Immune Gene Rearrangements: Unique Signatures for Tracing Physiological Lymphocytes and Leukemic Cells
Abstract
1. Immunoglobulin and T-Cell Receptor Rearrangements
1.1. Structure of the Immune Receptors
1.2. Somatic (V-D-J) Recombination
1.3. Junctional Diversity
1.4. Affinity Maturation
1.5. Rearrangement Process during Lymphocyte Development
2. IG/TR Rearrangements in Leukemia
2.1. Leukemic Clones & Oligoclonality
2.2. IG/TR Rearrangement Profiles in Leukemias
2.3. Stability and Sensitivity of IG/TR Rearrangements as MRD Targets
3. Minimal Residual Disease
3.1. MRD as an Important Prognostic Factor in Lymphoid Malignancies
3.2. MRD Detection Techniques
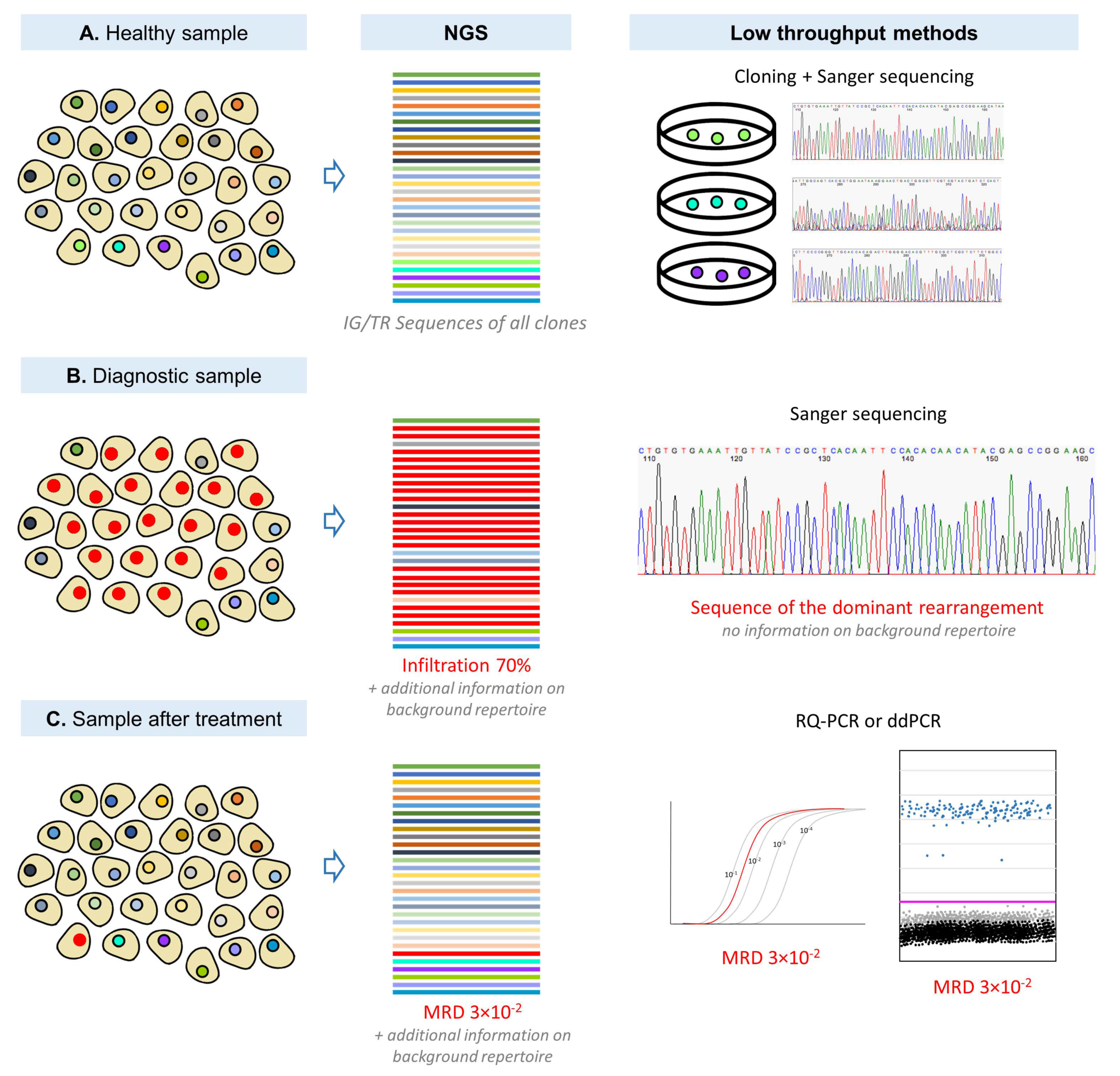
3.3. IG/TR-Based MRD Detection by Low and High Throughput Molecular Methods
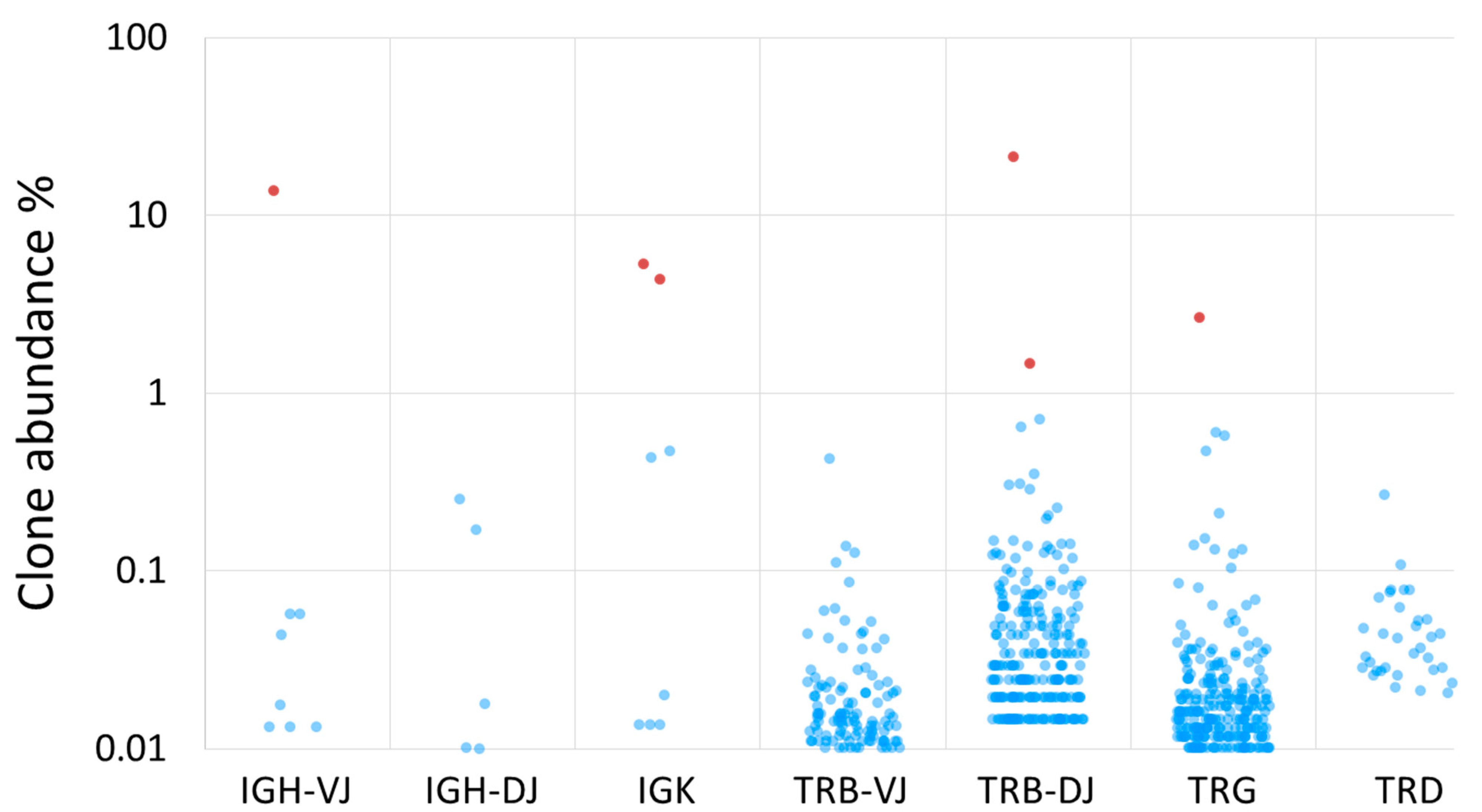
4. Summary
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Murphy, K. Janeway’s Immunobiology, 8th ed.; Garland Science, Taylor & Francis Group LLC: New York, NY, USA, 2012; ISBN 978-0-8153-4243-4. [Google Scholar]
- Jung, D.; Giallourakis, C.; Mostoslavsky, R.; Alt, F.W. Mechanism and control of V(D)J recombination at the immunoglobulin heavy chain locus. Annu. Rev. Immunol. 2006, 24, 541–570. [Google Scholar] [CrossRef]
- Tonegawa, S. Somatic generation of antibody diversity. Nature 1983, 302, 575–581. [Google Scholar] [CrossRef]
- Van Dongen, J.J.M.; Comans-Bitter, W.M.; Wolvers-Tettero, I.L.M.; Borst, J. Development of human T lymphocytes and their thymus-dependency. Thymus 1990, 16, 207–234. [Google Scholar] [PubMed]
- Davis, M.M.; Bjorkman, P.J. T-cell antigen receptor genes and T-cell recognition. Nature 1988, 334, 395–402. [Google Scholar] [CrossRef] [PubMed]
- Szczepański, T.; Van Der Velden, V.H.J.; Hoogeveen, P.G.; De Bie, M.; Jacobs, D.C.H.; Van Wering, E.R.; Van Dongen, J.J.M. Vδ2-Jα rearrangements are frequent in precursor-B-acute lymphoblastic leukemia but rare in normal lymphoid cells. Blood 2004, 103, 3798–3804. [Google Scholar] [CrossRef]
- Fröland, S.S.; Natvig, J.B. Class, subclass, and allelic exclusion of membrane-bound Ig of human B lymphocytes. J. Exp. Med. 1972, 136, 409–414. [Google Scholar] [CrossRef]
- Aifantis, I.; Buer, J.; Von Boehmer, H.; Azogui, O. Essential Role of the Pre-T Cell Receptor in Allelic Exclusion of the T Cell Receptor β locus. Immunity 1997, 7, 601–607. [Google Scholar] [CrossRef]
