i4mC-Deep: An Intelligent Predictor of N4-Methylcytosine Sites Using a Deep Learning Approach with Chemical Properties
Abstract
:1. Introduction
2. Materials and Methods
2.1. Benchmark Dataset
2.2. Deep Learning Approach
2.3. Evaluation Measures
3. Result and Discussion
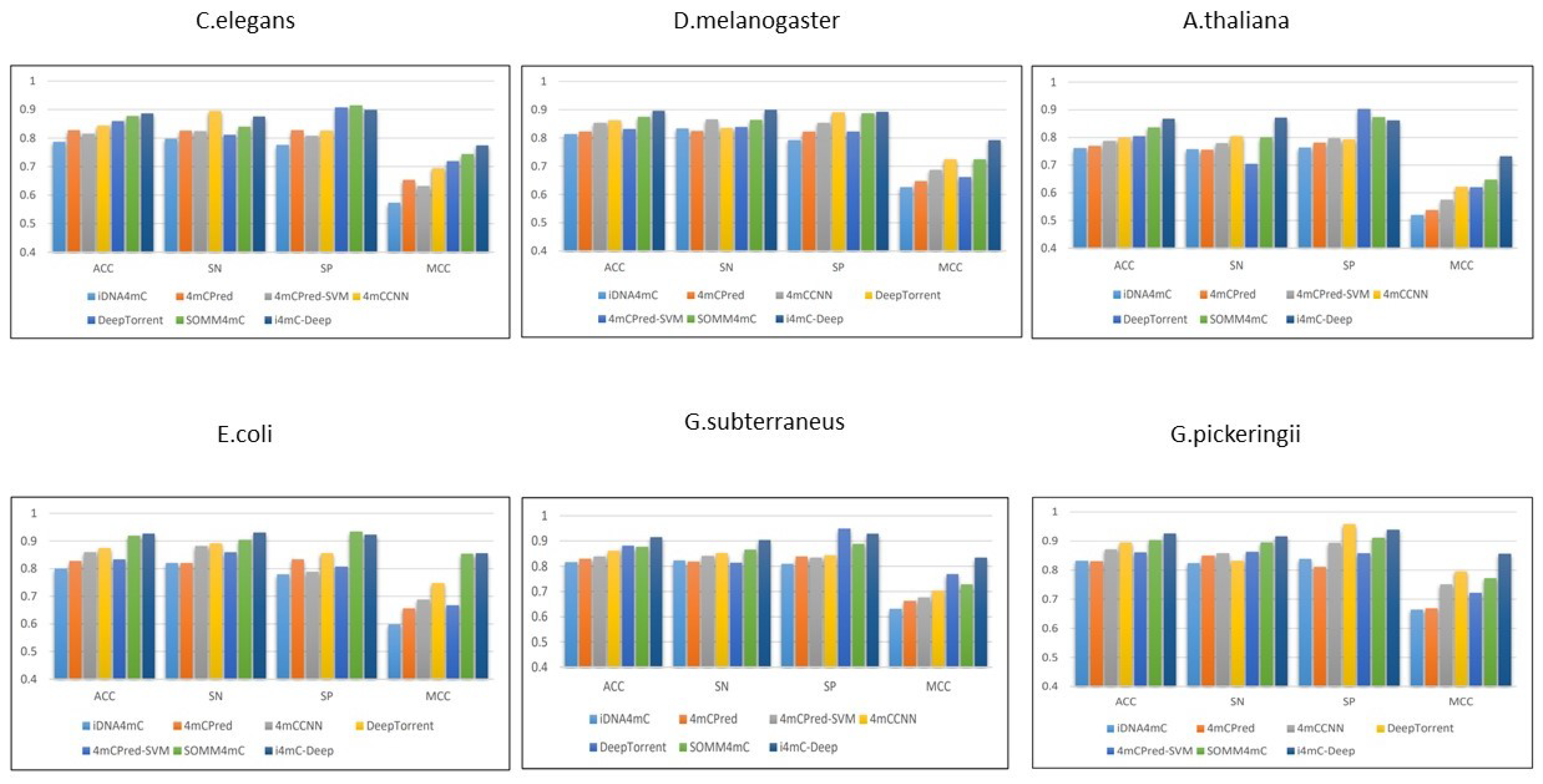
3.1. Comparison with Other State-of-the-Art Tools
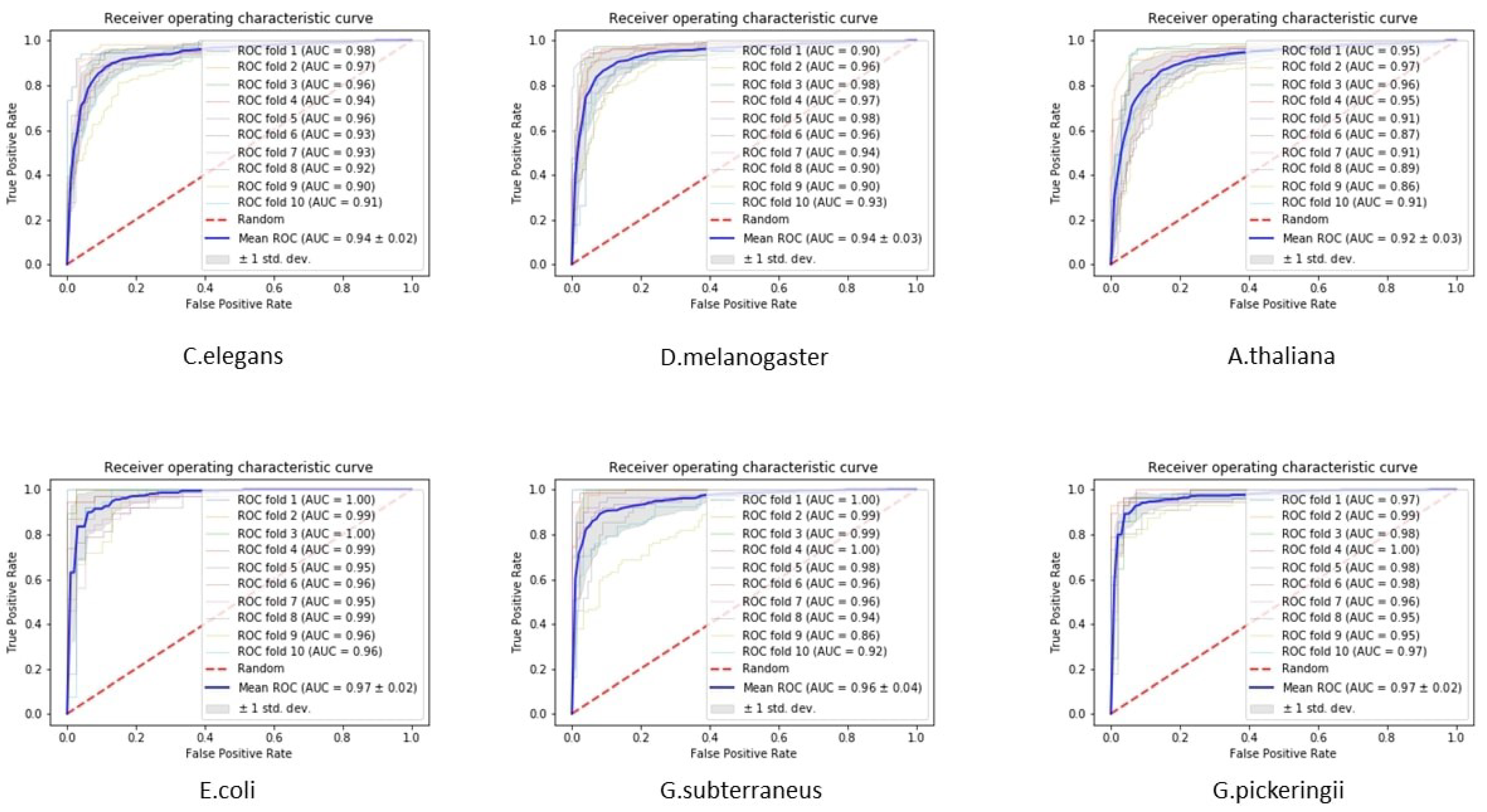
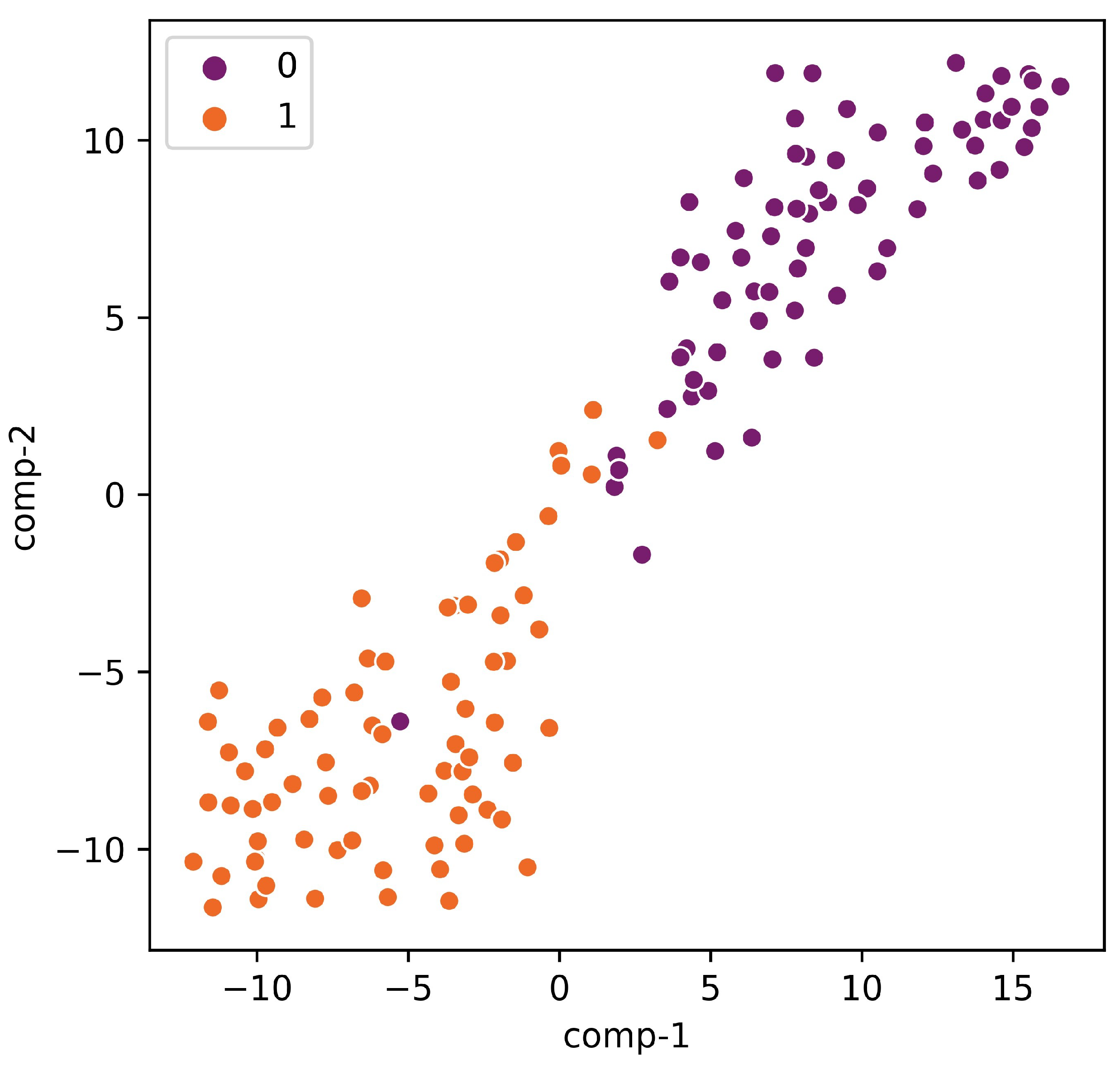
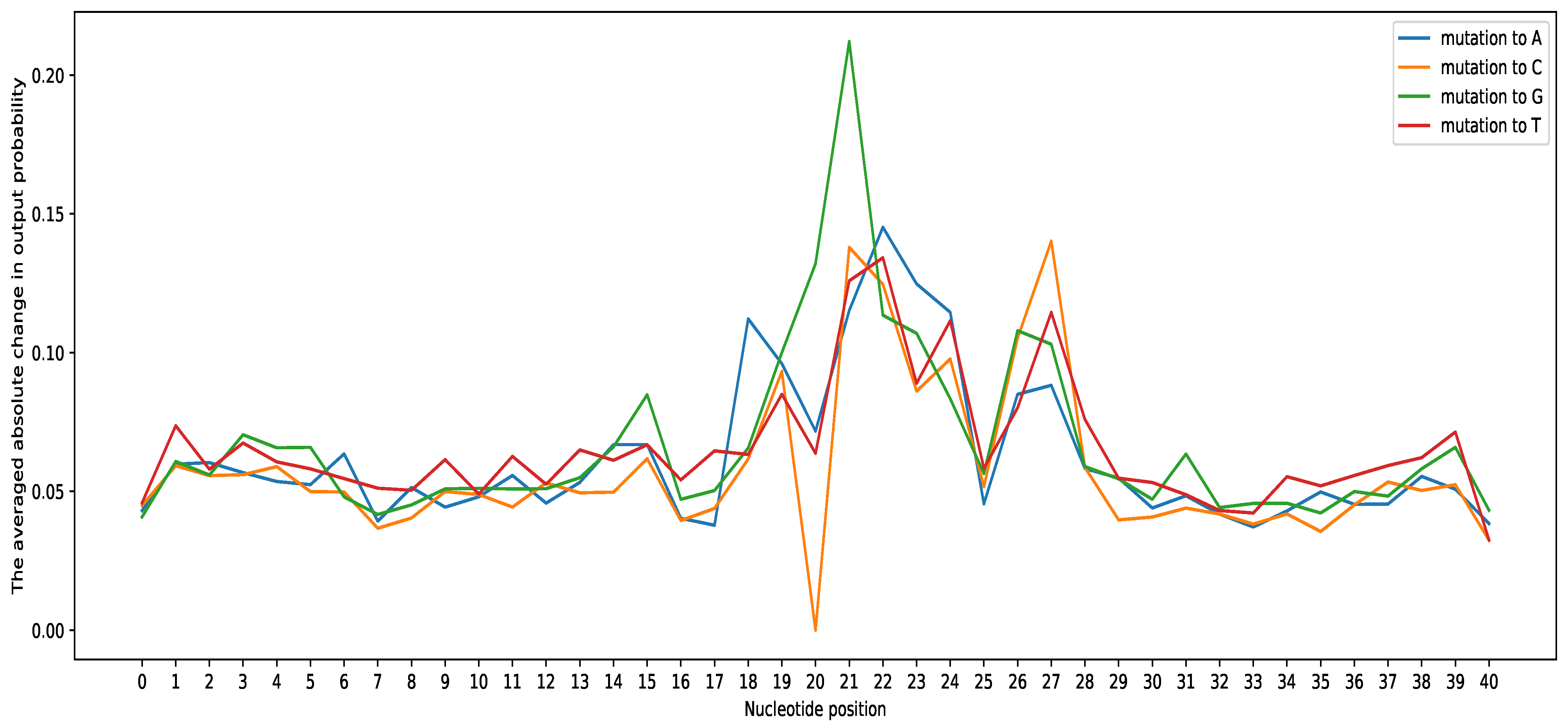
3.2. Interpretation of the Proposed Tool
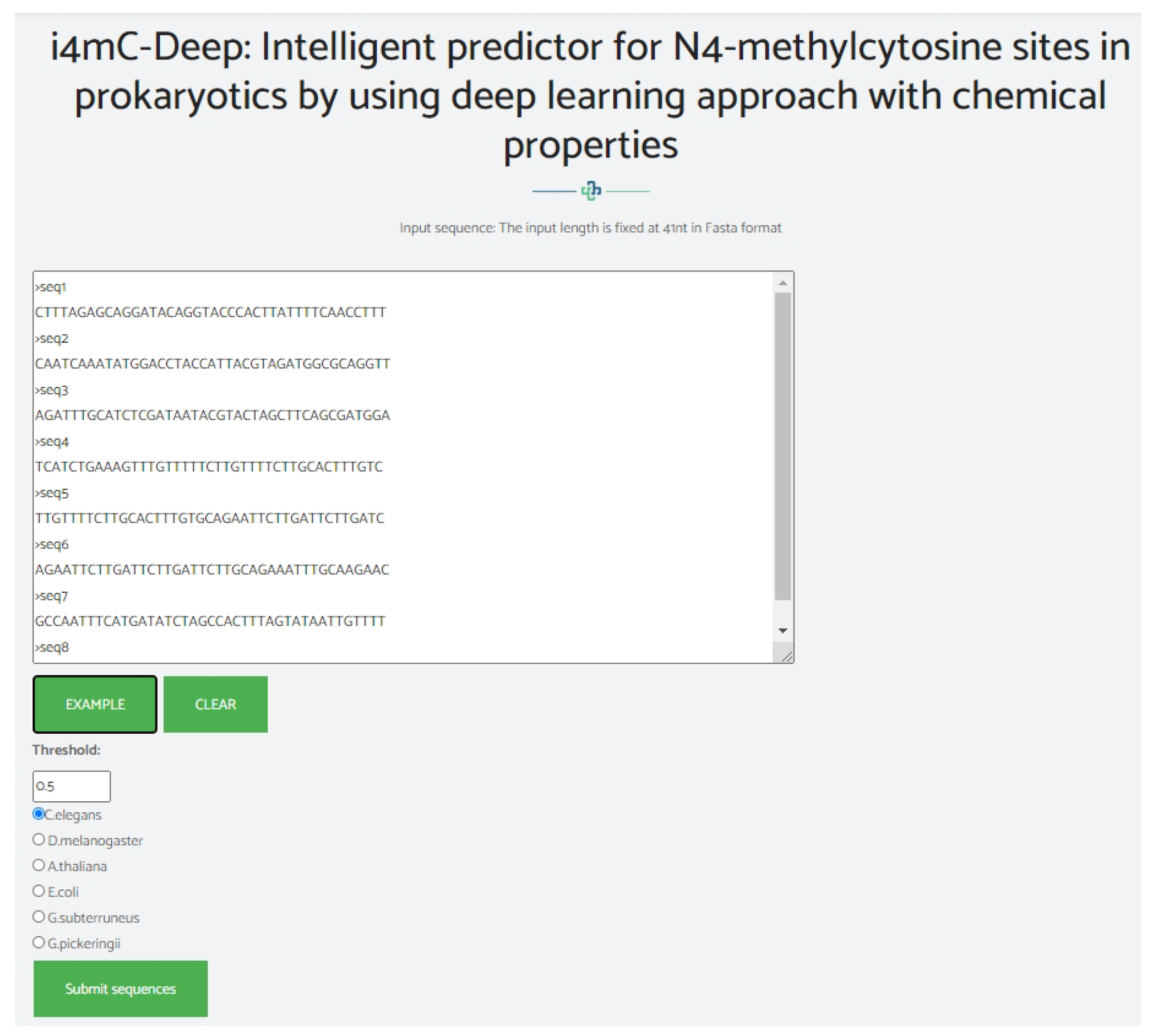
4. Web-Server
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Schübeler, D. Function and information content of DNA methylation. Nature 2015, 517, 321–326. [Google Scholar] [CrossRef] [PubMed]
- Rathi, P.; Maurer, S.; Summerer, D. Selective recognition of N 4-methylcytosine in DNA by engineered transcription-activator-like effectors. Philos. Trans. R. Soc. B Biol. Sci. 2018, 373, 20170078. [Google Scholar] [CrossRef] [Green Version]
- Pataillot-Meakin, T.; Pillay, N.; Beck, S. 3-methylcytosine in cancer: An underappreciated methyl lesion? Epigenomics 2016, 8, 451–454. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davis, B.M.; Chao, M.C.; Waldor, M.K. Entering the era of bacterial epigenomics with single molecule real time DNA sequencing. Curr. Opin. Microbiol. 2013, 16, 192–198. [Google Scholar] [CrossRef] [Green Version]
- Gu, J.; Stevens, M.; Xing, X.; Li, D.; Zhang, B.; Payton, J.E.; Oltz, E.M.; Jarvis, J.N.; Jiang, K.; Cicero, T.; et al. Mapping of variable DNA methylation across multiple cell types defines a dynamic regulatory landscape of the human genome. G3 Genes Genomes Genet. 2016, 6, 973–986. [Google Scholar] [CrossRef] [Green Version]
