhaltung des Alten Schwarzbunten Niederungsrindes. Bundesprogramm Ökologischer Landbau und Andere Formen Nachhaltiger Landwirtschaft. Available online: https://service.ble.de/ptdb/index2.php?detail_id=84928&site_key=141 (accessed on 17 May 2021).
- Van Dijk, J.; Sargison, N.D.; Kenyon, F.; Skuce, P.J. Climate change and infectious disease: Helminthological challenges to farmed ruminants in temperate regions. Animal 2010, 4, 377–392. [Google Scholar] [CrossRef] [PubMed]
- Charlier, J.; Rinaldi, L.; Musella, V.; Ploeger, H.W.; Chartier, C.; Vineer, H.R.; Hinney, B.; von Samson-Himmelstjerna, G.; Băcescu, B.; Mickiewicz, M.; et al. Initial assessment of the economic burden of major helminth infections to the ruminant livestock industry in Europe. Prev. Vet. Med. 2020, 182, 105103. [Google Scholar] [CrossRef]
- May, K.; Brügemann, K.; Yin, T.; Scheper, C.; Strube, C.; König, S. Genetic line comparisons and genetic parameters for endoparasite infections and test-day milk production traits. J. Dairy Sci. 2017, 100, 7330–7344. [Google Scholar] [CrossRef] [PubMed]
- May, K.; Scheper, S.; Brügemann, K.; Yin, T.; Strube, C.; Korkuc, P.; Brockmann, G.A.; König, S. Genome-wide associations and functional gene analyses for endoparasite resistance in an endangered population of native German Black Pied cattle. BMC Genom. 2019, 20, 277. [Google Scholar] [CrossRef] [PubMed]
- Twomey, A.J.; Carroll, R.I.; Doherty, M.L.; Byrne, N.; Graham, D.A.; Sayers, R.G.; Blom, A.; Berry, D.P. Genetic correlations between endo-parasite phenotypes and economically important traits in dairy and beef cattle. J. Anim. Sci. 2018, 96, 407–421. [Google Scholar] [CrossRef] [PubMed]
- Korkuć, P.; Arends, D.; May, K.; König, S.; Brockmann, G.A. Genomic loci affecting milk production in German Black Pied cattle (DSN). Front. Genet. 2021, 12, 640039. [Google Scholar] [CrossRef] [PubMed]
- Meier, S.; Arends, D.; Korkuć, P.; Neumann, G.B.; Brockmann, G.A. A genome-wide association study for clinical mastitis in the dual-purpose German Black Pied cattle breed. J. Dairy Sci. 2020, 103, 10289–10298. [Google Scholar] [CrossRef]
- Buttchereit, N.; Stamer, E.; Junge, W.; Thaller, G. Genetic parameters for energy balance, fat/protein ratio, body condition score and disease traits in German Holstein cows. J. Anim. Breed. Genet. 2012, 129, 280–288. [Google Scholar] [CrossRef] [PubMed]
- Klein, S.-L.; Scheper, C.; Brügemann, K.; Swalve, H.H.; König, S. Phenotypic relationships, genetic parameters, genome-wide associations, and identification of potential candidate genes for ketosis and fat-to-protein ratio in German Holstein cows. J. Dairy Sci. 2019, 102, 6276–6287. [Google Scholar] [CrossRef] [PubMed]
- Al-Kanaan, A. Heat Stress Response for Physiological Traits in Dairy and Dual Purpose Cattle Populations on Phenotypic and Genetic Scales. Ph.D. Thesis, Faculty of Organic Agriculture, University of Kassel, Kassel, Germany, January 2016. [Google Scholar]
- Ali, A.K.A.; Shook, G.E. An optimum transformation for somatic cell concentration in milk. J. Dairy Sci. 1980, 63, 487–490. [Google Scholar] [CrossRef]
- Jaeger, M.; Scheper, C.; König, S.; Brügemann, K. Inbreeding and genetic relationships of the endangered dual-purpose black and white cattle breed (DSN) based on own genetic breed percentage calculations. Züchtungskunde 2018, 90, 262–279. [Google Scholar]
- Thienpont, D.; Rochette, F.; van Parijs, O.F.J. Diagnosing Helminthiasis by Coproscopical Examination; Janssen Research Foundation: Beerse, Belgium, 1979; p. 187. [Google Scholar]
- Baermann, G. Eine einfache Methode zur Auffindung von Ankylostoma (Nematoden)-Larven in Erdproben. A simple method for isolation of Ancylostoma (nematode) larvae in soil samples. Geneeskd. Tijdschr. Voor Ned. Indie 1917, 51, 13–137. [Google Scholar]
- SAS Institute, version 9.4; SAS Insitute, Inc.: Cary, NC, USA, 2013.