- Di Noia, J.M.; Neuberger, M.S. Molecular Mechanisms of Antibody Somatic Hypermutation. Annu. Rev. Biochem. 2007, 76, 1–22. [Google Scholar] [CrossRef]
- Tomlinson, I.M.; Walter, G.; Jones, P.T.; Dear, P.H.; Sonnhammer, E.L.; Winter, G. The imprint of somatic hypermutation on the repertoire of human germline V genes. J. Mol. Biol. 1996, 256, 813–817. [Google Scholar] [CrossRef]
- Neuberger, M.S.; Milstein, C. Somatic hypermutation. Curr. Opin. Immunol. 1995, 7, 248–254. [Google Scholar] [CrossRef]
- Bye, J.M.; Carter, C.; Cui, Y.; Gorick, B.D.; Songsivilai, S.; Winter, G.; Hughes-Jones, N.C.; Marks, J.D. Germline variable region gene segment derivation of human monoclonal anti-Rh(D) antibodies: Evidence for affinity maturation by somatic hypermutation and repertoire shift. J. Clin. Investig. 1992, 90, 2481–2490. [Google Scholar] [CrossRef]
- Kosak, S.T.; Skok, J.A.; Medina, K.L.; Riblet, R.; Le Beau, M.M.; Fisher, A.G.; Singh, H. Subnuclear compartmentalization of immunoglobulin loci during lymphocyte development. Science 2002, 296, 158–162. [Google Scholar] [CrossRef]
- Stanhope-Baker, P.; Hudson, K.M.; Shaffer, A.L.; Constantinescu, A.; Schlissel, M.S. Cell Type-Specific Chromatin Structure Determines the Targeting of V(D)J Recombinase Activity In Vitro. Cell 1996, 85, 887–897. [Google Scholar] [CrossRef]
- Miyazaki, K.; Miyazaki, M.; Murre, C. The establishment of B versus T cell identity. Trends Immunol. 2014, 35, 205–210. [Google Scholar] [CrossRef] [PubMed]
- Van Zelm, M.C.; van der Burg, M.; de Ridder, D.; Barendregt, B.H.; de Haas, E.F.E.; Reinders, M.J.T.; Lankester, A.C.; Révész, T.; Staal, F.J.T.; van Dongen, J.J.M. Ig Gene Rearrangement Steps Are Initiated in Early Human Precursor B Cell Subsets and Correlate with Specific Transcription Factor Expression. J. Immunol. 2005, 175, 5912–5922. [Google Scholar] [CrossRef] [PubMed]
- Constantinescu, A.; Schlissel, M.S. Changes in locus-specific V(D)J recombinase activity induced by immunoglobulin gene products during B cell development. J. Exp. Med. 1997, 185, 609–620. [Google Scholar] [CrossRef]
- Mombaerts, P.; Iacomini, J.; Johnson, R.S.; Herrup, K.; Tonegawa, S.; Papaioannou, V.E. RAG-1-deficient mice have no mature B and T lymphocytes. Cell 1992, 68, 869–877. [Google Scholar] [CrossRef]