- Robertson, K.D. DNA methylation and human disease. Nat. Rev. Genet. 2005, 6, 597–610. [Google Scholar] [CrossRef]
- Jones, P.A. Functions of DNA methylation: Islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 2012, 13, 484–492. [Google Scholar] [CrossRef]
- Yao, B.; Jin, P. Cytosine modifications in neurodevelopment and diseases. Cell. Mol. Life Sci. 2014, 71, 405–418. [Google Scholar] [CrossRef] [Green Version]
- Ling, C.; Groop, L. Epigenetics: A molecular link between environmental factors and type 2 diabetes. Diabetes 2009, 58, 2718–2725. [Google Scholar] [CrossRef] [Green Version]
- Cheng, X. DNA modification by methyltransferases. Curr. Opin. Struct. Biol. 1995, 5, 4–10. [Google Scholar] [CrossRef]
- Chen, K.; Zhao, B.S.; He, C. Nucleic acid modifications in regulation of gene expression. Cell Chem. Biol. 2016, 23, 74–85. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doherty, R.; Couldrey, C. Exploring genome wide bisulfite sequencing for DNA methylation analysis in livestock: A technical assessment. Front. Genet. 2014, 5, 126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flusberg, B.A.; Webster, D.R.; Lee, J.H.; Travers, K.J.; Olivares, E.C.; Clark, T.A.; Korlach, J.; Turner, S.W. Direct detection of DNA methylation during single-molecule, real-time sequencing. Nat. Methods 2010, 7, 461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boch, J.; Bonas, U. Xanthomonas AvrBs3 family-type III effectors: Discovery and function. Annu. Rev. Phytopathol. 2010, 48, 419–436. [Google Scholar] [CrossRef] [PubMed]
- Ilyas, T.; Khan, A.; Umraiz, M.; Kim, H. SEEK: A Framework of Superpixel Learning with CNN Features for Unsupervised Segmentation. Electronics 2020, 9, 383. [Google Scholar] [CrossRef] [Green Version]