- Yang, J.; Lee, S.H.; Goddard, M.E.; Visscher, P.M. GCTA: A tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 2011, 88, 76–82. [Google Scholar] [CrossRef]
- Van Raden, P.M. Efficient methods to compute genomic predictions. J. Dairy Sci. 2008, 91, 4414–4423. [Google Scholar] [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M. The genome analysis toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- McLaren, W.; Gil, L.; Hunt, S.E.; Riat, H.S.; Ritchie, G.R.S.; Thormann, A.; Flicek, P.; Cunningham, F. The Ensembl Variant Effect Predictor. Genome Biol. 2016, 17, 122. [Google Scholar] [CrossRef]
- Browning, B.L.; Zhou, Y.; Browning, S.R. A one-penny imputed genome from next-generation reference panels. Am. J. Hum. Genet. 2018, 103, 338–348. [Google Scholar] [CrossRef]
- Korkuć, P.; Arends, D.; Brockmann, G.A. Finding the optimal imputation strategy for small cattle populations. Front. Genet. 2019, 10, 52. [Google Scholar] [CrossRef] [PubMed]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- The Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3000 shared controls. Nature 2007, 447, 661–678. [Google Scholar] [CrossRef] [PubMed]
- Durinck, S.; Moreau, Y.; Kasprzyk, A.; Davis, S.; De Moor, B.; Brazma, A.; Huber, W. BioMart and Bioconductor: A powerful link between biological databases and microarray data analysis. Bioinformatics 2005, 21, 3439–3440. [Google Scholar] [CrossRef] [PubMed]
- Durinck, S.; Spellman, P.; Birney, E.; Huber, W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat. Protoc. 2009, 4, 1184–1191. [Google Scholar] [CrossRef]
- ENSEMBL. Genome Browser. Available online: http://www.ensembl.org/index.html (accessed on 18 April 2021).
- Yates, A.D.; Achuthan, P.; Akanni, W.; Allen, J.; Allen, J.; Alvarez-Jarreta, J.; Amode, M.R.; Armean, I.M.; Azov, A.G.; Bennett, R.; et al. Ensembl 2020. Nucleic Acids Res. 2020, 48, D682–D688. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S.; Sato, Y.; Kawashima, M.; Furmichi, M.; Tanabe, M. Data, information, knowledge and principle: Back to metabolism in KEGG. Nucleic Acids Res. 2014, 42, 199–205. [Google Scholar] [CrossRef]
- Hang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2008, 4, 44–57. [Google Scholar] [CrossRef] [PubMed]
- McRae, K.M.; Stear, M.J.; Good, B.; Keane, O.M. The host immune response to gastrointestinal nematode infection in sheep. Parasite Immunol. 2015, 37, 605–613. [Google Scholar] [CrossRef]
- Liu, L.; Zhou, J.; Chen, C.J.; Zhang, J.; Wen, W.; Tian, J.; Zhang, Z.; Gu, Y. GWAS-based identification of new loci for milk yield, fat, and protein in Holstein cattle. Animals 2020, 10, 2048. [Google Scholar] [CrossRef] [PubMed]
- Luoreng, Z.-M.; Wang, X.-P.; Mei, C.-G.; Zan, L.-S. Expression profiling of peripheral blood miRNA using RNAseq technology in dairy cows with Escherichia coli-induced mastitis. Sci. Rep. 2018, 8, 12693. [Google Scholar] [CrossRef] [PubMed]
- Kolodziejczyk, L.; Siemieniuk, E.; Skrzydlewska, E. Fasciola hepatica: Effects on the antioxidative properties and lipid peroxidation of rat serum. Exp. Parasitol. 2006, 113, 43–48. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.-S.; Sonstegard, T.S.; da Silva, M.V.G.B.; Gasbarre, L.C.; Van Tassel, C.P. Genome-wide scan of gastrointestinal nematode resistance in closed Angus population selected for minimized influence of MHC. PLoS ONE 2015, 10, e0119380. [Google Scholar] [CrossRef]
- Hu, R.-S.; Zhang, F.-K.; Esheikkha, H.M.; Ma, Q.-N.; Ehsan, M.; Zhao, Q.; Zhu, X.-Q. Proteomic profiling of the liver, hepatic lymph nodes, and spleen of buffaloes infected with Fasciola gigantica. Pathogens 2020, 9, 982. [Google Scholar] [CrossRef]
- Rojas-Caraballo, J.; López-Abán, J.; Moreno-Pérez, D.A.; Vicente, B.; Fernández-Soto, P.; del Olmo, E.; Patarroyo, M.A.; Muro, A. Transcriptomic profiling of gene expression during immunization trial against Fasciola hepatica: Identification of genes and pathways involved in conferring immunoprotection in a murine model. BMC Infect. Dis. 2017, 17, 94. [Google Scholar] [CrossRef] [PubMed]
- Loukas, A.; Maizels, R.M. Helminth C-type lectins and host-parasite interactions. Parasitol. Today 2000, 16, 333–339. [Google Scholar] [CrossRef]
- Zhang, X.-X.; Cong, W.; Elsheikha, H.M.; Li, G.-H.; Ma, J.-G.; Huang, W.-Y.; Zhao, Q.; Zhu, X.-Q. De novo transcriptome sequencing and analysis of the juvenile and adult sages of Fasciola gigantica. Infect. Genet. Evol. 2017, 51, 33–40. [Google Scholar] [CrossRef]