- Shinkai, Y.; Rathbun, G.; Lam, K.P.; Oltz, E.M.; Stewart, V.; Mendelsohn, M.; Charron, J.; Datta, M.; Young, F.; Stall, A.M.; et al. RAG-2-deficient mice lack mature lymphocytes owing to inability to initiate V(D)J rearrangement. Cell 1992, 68, 855–867. [Google Scholar] [CrossRef]
- Spanopoulou, E.; Roman, C.A.J.; Corcoran, L.M.; Schlissel, M.S.; Silver, D.P.; Nemazee, D.; Nussenzweig, M.C.; Shinton, S.A.; Hardy, R.R.; Baltimore, D. Functional immunoglobulin transgenes guide ordered B-cell differentiation in Rag-1-deficient mice. Genes Dev. 1994, 8, 1030–1042. [Google Scholar] [CrossRef]
- Young, F.; Ardman, B.; Shinkai, Y.; Lansford, R.; Blackwell, T.K.; Mendelsohn, M.; Rolink, A.; Melchers, F.; Alt, F.W. Influence of immunoglobulin heavy- and light-chain expression on B-cell differentiation. Genes Dev. 1994, 8, 1043–1057. [Google Scholar] [CrossRef]
- Capone, M.; Hockett, R.D.; Zlotnik, A. Kinetics of T cell receptor β, γ, and δ rearrangements during adult thymic development: T cell receptor rearrangements are present in CD44+CD25+ Pro-T thymocytes. Proc. Natl. Acad. Sci. USA 1998, 95, 12522–12527. [Google Scholar] [CrossRef] [PubMed]
- Staal, F.J.T.; Weerkamp, F.; Langerak, A.W.; Hendriks, R.W.; Clevers, H.C. Transcriptional Control of T Lymphocyte Differentiation. Stem Cells 2001, 19, 165–179. [Google Scholar] [CrossRef]
- Von Boehmer, H. Selection of the T-cell repertoire: Receptor-controlled checkpoints in T-cell development. Adv. Immunol. 2004, 84, 201–238. [Google Scholar] [CrossRef]
- Dik, W.A.; Pike-Overzet, K.; Weerkamp, F.; De Ridder, D.; De Haas, E.F.E.; Baert, M.R.M.; Van Der Spek, P.; Koster, E.E.L.; Reinders, M.J.T.; Van Dongen, J.J.M.; et al. New insights on human T cell development by quantitative T cell receptor gene rearrangement studies and gene expression profiling. J. Exp. Med. 2005, 201, 1715–1723. [Google Scholar] [CrossRef] [PubMed]
- Imkeller, K.; Wardemann, H. Assessing human B cell repertoire diversity and convergence. Immunol. Rev. 2018, 284, 51–66. [Google Scholar] [CrossRef]
- Szczepański, T.; Flohr, T.; Van Der Velden, V.H.J.; Bartram, C.R.; Van Dongen, J.J.M. Molecular monitoring of residual disease using antigen receptor genes in childhood acute lymphoblastic leukaemia. Best Pract. Res. Clin. Haematol. 2002, 15, 37–57. [Google Scholar] [CrossRef] [PubMed]