- Khan, A.; Ilyas, T.; Umraiz, M.; Mannan, Z.I.; Kim, H. CED-Net: Crops and Weeds Segmentation for Smart Farming Using a Small Cascaded Encoder-Decoder Architecture. Electronics 2020, 9, 1602. [Google Scholar] [CrossRef]
- Nizami, I.F.; ur Rehman, M.; Majid, M.; Anwar, S.M. Natural scene statistics model independent no-reference image quality assessment using patch based discrete cosine transform. Multimed. Tools Appl. 2020, 79, 26285–26304. [Google Scholar] [CrossRef]
- Graves, A.; Schmidhuber, J. Framewise phoneme classification with bidirectional LSTM and other neural network architectures. Neural Networks 2005, 18, 602–610. [Google Scholar] [CrossRef]
- Sundermeyer, M.; Alkhouli, T.; Wuebker, J.; Ney, H. Translation modeling with bidirectional recurrent neural networks. In Proceedings of the 2014 Conference on Empirical Methods in Natural Language Processing (EMNLP), Doha, Qatar, 25–29 October 2014; pp. 14–25. [Google Scholar]
- Tayara, H.; Chong, K. Improved Predicting of The Sequence Specificities of RNA Binding Proteins by Deep Learning. IEEE Acm Trans. Comput. Biol. Bioinform. 2020. [Google Scholar] [CrossRef]
- Rehman, M.U.; Chong, K.T. DNA6mA-MINT: DNA-6mA modification identification neural tool. Genes 2020, 11, 898. [Google Scholar] [CrossRef]
- Tayara, H.; Tahir, M.; Chong, K.T. iSS-CNN: Identifying splicing sites using convolution neural network. Chemom. Intell. Lab. Syst. 2019, 188, 63–69. [Google Scholar] [CrossRef]
- Alam, W.; Ali, S.D.; Tayara, H.; Chong, K.T. A CNN-Based RNA N6-Methyladenosine Site Predictor for Multiple Species Using Heterogeneous Features Representation. IEEE Access 2020, 8, 138203–138209. [Google Scholar] [CrossRef]