- Hagberg, M. Immune Cell Responses to the Cattle Lungworm, Dictyocaulus viviparus. Ph.D. Thesis, University Uppsala, Uppsala, Sweden, 2008. [Google Scholar]
- Escobedo, G.; Roberts, C.W.; Carrero, J.C.; Morales-Montor, J. Parasite regulation by host hormones: An old mechanism of host exploitation? Trends Parasitol. 2005, 21, 588–593. [Google Scholar] [CrossRef]
- Aguayo, V.; Fernandez, B.N.V.; Rodríguez-Valentín, M.; Riz-Jiménez, C.; Ramos-Benítez, M.J.; Méndez, L.B.; Espino, A.M. Fasciola hepatica GST downregulates NF-κB pathway effectors and inflammatory cytokines while promoting survival in a mouse septic shock model. Sci. Rep. 2019, 9, 2275. [Google Scholar] [CrossRef] [PubMed]
- Johnson, D.R.; Saes, J.; Matthews, J.B. Local cytokine responses in Dictyocaulus viviparus infection. Vet. Parsitol. 2005, 128, 309–318. [Google Scholar] [CrossRef]
- Laodim, T.; Elzo, M.A.; Koonawootrittriron, S.; Suwanasopee, T.; Jattawa, D. Pathway enrichment and protein interaction network analysis for milk yield, fat yield and age at first calving in a Thai multibreed dairy population. Asian Australas. J. Anim. Sci. 2019, 32, 508–518. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Shen, D.; Li, C.; Cai, W.; Liu, S.; Yin, H.; Shi, S.; Cao, M.; Zhang, S. Comparative transcriptomic and proteomic analyses identify key genes associated with milk fat traits in Chinese Holstein cows. Front. Genet. 2019, 10, 672. [Google Scholar] [CrossRef] [PubMed]
- Jedlina, L.; Kozak-Ljunggren, M.; Wedrychowicz, H. In vivo studies of the early, peritoneal, cellular and free radical responses in rats infected with Fasciola hepatica by flow cytometric analysis. Exp. Parasitol. 2011, 128, 291–297. [Google Scholar] [CrossRef]
- Fu, Y.; Browne, J.A.; Killick, K.; Mulcahy, G. Network analysis of the systemic response to Fasciola hepatica infection in sheep reveals changes in fibrosis, apoptosis, toll-like receptors 3/4 and B cell function. Front. Immunol. 2017, 8, 485. [Google Scholar] [CrossRef] [PubMed]
- Foster, N.; Elsheikha, H.M. The immune response to parasitic helminths of veterinary importance and its potential manipulation for futre vaccine control strategies. Parasitol. Res. 2012, 110, 1587–1599. [Google Scholar] [CrossRef] [PubMed]
- Walsh, K.P.; Brady, M.T.; Finlay, C.M.; Boon, L.; Mills, K.H.G. Infection with a helminth parasite attenuates autoimmunity through TGF-beta-mediated suppression of Th17 and Th1 responses. J. Immunol. 2009, 183, 1577–1586. [Google Scholar] [CrossRef]
- Brondum, R.F.; Guldbrandtsen, B.; Sahana, G.; Lund, M.S.; Su, G. Strategies for imputation to whole genome sequence using a single or multi-breed reference population in cattle. BMC Genom. 2014, 15, 728. [Google Scholar] [CrossRef] [PubMed]
- Mao, X.; Sahana, G.; De Koning, D.-J.; Guldbrandtsen, B. Genome-wide association studies of growth traits in three dairy cattle breeds using whole-genome sequence data. J. Anim. Sci. 2016, 94, 1426–1437. [Google Scholar] [CrossRef]
- Iso-Touru, T.; Sahana, G.; Guldbrandtsen, B.; Lund, M.S.; Vilkki, J. Genome-wide association analysis of milk yield traits in Nordic Red cattle using imputed whole genome sequence variants. BMC Genet. 2016, 17, 55. [Google Scholar] [CrossRef] [PubMed]
- Hoglund, J.K.; Guldbrandtsen, B.; Su, G.; Thomsen, B.; Lund, M.S. Genome scan detects quantitative trait loci affecting female fertility traits in Danish and Swedish Holstein cattle. J. Dairy Sci. 2009, 92, 2136–2143. [Google Scholar] [CrossRef]
- Fortes, M.R.S.; Suhaimi, A.H.M.S.; Porto-Neto, L.R.; McWilliam, S.M.; Flatscher-Bader, T.; Moore, S.S.; D’Occhio, M.J.; Meira, C.T.; Thomas, M.G.; Snelling, W.M. Post-partum anoestrus in tropical beef cattle: A systems approach combining gene expression and genome-wide association results. Livest. Sci. 2014, 166, 158–166. [Google Scholar] [CrossRef]
- Rosa, L.R.O.; Soares, G.M.; Silveira, L.R.; Boschero, A.C.; Barbosa-Sampaio, H.C.L. ARHGAP21 as a regulator of multiple cellular processes. J. Cell. Physiol. 2018, 233, 8477–8481. [Google Scholar] [CrossRef]
- Laskowski, D.; Sjunnesson, Y.; Humblot, P.; Andersson, G.; Gustafsson, H.; Båge, R. The functional role of insulin in fertility and embryonic development—What can we learn from bovine model? Theriogenology 2016, 86, 457–464. [Google Scholar] [CrossRef]
- Gong, J.; Lee, W.; Garnswothy, P.; Webb, R. Effect of dietary-induced increases in circulating insulin concentrations during the early postpartum period on reproductive function in dairy cows. Reproduction 2002, 123, 419–427. [Google Scholar] [CrossRef] [PubMed]