- Beishuizen, A.; Verhoeven, M.A.; van Wering, E.R.; Hählen, K.; Hooijkaas, H.; van Dongen, J.J. Analysis of Ig and T-cell receptor genes in 40 childhood acute lymphoblastic leukemias at diagnosis and subsequent relapse: Implications for the detection of minimal residual disease by polymerase chain reaction analysis. Blood 1994, 83, 2238–2247. [Google Scholar] [CrossRef]
- Beishuizen, A.; Hählen, K.; Hagemeijer, A.; Verhoeven, M.A.; Hooijkaas, H.; Adriaansen, H.J.; Wolvers-Tettero, I.L.; van Wering, E.R.; van Dongen, J.J. Multiple rearranged immunoglobulin genes in childhood acute lymphoblastic leukemia of precursor B-cell origin. Leukemia 1991, 5, 657–667. [Google Scholar] [PubMed]
- Steenbergen, E.J.; Verhagen, O.J.; van Leeuwen, E.F.; von dem Borne, A.E.; van der Schoot, C.E. Distinct ongoing Ig heavy chain rearrangement processes in childhood B-precursor acute lymphoblastic leukemia. Blood 1993, 82, 581–589. [Google Scholar] [CrossRef]
- Kitchingman, G.R. Immunoglobulin heavy chain gene VH-D junctional diversity at diagnosis in patients with acute lymphoblastic leukemia. Blood 1993, 81, 775–782. [Google Scholar] [CrossRef]
- Van Der Velden, V.H.J.; Van Dongen, J.J.M. MRD detection in acute lymphoblastic leukemia patients using Ig/TCR gene rearrangements as targets for real-time quantitative PCR. Methods Mol. Biol. 2009, 538, 115–150. [Google Scholar] [CrossRef] [PubMed]
- Theunissen, P.M.J.; van Zessen, D.; Stubbs, A.P.; Faham, M.; Zwaan, C.M.; van Dongen, J.J.M.; Van Der Velden, V.H.J. Antigen receptor sequencing of paired bone marrow samples shows homogeneous distribution of acute lymphoblastic leukemia subclones. Haematologica 2017, 102, 1869–1877. [Google Scholar] [CrossRef][Green Version]
- Gawad, C.; Pepin, F.; Carlton, V.E.H.; Klinger, M.; Logan, A.C.; Miklos, D.B.; Faham, M.; Dahl, G.; Lacayo, N. Massive evolution of the immunoglobulin heavy chain locus in children with B precursor acute lymphoblastic leukemia. Blood 2012, 120, 4407–4417. [Google Scholar] [CrossRef] [PubMed]
- Szczepański, T.; Pongers-Willemse, M.J.; Langerak, A.W.; Harts, W.A.; Wijkhuijs, A.J.M.; van Wering, E.R.; van Dongen, J.J.M. Ig Heavy Chain Gene Rearrangements in T-Cell Acute Lymphoblastic Leukemia Exhibit Predominant Dh6-19 and Dh7-27 Gene Usage, Can Result in Complete V-D-J Rearrangements, and Are Rare in T-Cell Receptor αβ Lineage. Blood 1999, 93, 4079–4085. [Google Scholar] [CrossRef]