- Ali, S.D.; Alam, W.; Tayara, H.; Chong, K. Identification of Functional piRNAs Using a Convolutional Neural Network. IEEE Acm Trans. Comput. Biol. Bioinform. 2020. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Yang, H.; Feng, P.; Ding, H.; Lin, H. iDNA4mC: Identifying DNA N4-methylcytosine sites based on nucleotide chemical properties. Bioinformatics 2017, 33, 3518–3523. [Google Scholar] [CrossRef]
- He, W.; Jia, C.; Zou, Q. 4mCPred: Machine learning methods for DNA N4-methylcytosine sites prediction. Bioinformatics 2019, 35, 593–601. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.; Luan, S.; Nagai, L.A.E.; Su, R.; Zou, Q. Exploring sequence-based features for the improved prediction of DNA N4-methylcytosine sites in multiple species. Bioinformatics 2019, 35, 1326–1333. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Lang, K.; Zhang, G.; Fan, X.; Chen, Y.; Pian, C. SOMM4mC: A second-order Markov model for DNA N4-methylcytosine site prediction in six species. Bioinformatics 2020, 36, 4103–4105. [Google Scholar] [CrossRef] [PubMed]
- Khanal, J.; Nazari, I.; Tayara, H.; Chong, K.T. 4mCCNN: Identification of N4-methylcytosine sites in prokaryotes using convolutional neural network. IEEE Access 2019, 7, 145455–145461. [Google Scholar] [CrossRef]
- Liu, Q.; Chen, J.; Wang, Y.; Li, S.; Jia, C.; Song, J.; Li, F. DeepTorrent: A deep learning-based approach for predicting DNA N4-methylcytosine sites. Briefings Bioinform. 2020, 22, bbaa124. [Google Scholar] [CrossRef]
- Bari, A.G.; Reaz, M.R.; Choi, H.J.; Jeong, B.S. DNA encoding for splice site prediction in large DNA sequence. In Proceedings of the International Conference on Database Systems for Advanced Applications; Springer: Berlin/Heidelberg, Germany, 2013; pp. 46–58. [Google Scholar]
- Alam, W.; Tayara, H.; Chong, K.T. XG-ac4C: Identification of N4-acetylcytidine (ac4C) in mRNA using eXtreme gradient boosting with electron-ion interaction pseudopotentials. Sci. Rep. 2020, 10, 20942. [Google Scholar] [CrossRef]