- Holmberg, M.; Andersson-Eklund, L. Quantitative trait loci affecting fertility and calving traits in Swedish dairy cattle. J. Dairy Sci. 2006, 89, 3664–3671. [Google Scholar] [CrossRef]
- Fortes, M.R.S.; Sneling, W.M.; Reverter, A.; Nagaraj, S.H.; Lehnert, S.; Hawken, R.; DeAtley, K.L.; Peters, K.E.; Silver, G.A.; Rincon, G.; et al. Gene network analyses of first service conception in Brangus heifers: Use of genome and traits associations, hypothalamic-transcriptome information, and transcription factors. J. Anim. Sci. 2012, 90, 2894–2906. [Google Scholar] [CrossRef] [PubMed]
- Moore, S.G.; Pryce, J.E.; Hayes, B.J.; Chamberlain, A.J.; Kemper, K.E.; Berry, D.P.; McCabe, M.; Cormican, P.; Lonergan, P.; Fair, T.; et al. Differentially Expressed Genes in Endometrium and Corpus Luteum of Holstein Cows Selected for High and Low Fertility Are Enriched for Sequence Variants Associated with Fertility. Biol. Reprod. 2016, 94, 1–11. [Google Scholar] [CrossRef]
- Kiser, J.N.; Keuter, E.M.; Seabury, C.M.; Neupane, M.; Moraes, J.G.N.; Dalton, J.; Burns, G.W.; Spencer, T.E.; Neibergs, H.L. Validation of 46 loci associated with female fertility traits in cattle. BMC Genom. 2019, 20, 576. [Google Scholar] [CrossRef]
- May, K.; Sames, L.; Scheper, C.; König, S. Genomic loci for uterine diseases in Holsten cows and impact on milk production and fertility. In Proceedings of the 72nd Annual Meeting of the European Association for Animal Production, Davos, Switzerland, 30 August–3 September 2021; Wageningen Academic Publishers: Wageningen, The Netherlands, 2021. [Google Scholar]
- Timperio, A.M.; D’Alessandro, A.; Pariset, L.; D’Amici, G.M.; Valentini, A.; Zolla, L. Comparative proteomics and transcriptomics analyses of livers from two different Bos taurus breeds: “Chianina and Holstein Friesian”. J. Proteom. 2009, 73, 309–322. [Google Scholar] [CrossRef] [PubMed]
- Lehnert, S.A.; Reverter, A.; Byrne, K.A.; Wang, Y.; Nattrass, G.S.; Hudson, N.J.; Greenwood, P.L. Gene expression studies of developing bovine longissimus muscle from two different beef cattle breeds. BMC Dev. Biol. 2007, 7, 95. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wurmser, C.; Pausch, H.; Jung, S.; Reinhardt, F.; Tetens, J.; Thaller, G.; Fries, R. Identification and dissection of four major QTL affecting milk fat content in the German Holstein-Friesian population. PLoS ONE 2012, 7, e40711. [Google Scholar] [CrossRef]
- Bhat, S.A.; Ahmad, S.M.; Ibeagha-Awemu, E.I.; Bhat, B.A.; Dar, M.A.; Mumtaz, P.T.; Shah, R.A.; Ganai, N.A. Comparative transcriptome analysis of mammary epithelial cells at different stages of lactation reveals wide differences in gene expression and pathways regulating milk synthesis between Jersey and Kashmiri cattle. PLoS ONE 2019, 14, e011773. [Google Scholar] [CrossRef] [PubMed]
- Braz, C.U.; Rowan, T.N.; Schnabel, R.D.; Decker, J.E. Extensive genome-wide association analyses identify genotype-by-environment interactions of growth traits in Simmental cattle. bioRxiv 2020. [Google Scholar] [CrossRef]
- AnimalQTLdb. Available online: https://www.animalgenome.org/cgi-bin/QTLdb/index (accessed on 14 May 2021).
- Liu, Y.; Xu, L.; Wang, Z.; Xu, L.; Chen, Y.; Zhang, L.; Xu, L.; Gao, X.; Zhu, B.; Li, J. Genomic prediction and association analysis with models including dominance effects for important traits in Chinese Simmental beef cattle. Animal 2019, 9, 1055. [Google Scholar] [CrossRef]
- Howard, J.T.; Pryce, J.E.; Baes, C.; Maltecca, C. Invited review: Inbreeding in the genomics era: Inbreeding, inbreeding depression, and management of genomic variability. J. Dairy Sci. 2017, 100, 6009–6024. [Google Scholar] [CrossRef] [PubMed]
- Sorensen, A.C.; Madsen, P.; Sorenzen, M.K.; Berg, P. Udder health shows inbreeding depression in Danish Holsteins. J. Dairy Sci. 2006, 89, 4077–4082. [Google Scholar] [CrossRef]
- May, K.; Weimann, C.; Scheper, C.; Strube, S.; König, S. Allele substitution and dominance effects of CD166/ALCAM gene polymorphisms for endoparasite resistance and test-day traits in a small cattle population using logistic regression analyses. Mamm. Genome 2019, 30, 301–317. [Google Scholar] [CrossRef] [PubMed]
- Fugate, R.T.; Dauten, L.H.; Wiggans, G.R.; White, H.M. Determination of Single Nucleotide Polymorphisms Associated with Subclinical Ketosis in Jersey Cattle. Available online: https://heatherwhite.dysci.wisc.edu/wp-content/uploads/sites/164/2014/07/ADSA-Jersey-SNP-poster-FINAL.pdf (accessed on 19 May 2021).