- Stamatopoulos, B.; Timbs, A.; Bruce, D.; Smith, T.; Clifford, R.; Robbe, P.; Burns, A.; Vavoulis, D.V.; Lopez, L.; Antoniou, P.; et al. Targeted deep sequencing reveals clinically relevant subclonal IgHV rearrangements in chronic lymphocytic leukemia. Leukemia 2017, 31, 837–845. [Google Scholar] [CrossRef]
- Langerak, A.W.; Davi, F.; Ghia, P.; Hadzidimitriou, A.; Murray, F.; Potter, K.N.; Rosenquist, R.; Stamatopoulos, K.; Belessi, C. Immunoglobulin sequence analysis and prognostication in CLL: Guidelines from the ERIC review board for reliable interpretation of problematic cases. Leukemia 2011, 25, 979–984. [Google Scholar] [CrossRef]
- Plevova, K.; Francova, H.S.; Burckova, K.; Brychtova, Y.; Doubek, M.; Pavlova, S.; Malcikova, J.; Mayer, J.; Tichy, B.; Pospisilova, S. Multiple productive immunoglobulin heavy chain gene rearrangements in chronic lymphocytic leukemia are mostly derived from independent clones. Haematologica 2014, 99, 329–338. [Google Scholar] [CrossRef] [PubMed]
- Brazdilova, K.; Plevova, K.; Skuhrova Francova, H.; Kockova, H.; Borsky, M.; Bikos, V.; Malcikova, J.; Oltova, A.; Kotaskova, J.; Tichy, B.; et al. Multiple productive IGH rearrangements denote oligoclonality even in immunophenotypically monoclonal CLL. Leukemia 2018, 32, 234–236. [Google Scholar] [CrossRef]
- Hübner, S.; Cazzaniga, G.; Flohr, T.; van der Velden, V.H.J.; Konrad, M.; Pötschger, U.; Basso, G.; Schrappe, M.; van Dongen, J.J.M.; Bartram, C.R.; et al. High incidence and unique features of antigen receptor gene rearrangements in TEL–AML1-positive leukemias. Leukemia 2004, 18, 84–91. [Google Scholar] [CrossRef][Green Version]
- Brumpt, C.; Delabesse, E.; Beldjord, K.; Davi, F.; Cayuela, J.M.; Millien, C.; Villarese, P.; Quartier, P.; Buzyn, A.; Valensi, F.; et al. The incidence of clonal T-cell receptor rearrangements in B-cell precursor acute lymphoblastic leukemia varies with age and genotype. Blood 2000, 96, 2254–2261. [Google Scholar] [CrossRef]
- Van der Velden, V.H.J.; Szczepanski, T.; Wijkhuijs, J.M.; Hart, P.G.; Hoogeveen, P.G.; Hop, W.C.J.; van Wering, E.R.; van Dongen, J.J.M. Age-related patterns of immunoglobulin and T-cell receptor gene rearrangements in precursor-B-ALL: Implications for detection of minimal residual disease. Leukemia 2003, 17, 1834–1844. [Google Scholar] [CrossRef] [PubMed]