- Tahir, M.; Hayat, M. iNuc-STNC: A sequence-based predictor for identification of nucleosome positioning in genomes by extending the concept of SAAC and Chou’s PseAAC. Mol. BioSystems 2016, 12, 2587–2593. [Google Scholar] [CrossRef]
- Tahir, M.; Hayat, M. Machine learning based identification of protein–protein interactions using derived features of physiochemical properties and evolutionary profiles. Artif. Intell. Med. 2017, 78, 61–71. [Google Scholar] [CrossRef]
- Tahir, M.; Hayat, M.; Kabir, M. Sequence based predictor for discrimination of enhancer and their types by applying general form of Chou’s trinucleotide composition. Comput. Methods Programs Biomed. 2017, 146, 69–75. [Google Scholar] [CrossRef]
- Ye, P.; Luan, Y.; Chen, K.; Liu, Y.; Xiao, C.; Xie, Z. MethSMRT: An integrative database for DNA N6-methyladenine and N4-methylcytosine generated by single-molecular real-time sequencing. Nucleic Acids Res. 2016, 45, gkw950. [Google Scholar] [CrossRef] [Green Version]
- Wahab, A.; Ali, S.D.; Tayara, H.; Chong, K.T. iIM-CNN: Intelligent identifier of 6mA sites on different species by using convolution neural network. IEEE Access 2019, 7, 178577–178583. [Google Scholar] [CrossRef]
- Chantsalnyam, T.; Lim, D.Y.; Tayara, H.; Chong, K.T. ncRDeep: Non-coding RNA classification with convolutional neural network. Comput. Biol. Chem. 2020, 88, 107364. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.D.; Kim, J.H.; Tayara, H.; Chong, K.T. Prediction of RNA 5-Hydroxymethylcytosine Modifications Using Deep Learning. IEEE Access 2021, 9, 8491–8496. [Google Scholar] [CrossRef]