- Littlejohn, M.D.; Walker, C.G.; Ward, H.E.; Lehnert, K.B.; Snell, R.G.; Verkerk, G.A.; Spelman, R.J.; Clark, D.A.; Davis, S.R. Effects of reduced frequency of milk removal on gene expression in the bovine mammary gland. Physiol. Genom. 2010, 41, 21–32. [Google Scholar] [CrossRef]
- Buitenhuis, B.; Rontved, C.M.; Edwards, S.M.; Ingvartsen, K.; Sorensen, P. In depth analysis of genes and pathways of the mammary gland involve in the pathogenesis of bovine Escherichia coli-mastitis. BMC Genom. 2011, 12, 130. [Google Scholar] [CrossRef]
- Yu, C.; Wang, F.; Jin, C.; Huang, X.; Miller, D.L.; Basilico, C.; McKeehan, W.L. Role of fibroblast growth factor type 1 and 2 in carbon tetrachloride-induced hepatic injury and fibrogenesis. Am. J. Pathol. 2003, 163, 1653–1662. [Google Scholar] [CrossRef]
- Sulaiman, A.A.; Zolnierczyk, K.; Japa, O.; Owen, J.P.; Maddison, B.C.; Emes, R.D.; Hodgkinson, J.E.; Gough, K.C.; Flynn, R.J. A trematode parasite derived growth factor binds and exerts influences on host immune functions via host cytokine receptor complexes. PLoS Pathog. 2016, 12, e1005991. [Google Scholar] [CrossRef] [PubMed]

| Trait 1 | No. of Records | No. of Cows | No. of Cows with Sequence Level Genotypes | Mean 2 | SD 2 | Min. 2 | Max. 2 |
|---|---|---|---|---|---|---|---|
| CTFS | 1683 | 1683 | 1683 | 78.31 | 27.44 | 26.0 | 241.0 |
| NR56 | 1683 | 1683 | 1683 | 0.64 | 0.48 | 0 | 1.0 |
| SCS | 1638 | 1638 | 1638 | 2.58 | 1.48 | −1.06 | 8.88 |
| FPR | 1638 | 1638 | 1638 | 1.18 | 0.18 | 0.45 | 2.42 |
| FEC-GIN | 1997 | 1166 | 142 | 11.35 | 22.57 | 0 | 225.0 |
| FEC-FH | 2006 | 1166 | 142 | 0.61 | 3.64 | 0 | 89.0 |
| FLC-DV | 1988 | 1163 | 142 | 0.17 | 2.14 | 0 | 46.0 |
| BTA | Gene Position 1 | No. of SVs within/Close to Gene 2 | Position of Maximum Association (p-Value) | rs Number of Maximum Association | Gene Name |
|---|---|---|---|---|---|
| CTFS | |||||
| 12 | 43,998,992–44,525,321 | 3/0 | 44,156,560 (2.93 × 10−6) * | rs136060929 | KLHL1 |
| 13 | 25,463,655–25,592,062 | 5/10 | 25,577,261 (2.25 × 10−7) * | rs384930569 | ARHGAP21 |
| 15 | 58,214,580–58,224,553 | 0/1 | 58,258,453 (1.32 × 10−6) * | rs41777070 | LIN7C |
| 78,914,154–78,915,587 | 0/1 | 78,929,182 (2.08 × 10−6) * | rs379801720 | ENSBTAG00000052005 | |
| 79,013,311–79,014,249 | 0/1 | 78,929,182 (2.08 × 10−6) * | rs379801720 | OR5BE5 | |
| 28 | 43,760,392–43,804,579 | 10/0 | 43,796,638 (1.37 × 10−6) * | rs42155599 | CHAT |
| NR56 | |||||
| 8 | 96,644,114–96,759,752 | 2/0 | 96,724,190 (4.07 × 10−7) * | rs110809463 | ZNF462 |
| 11 | 74,126,883–74,224,519 | 1/0 | 74,190,369 (8.30 × 10−7) * | rs383197946 | EFR3B |
| 20 | 56,953,217–57,071,806 | 0/1 | 56,867,118 (1.34 × 10−6) * | rs207515592 | MARCH11 |
| BTA | Gene Position 1 | No. of SVs within/Close to Gene 2 | Position of Maximum Association (p-Value) | rs Number of Maximum Association | Gene Name |
|---|---|---|---|---|---|
| SCS | |||||
| 1 | 5,689,903–5,690,709 | 0/1 | 5,693,321 (2.49 × 10−6) * | - | KRTAP24-1 |
| 5,770,225–5,772,218 | 0/1 | 5,693,321 (2.49 × 10−6) * | - | CLDN8 | |
| 9 | 15,242,718–15,373,861 | 1/0 | 15,301,365 (2.76 × 10−6) * | - | SENP6 |
| 11 | 101,323,401–101,414,345 | 0/1 | 101,435,611 (2.06 × 10−6) * | rs211669575 | NUP214 |
| 101,450,754–101,465,047 | 0/1 | 101,435,611 (2.06 × 10−6) * | rs211669575 | FAM78A | |
| 101,696,993–101,834,040 | 2/0 | 101,761,967 (1.30 × 10−7) ** | rs137783421 | RAPGEF1 | |
| 13 | 61,874,277–61,955,381 | 0/1 | 61,959,686 (1.04 × 10−6) * | rs211178277 | NOL4L |
| 23 | 42,945,450–43,017,711 | 1/0 | 42,9749,92 (4.20 × 10−7) * | rs876215027 | RANBP9 |
| 25 | 6,224,841–6,638,491 | 2/45 | 6,654,595 (2.58 × 10−7) * | rs136166815 | RBFOX1 |