- Flohr, T.; Schrauder, A.; Cazzaniga, G.; Panzer-Grümayer, R.; van der Velden, V.; Fischer, S.; Stanulla, M.; Basso, G.; Niggli, F.K.; Schäfer, B.W.; et al. Minimal residual disease-directed risk stratification using real-time quantitative PCR analysis of immunoglobulin and T-cell receptor gene rearrangements in the international multicenter trial AIEOP-BFM ALL 2000 for childhood acute lymphoblastic leukemia. Leukemia 2008, 22, 771–782. [Google Scholar] [CrossRef] [PubMed]
- Szczepański, T.; Langerak, A.W.; Wolvers-Tettero, I.L.M.; Ossenkoppele, G.J.; Verhoef, G.; Stul, M.; Petersen, E.J.; De Bruijn, M.A.C.; Van’t Veer, M.B.; Van Dongen, J.J.M. Immunoglobulin and T cell receptor gene rearrangement patterns in acute lymphoblastic leukemia are less mature in adults than in children: Implications for selection of PCR targets for detection of minimal residual disease. Leukemia 1998, 12, 1081–1088. [Google Scholar] [CrossRef] [PubMed]
- Brüggemann, M.; van der Velden, V.H.J.; Raff, T.; Droese, J.; Ritgen, M.; Pott, C.; Wijkhuijs, A.J.; Gökbuget, N.; Hoelzer, D.; van Wering, E.R.; et al. Rearranged T-cell receptor beta genes represent powerful targets for quantification of minimal residual disease in childhood and adult T-cell acute lymphoblastic leukemia. Leukemia 2004, 18, 709–719. [Google Scholar] [CrossRef] [PubMed]
- Greaves, M.F.; Chan, L.C.; Furley, A.J.; Watt, S.M.; Molgaard, H.V. Lineage promiscuity in hemopoietic differentiation and leukemia. Blood 1986, 67, 1–11. [Google Scholar] [CrossRef]
- Bertrand, F.E.; Billips, L.G.; Burrows, P.D.; Gartland, G.L.; Kubagawa, H.; Schroeder, H.W. Ig D(H) gene segment transcription and rearrangement before surface expression of the pan-B-cell marker CD19 in normal human bone marrow. Blood 1997, 90, 736–744. [Google Scholar] [CrossRef]
- Szczepański, T.; Beishuizen, A.; Pongers-Willemse, M.J.; Hählen, K.; Van Wering, E.R.; Wijkhuijs, A.J.M.; Tibbe, G.J.M.; De Bruijn, M.A.C.; Van Dongen, J.J.M. Cross-lineage T cell receptor gene rearrangements occur in more than ninety percent of childhood precursor-B acute lymphoblastic leukemias: Alternative PCR targets for detection of minimal residual disease. Leukemia 1999, 13, 196–205. [Google Scholar] [CrossRef]