- Siraj, A.; Lim, D.Y.; Tayara, H.; Chong, K.T. UbiComb: A Hybrid Deep Learning Model for Predicting Plant-Specific Protein Ubiquitylation Sites. Genes 2021, 12, 717. [Google Scholar] [CrossRef]
- Tahir, M.; Tayara, H.; Chong, K.T. iDNA6mA (5-step rule): Identification of DNA N6-methyladenine sites in the rice genome by intelligent computational model via Chou’s 5-step rule. Chemom. Intell. Lab. Syst. 2019, 189, 96–101. [Google Scholar] [CrossRef]
- Tahir, M.; Hayat, M.; Ullah, I.; Chong, K.T. A deep learning-based computational approach for discrimination of DNA N6-methyladenosine sites by fusing heterogeneous features. Chemom. Intell. Lab. Syst. 2020, 206, 104151. [Google Scholar] [CrossRef]
- Siraj, A.; Chantsalnyam, T.; Tayara, H.; Chong, K.T. Recsno: Prediction of protein s-nitrosylation sites using a recurrent neural network. IEEE Access 2021, 9, 6674–6682. [Google Scholar] [CrossRef]
- Wahab, A.; Mahmoudi, O.; Kim, J.; Chong, K.T. DNC4mC-Deep: Identification and analysis of DNA N4-methylcytosine sites based on different encoding schemes by using deep learning. Cells 2020, 9, 1756. [Google Scholar] [CrossRef] [PubMed]
- Zou, J.; Huss, M.; Abid, A.; Mohammadi, P.; Torkamani, A.; Telenti, A. A primer on deep learning in genomics. Nat. Genet. 2019, 51, 12–18. [Google Scholar] [CrossRef] [PubMed]
- Raimondi, D.; Orlando, G.; Tabaro, F.; Lenaerts, T.; Rooman, M.; Moreau, Y.; Vranken, W.F. Large-scale in-silico statistical mutagenesis analysis sheds light on the deleteriousness landscape of the human proteome. Sci. Rep. 2018, 8, 16980. [Google Scholar] [CrossRef] [PubMed]
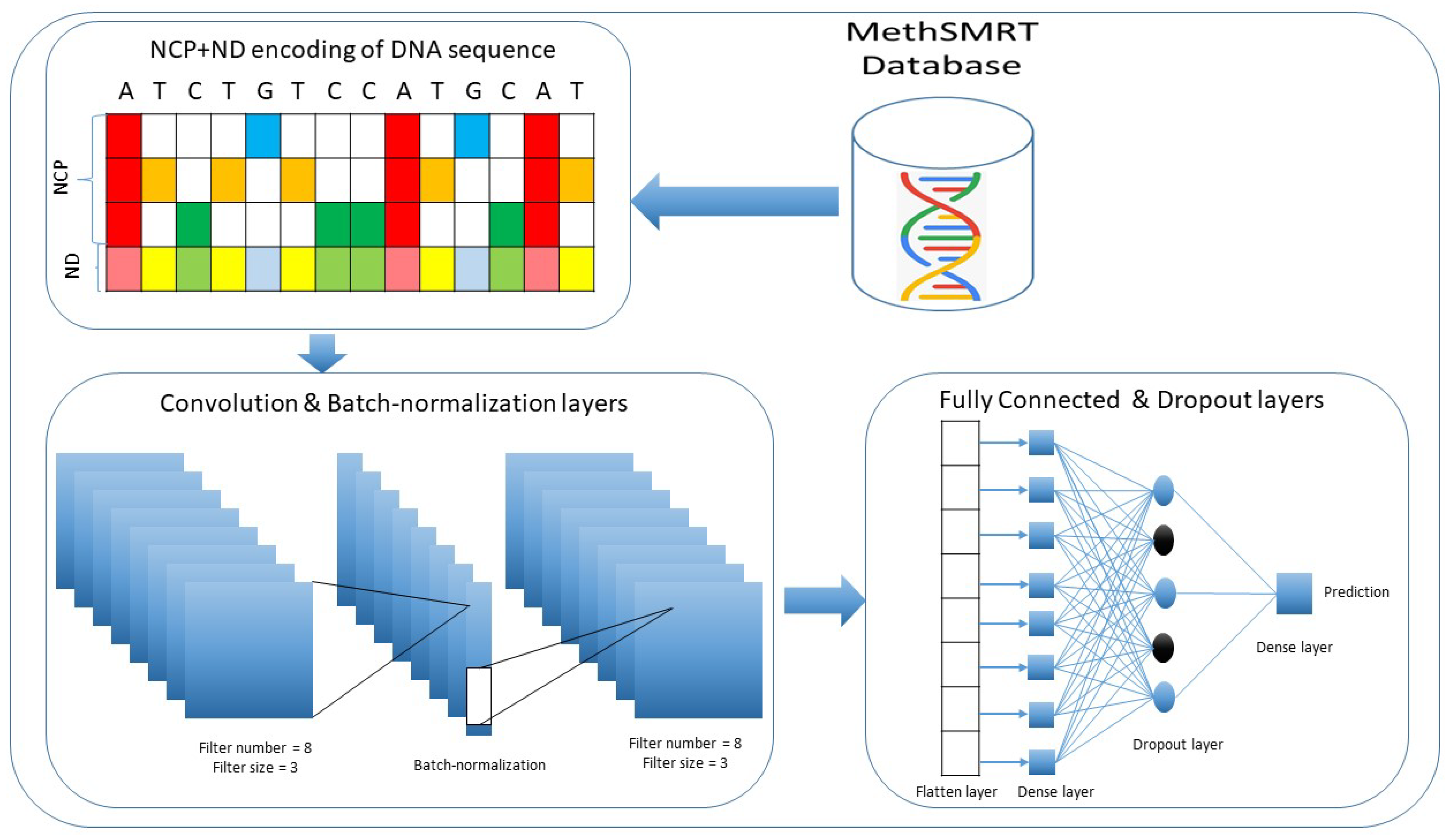


| Species | Sequences | Total | |
|---|---|---|---|
| C. elegans | Positive | 1554 | 3108 |
| Negative | 1554 | ||
| D. melanogaster | Positive | 1769 | 3538 |
| Negative | 1769 | ||