| FPR | |||||
| 9 | 83,287,300–83,713,833 | 1/0 | 83,557,515 (3.16 × 10−7) * | rs467161057 | GRM1 |
| 11 | 6,481,883–6,555,244 | 0/4 | 6,464,004 (3.06 × 10−7) * | rs134892674 | ENSBTAG00000054755 |
| 12 | 11,675,264–12,071,822 | 15/1 | 11,736,920 (1.39 × 10−7) ** | rs439994366 | VWA8 |
| 12,256,326–12,294,096 | 0/1 | 12,195,520 (2.71 × 10−6) * | rs209487147 | ENSBTAG00000053271 | |
| 12,410,713–12,461,551 | 0/9 | 12,496,948 (1.49 × 10−6) * | rs208793423 | AKAP11 | |
| 13 | 58,456,060–58,457,612 | 0/2 | 58,382,430 (2.12 × 10−6) * | rs42020993 | ENSBTAG00000054668 |
| 60,191,141–60,234,885 | 1/0 | 60,228,471 (2.65 × 10−6) * | - | ANGPT4 | |
| 61,107,684–61,147,486 | 2/0 | 61,145,994 (8.51 × 10−7) * | - | HM13 | |
| 62,381,695–62,408,299 | 0/1 | 62,434,039 (1.91 × 10−6) * | - | BPIFB4 | |
| 62,503,682–62,514,220 | 0/1 | 62,434,039 (1.91 × 10−6) * | - | BPIFA2A | |
| 62,645,078–62,655,447 | 0/1 | 62,659,594 (2.82 × 10−6) * | - | BPIFA2B | |
| 62,676,036–62,685,906 | 0/1 | 62,659,594 (2.82 × 10−6) * | - | ENSBTAG00000031373 | |
| 65,100,388–65,148,653 | 2/0 | 65,123,915 (1.50 × 10−6) * | rs109380861 | CNBD2 | |
| 72,891,431–72,905,729 | 1/0 | 72,897,116 (1.46 × 10−6) * | rs137243257 | TTPAL | |
| 75,255,202–75,339,020 | 2/0 | 75,261,912 (2.42 × 10−6) * | rs137115876 | SLC13A3 | |
| 19 | 30,045,106–30,177,093 | 0/1 | 30,186,388 (2.15 × 10−6) * | - | ENSBTAG00000049618 |
| 27 | 36,454,819–36,470,324 | 0/12 | 36,520,069 (6.89 × 10−7) * | rs211250281 | GINS4 |
| 36,522,605–36,539,773 | 1/12 | 36,520,069 (6.89 × 10−7) * | rs211250281 | GPAT4 | |
| 41,461,703–41,896,044 | 1/0 | 41,614,645 (2.30 × 10−6) * | rs135231909 | THRB |
| Pathway | KEGG Entry | Trait 1 | Candidate Gene (BTA) | Possible Association of Pathway with Trait according to Literature |
|---|---|---|---|---|
| B cell receptor signaling pathway | bta04662 | RES-GIN | MALT1 (24) | Involvement of B cells in immune response to gastrointestinal nematodes in ruminants [48]. |
| Calcium signaling pathway | FPR | GRM1 (9) | Pathway associated with milk fat content in Holstein cattle [49]. | |
| Cell adhesion molecules | bta04514 | SCS | CLDN8 (1) | Identification of the cell adhesin molecules pathway for Escherichia coli-induced mastitis in RNAseq analysis [50]. |
| RES-DV | CD226 (24) | Cell adhesion molecules pathway identified for F. hepatica infections in cattle [22]. | ||
| cGMP-PKG signaling pathway | bta04022 | RES-FH | PLCB1 (13), PRKG1 (26), ADRB3 (27) | Cyclic GMP (cGMP) is an intracellular messenger that mediates the action of nitric oxide, which is increasingly produced by leukocytes during F. hepatica infections [51]. |
| Chemokine signaling pathway | bta04062 | RES-GIN | ADCY1 (4) | Pathway was associated with resistance to gastrointestinal nematode infections in Angus cattle [52]. |
| RES-FH | PLCB1 (13), PTK2 (14) | Pathway was associated with Fasciola gigantica infections in Buffalo via proteomics analysis [53]; pathway associated with F. hepatica infections in mice [54]. | ||
| C-type lectin receptor signaling pathway | bta04625 | RES-GIN | MALT1 (24) | C-type lectin receptors are involved in innate and adaptive immunity to pathogens [46]; helminth C-type lectins are involved in host–parasite interactions [55]. |
| Cytokine–cytokine receptor interaction | bta04060 | RES-GIN | ACVR1C (2) | Pathway was associated with resistance to GIN infections in Angus cattle [52] and in German Black Pied dairy cattle [22]; up- and downregulation of cytokines as immune mechanism in cattle in response to helminth infections [56,57]. |
| RES-DV | BMPR1B (6) | |||