- Meleshko, A.N.; Belevtsev, M.V.; Savitskaja, T.V.; Potapnev, M.P. The incidence of T-cell receptor gene rearrangements in childhood B-lineage acute lymphoblastic leukemia is related to immunophenotype and fusion oncogene expression. Leuk. Res. 2006, 30, 795–800. [Google Scholar] [CrossRef]
- Biondi, A.; Francia di Celle, P.; Rossi, V.; Casorati, G.; Matullo, G.; Giudici, G.; Foa, R.; Migone, N. High prevalence of T-cell receptor V delta 2-(D)-D delta 3 or D delta 1/2-D delta 3 rearrangements in B-precursor acute lymphoblastic leukemias. Blood 1990, 75, 1834–1840. [Google Scholar] [CrossRef]
- Hara, J.; Benedict, S.H.; Champagne, E.; Takihara, Y.; Mak, T.W.; Minden, M.; Gelfand, E.W. T cell receptor δ gene rearrangements in acute lymphoblastic leukemia. J. Clin. Investig. 1988, 82, 1974–1982. [Google Scholar] [CrossRef]
- Felix, C.A.; Poplack, D.G. Characterization of acute lymphoblastic leukemia of childhood by immunoglobulin and T-cell receptor gene patterns. Leukemia 1991, 5, 1015–1025. [Google Scholar] [CrossRef] [PubMed]
- Van der Velden, V.H.J.; Brüggemann, M.; Hoogeveen, P.G.; de Bie, M.; Hart, P.G.; Raff, T.; Pfeifer, H.; Lüschen, S.; Szczepański, T.; van Wering, E.R.; et al. TCRB gene rearrangements in childhood and adult precursor-B-ALL: Frequency, applicability as MRD-PCR target, and stability between diagnosis and relapse. Leukemia 2004, 18, 1971–1980. [Google Scholar] [CrossRef]
- Szczepański, T.; Pongers-Willemse, M.J.; Langerak, A.W.; Van Dongen, J.J.M. Unusual immunoglobulin and T-cell receptor gene rearrangement patterns in acute lymphoblastic leukemias. Curr. Top. Microbiol. Immunol. 1999, 246, 205–215. [Google Scholar] [PubMed]
- Foroni, L.; Foldi, J.; Matutes, E.; Catovsky, D.; O’Connor, N.J.O.; Baer, R.; Forster, A.; Rabbitts, T.H.; Luzzatto, L. α, β and γ T-cell receptor genes: Rearrangements correlate with haematological phenotype in T cell leukaemias. Br. J. Haematol. 1987, 67, 307–318. [Google Scholar] [CrossRef] [PubMed]
- Kneba, M.; Bergholz, M.; Bolz, I.; Hulpke, M.; Bätge, R.; Schauer, A.; Krieger, G. Heterogeneity of immunoglobulin gene rearrangements in B-cell lymphomas. Int. J. Cancer 1990, 45, 609–613. [Google Scholar] [CrossRef] [PubMed]