| A. thaliana | Positive | 1978 | 3956 |
| Negative | 1978 | ||
| E. coli | Positive | 388 | 776 |
| Negative | 388 | ||
| G. subterraneus | Positive | 906 | 1812 |
| Negative | 906 | ||
| G. pickeringii | Positive | 569 | 1138 |
| Negative | 569 | ||
| Hyper-Parameters | Range |
|---|---|
| Filters of Conv1D | [8,16,32] |
| Conv1D kernel size | [3,5,7] |
| Conv1D Strides | [2,3] |
| Dropout | [0.2,0.3,0.4,0.5] |
| Dense layer units | [8,16,32] |
| Datasets | Methods | ACC | SN | SP | MCC |
|---|---|---|---|---|---|
| C. elegans | iDNA4mC | 0.786 | 0.797 | 0.775 | 0.572 |
| 4mCPred | 0.826 | 0.825 | 0.826 | 0.652 | |
| 4mCPred-SVM | 0.815 | 0.824 | 0.807 | 0.631 | |
| 4mCCNN | 0.842 | 0.894 | 0.825 | 0.694 | |
| DeepTorrent | 0.858 | 0.810 | 0.906 | 0.719 | |
| SOMM4mC | 0.876 | 0.839 | 0.913 | 0.743 | |
| i4mC-Deep | 0.886 | 0.874 | 0.898 | 0.774 | |
| D. melanogaster | iDNA4mC | 0.812 | 0.833 | 0.791 | 0.625 |
| 4mCPred | 0.822 | 0.824 | 0.821 | 0.646 | |
| 4mCPred-SVM | 0.830 | 0.838 | 0.822 | 0.661 | |
| 4mCCNN | 0.853 | 0.864 | 0.853 | 0.686 | |
| DeepTorrent | 0.861 | 0.834 | 0.889 | 0.724 | |
| SOMM4mC | 0.874 | 0.862 | 0.886 | 0.724 | |
| i4mC-Deep | 0.895 | 0.898 | 0.892 | 0.791 | |
| A. thaliana | iDNA4mC | 0.760 | 0.757 | 0.762 | 0.519 |
| 4mCPred | 0.768 | 0.755 | 0.780 | 0.536 | |
| 4mCPred-SVM | 0.787 | 0.778 | 0.796 | 0.573 | |
| 4mCCNN | 0.797 | 0.803 | 0.792 | 0.621 | |
| DeepTorrent | 0.803 | 0.703 | 0.903 | 0.620 | |
| SOMM4mC | 0.836 | 0.800 | 0.872 | 0.647 | |
| i4mC-Deep | 0.865 | 0.871 | 0.861 | 0.731 | |
| E. coli | iDNA4mC | 0.799 | 0.820 | 0.778 | 0.598 |
| 4mCPred | 0.826 | 0.819 | 0.832 | 0.655 | |
| 4mCPred-SVM | 0.833 | 0.858 | 0.807 | 0.666 | |
| 4mCCNN | 0.859 | 0.881 | 0.788 | 0.687 | |
| DeepTorrent | 0.873 | 0.891 | 0.855 | 0.747 | |