| RES-FH | IL21 (17), PRP9 (23), PRP-VII (23) | |||
| Estrogen signaling pathway | bta04915 | RES-GIN | ADCY1 (4) | Increase in reproduction rate of helminths as a result of increasing metabolism of 17-ß-estradiol in the host [58] |
| RES-FH | PLCB1 (13), KRT31 (19), KRT34 (19) | |||
| JAK-STAT signaling pathway | bta04630 | RES-FH | IL21 (17), PRP9 (23), PRP-VII (23) | F. hepatica excretory-secretory antigens suppress multiple proteins participating in the JAK-STAT signaling pathway in mice [59]. |
| RES-DV | CCND3 (23), SOCS6 (24) | JAK-STAT pathway is the principal signaling mechanism for cytokines involved in D. viviparus infections [46,60]. | ||
| Leukocyte transendothelial migration | bta04670 | RES-FH | PTK2 (14) | Pathway was associated with Fasciola gigantica infections in buffalo via proteomics analysis [53]. |
| SCS | CLDN8 (1) | Identification of the Leukocyte transendothelial migration pathway for Escherichia coli-induced mastitis in RNAseq analysis [50]. | ||
| MAPK signaling pathway | bta04010 | FPR | ANGPT4 (13) | Pathway associated with milk fat traits in dairy cattle [61,62]. |
| Natural killer cell mediated cytotoxicity | bta04650 | RES-FH | PTK2 (14) | Natural killer cells are lymphocytes of the innate immune response involved in host defense against infections with parasites [46]; cytotoxic natural killer cells were involved in early stage of infection by F. hepatica in rats [63]; pathway identified for F. hepatica infections in mice [54]. |
| Neuroactive ligand-receptor interaction | bta04080 | FPR | GRM1 (9), THRB (27) | Pathway associated with milk fat traits in dairy cattle [49] |
| NF-kappa B signaling pathway | bta04064 | RES-GIN | MALT1 (24) | Family of transcription factors regulating genes involved in immunity [46]; pathway associated with F. hepatica infections in sheep [64]. |
| NOD-like receptor signaling pathway | bta04621 | RES-FH | PLCB1 (13) | Family of pattern recognition receptors responsible for various pathogens and generating innate immune response [46]. |
| Phospholipase D signaling pathway | bta04072 | FPR | GRM1 (9) | Phospholipase D is an essential enzyme for the production of phosphatidic acid, a key intermediate in milk fat synthesis during lactation [61]. |
| PI3K-Akt signaling pathway | bta04151 | RES-GIN | PHLPP1 (24) | Pathway has important functions in cellular immune response [46]. |
| RES-FH | PTK2 (14), PRP9 (23), PRP-VII (23), EIF4EBP1 (27), FGFR1 (27) | |||
| RES-DV | CCND3 (23), LAMA3 (24) | |||
| FPR | ANGPT4 (13) | Pathway associated with milk fat traits in dairy cattle [62]. | ||
| Rap 1 signaling pathway | bta04015 | FPR | ANGPT4 (13) | Pathway associated with milk fat traits in dairy cattle [61]. |
| Ras signaling pathway | bta04014 | FPR | ANGPT4 (13) | Pathway associated with milk fat traits in dairy cattle [61]. |
| T cell receptor signaling pathway | bta04660 | RES-GIN | MALT1 (24) | Involvement of B cells in immune response to gastrointestinal nematodes in ruminants [48]. |
| TGF-β signaling pathway | bta04350 | RES-GIN | ACVR1C (2) | TGF-ß involved in host immune response during Ostertagia ostertagi (GIN species) infections [65]; pathway is associated in host–F. hepatica interactions: binding of F. hepatica growth factors to host TGF- ß receptors and triggering SMAD (Sma-and Mad-related proteins) in host leukocytes [56]. |
| RES-FH | SMAD4 (24) | |||
| RES-DV | BMPR1B (6) | |||
| Th17 cell differentiation | bta04659 | RES-FH | IL21 (17), SMAD4 (4) | F. hepatica-induced TGF-ß suppresses Th17 responses in infected mice [66]. |
| BTA | RES-GIN | RES-FH | RES-DV |
|---|---|---|---|