| SOMM4mC | 0.918 | 0.903 | 0.934 | 0.853 | |
| i4mC-Deep | 0.926 | 0.930 | 0.922 | 0.854 | |
| G. subterraneus | iDNA4mC | 0.815 | 0.822 | 0.808 | 0.630 |
| 4mCPred | 0.828 | 0.818 | 0.837 | 0.662 | |
| 4mCPred-SVM | 0.837 | 0.840 | 0.834 | 0.674 | |
| 4mCCNN | 0.860 | 0.851 | 0.843 | 0.703 | |
| DeepTorrent | 0.880 | 0.813 | 0.948 | 0.768 | |
| SOMM4mC | 0.876 | 0.864 | 0.888 | 0.728 | |
| i4mC-Deep | 0.915 | 0.904 | 0.926 | 0.833 | |
| G. pinckeringii | iDNA4mC | 0.831 | 0.824 | 0.838 | 0.663 |
| 4mCPred | 0.830 | 0.850 | 0.810 | 0.668 | |
| 4mCPred-SVM | 0.860 | 0.863 | 0.858 | 0.721 | |
| 4mCCNN | 0.871 | 0.857 | 0.893 | 0.750 | |
| DeepTorrent | 0.894 | 0.831 | 0.957 | 0.795 | |
| SOMM4mC | 0.903 | 0.895 | 0.911 | 0.772 | |
| i4mC-Deep | 0.926 | 0.915 | 0.938 | 0.855 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alam, W.; Tayara, H.; Chong, K.T. i4mC-Deep: An Intelligent Predictor of N4-Methylcytosine Sites Using a Deep Learning Approach with Chemical Properties. Genes 2021, 12, 1117. https://doi.org/10.3390/genes12081117
Alam W, Tayara H, Chong KT. i4mC-Deep: An Intelligent Predictor of N4-Methylcytosine Sites Using a Deep Learning Approach with Chemical Properties. Genes. 2021; 12(8):1117. https://doi.org/10.3390/genes12081117
Chicago/Turabian StyleAlam, Waleed, Hilal Tayara, and Kil To Chong. 2021. "i4mC-Deep: An Intelligent Predictor of N4-Methylcytosine Sites Using a Deep Learning Approach with Chemical Properties" Genes 12, no. 8: 1117. https://doi.org/10.3390/genes12081117
APA StyleAlam, W., Tayara, H., & Chong, K. T. (2021). i4mC-Deep: An Intelligent Predictor of N4-Methylcytosine Sites Using a Deep Learning Approach with Chemical Properties. Genes, 12(8), 1117. https://doi.org/10.3390/genes12081117