| 1 | ENSBTAG00000048985 | - | - |
| 2 | ACVR1, ACVR1C, CNTNAP5, CPS1, ENSBTAG00000054211, ENSBTAG00000040367, ENSBTAG00000051630, GPD2, KIF5C, LANCL1, LRP1B, LYPD6B, MAP2, NR4A2, UNC80 | CRYGA, CRYGB | ENSBTAG00000033143 *, ENSBTAG00000050185, ENSBTAG00000049959, ENSBTAG00000055116 FAM124B *, GALNT13, GPR55 *, SPATA3 * |
| 3 | PDE4B | CDC14A, ENSBTAG00000037539, KCNN3, TTC4 | - |
| 4 | ADCY1 | CNPY1*, HTR5A *, LMBR1, MNX1, NCAPG2 *, PTPRN2 *, RBM33 *, UBE3C | ESYT2 |
| 5 | - | - | ARHGDIB *, ASCL1, DRAM1, EMP1 *, FAM234B *, GNPTAB, GRIN2B *, LRP6, NELL2, PDE6H *, SLC38A1, TPH2 *, UTP20, YAF2, ZCRB1 |
| 6 | NDST4 | GPRIN3 | BMPR1B *, KCNIP4, PDLIM5 * |
| 7 | MEGF10 | ADAMTS19, AADAT, KIAA0825, MAN2A1, TMEM232 * | HAND1 |
| 8 | ENSBTAG00000052065 * | - | ALDH1A1, ENSBTAG00000052698 |
| 9 | - | LCA5, SH3BGRL2 | ENSBTAG00000055087 *, SASH1 *, TBXT, UST * |
| 10 | - | ENSBTAG00000050159 *, TMEM87A | ABCD4, DDHD1, FERMT2, VRTN |
| 11 | - | EXOC6B*, FAM98A *, REEP1* | ENSBTAG00000009599, LCN10, NELFB, RALGDS, TOR4A |
| 13 | - | PLCB1, HAO1 | SPINT3 |
| 14 | CNBD1 | PTK2 | RALYL |
| 15 | - | ARHGAP20*, ALX4*, FAM111B *, GLYATL2 * | CNTN5* |
| 16 | SYT14 | ENSBTAG00000053468 | SOX13 * |
| 17 | - | IL21 | ENSBTAG00000033967, ENSBTAG00000055004, RAB36, RSPH14 |
| 18 | APRT, CBFA2T3, DNAJA2, GALNS, ENSBTAG00000048593, ENSBTAG00000048735, NETO2, PHKB | - | - |
| 19 | HELZ | KRT31, KRT34 | |
| 20 | - | - | DAB2, ENSBTAG00000047333, ENSBTAG00000000617, ENSBTAG00000049964* |
| 21 | - | - | APOPT1, ENSBTAG00000035184, ENSBTAG00000052298, ENSBTAG00000003957, KLC1, MCTP2, PPP1R13B, SNRPA1, XRCC3, ZFYVE21 |
| 23 | ENSBTAG00000048946, ENSBTAG00000053227, KHDRBS2 *, PRP9, PRP-VII, TFAP2D, TRERF1 | CCND3, FOXP4, LRFN2, TAF8, TRERF1, UBR2 | |
| 24 | ALPK2*, MALT1, NEDD4L *, PHLPP1, TSHZ1, ZCCHC2, ZNF532 | ELAC1, MEX3C, PIK3C3, SMAD4 | CD226 *, CDH19, C24H18orf63, CYB5A *, DIPK1C, DOK6 *, DTNA *, ENSBTAG00000049503, LAMA3 *, NPC1, OSBPL1A, PIK3C3 *, RTTN *, RIOK3 *, SOCS6 *, TMEM241, WDR7 |
| 26 | BCL2, CRTAC1 *, ENSBTAG00000048707, IDE *, KIF11, PCDH15, TNKS2 | ATRNL1, EXOC6, HHEX, PCDH15, PRKG1 | - |
| 27 | - | ADRB3, EIF4EBP1, FGFR1*, ZNF385D | THRB |
| 28 | - | ACTA1, ENSBTAG00000048654 | ARHGAP22 |
| 29 | - | PRMT3, SLC6A5 | ENSBTAG00000008274, ENSBTAG00000050398 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wolf, M.J.; Yin, T.; Neumann, G.B.; Korkuć, P.; Brockmann, G.A.; König, S.; May, K. Genome-Wide Association Study Using Whole-Genome Sequence Data for Fertility, Health Indicator, and Endoparasite Infection Traits in German Black Pied Cattle. Genes 2021, 12, 1163. https://doi.org/10.3390/genes12081163
Wolf MJ, Yin T, Neumann GB, Korkuć P, Brockmann GA, König S, May K. Genome-Wide Association Study Using Whole-Genome Sequence Data for Fertility, Health Indicator, and Endoparasite Infection Traits in German Black Pied Cattle. Genes. 2021; 12(8):1163. https://doi.org/10.3390/genes12081163
Chicago/Turabian StyleWolf, Manuel J., Tong Yin, Guilherme B. Neumann, Paula Korkuć, Gudrun A. Brockmann, Sven König, and Katharina May. 2021. "Genome-Wide Association Study Using Whole-Genome Sequence Data for Fertility, Health Indicator, and Endoparasite Infection Traits in German Black Pied Cattle" Genes 12, no. 8: 1163. https://doi.org/10.3390/genes12081163
APA StyleWolf, M. J., Yin, T., Neumann, G. B., Korkuć, P., Brockmann, G. A., König, S., & May, K. (2021). Genome-Wide Association Study Using Whole-Genome Sequence Data for Fertility, Health Indicator, and Endoparasite Infection Traits in German Black Pied Cattle. Genes, 12(8), 1163. https://doi.org/10.3390/genes12081163