- Rai, L.; Casanova, A.; Moorman, A.V.; Richards, S.; Buck, G.; Goldstone, A.H.; Fielding, A.K.; Foroni, L. Antigen receptor gene rearrangements reflect on the heterogeneity of adult Acute Lymphoblastic Leukaemia (ALL) with implications of cell-origin of ALL subgroups-A UKALLXII study. Br. J. Haematol. 2010, 148, 394–401. [Google Scholar] [CrossRef]
- Stamatopoulos, K.; Belessi, C.; Moreno, C.; Boudjograh, M.; Guida, G.; Smilevska, T.; Belhoul, L.; Stella, S.; Stavroyianni, N.; Crespo, M.; et al. Over 20% of patients with chronic lymphocytic leukemia carry stereotyped receptors: Pathogenetic implications and clinical correlations. Blood 2007, 109, 259–270. [Google Scholar] [CrossRef]
- Landgren, O.; Albitar, M.; Ma, W.; Abbasi, F.; Hayes, R.B.; Ghia, P.; Marti, G.E.; Caporaso, N.E. B-Cell Clones as Early Markers for Chronic Lymphocytic Leukemia. N. Engl. J. Med. 2009, 360, 659–667. [Google Scholar] [CrossRef]
- Chiorazzi, N.; Ferrarini, M. B cell chronic lymphocytic leukemia: Lessons learned from studies of the B cell antigen receptor. Annu. Rev. Immunol. 2003, 21, 841–894. [Google Scholar] [CrossRef]
- Fais, F.; Ghiotto, F.; Hashimoto, S.; Sellars, B.; Valetto, A.; Allen, S.L.; Schulman, P.; Vinciguerra, V.P.; Rai, K.; Rassenti, L.Z.; et al. Chronic lymphocytic leukemia B cells express restricted sets of mutated and unmutated antigen receptors. J. Clin. Investig. 1998, 102, 1515–1525. [Google Scholar] [CrossRef]
- Stevenson, F.K.; Caligaris-Cappio, F. Chronic lymphocytic leukemia: Revelations from the B-cell receptor. Blood 2004, 103, 4389–4395. [Google Scholar] [CrossRef] [PubMed]
- Abusedra, A.; Joshi, R.; Bybee, A.; Apperley, J.F.; Wagner, S.D. Vλ genes in chronic lymphocytic leukaemia: Highly skewed V gene segment usage with similar CDR3 sequences. Leukemia 2008, 22, 1073–1075. [Google Scholar] [CrossRef] [PubMed]
- Hamblin, T.J.; Davis, Z.; Gardiner, A.; Oscier, D.G.; Stevenson, F.K. Unmutated Ig V(H) genes are associated with a more aggressive form of chronic lymphocytic leukemia. Blood 1999, 94, 1848–1854. [Google Scholar] [CrossRef] [PubMed]
- Damle, R.N.; Wasil, T.; Fais, F.; Ghiotto, F.; Valetto, A.; Allen, S.L.; Buchbinder, A.; Budman, D.; Dittmar, K.; Kolitz, J.; et al. Ig V Gene Mutation Status and CD38 Expression As Novel Prognostic Indicators in Chronic Lymphocytic Leukemia. Blood 1999, 94, 1840–1847. [Google Scholar] [CrossRef] [PubMed]
- Baruchel, A.; Cayuela, J.-M.; Macintyre, E.; Berger, I.R.; Sigaux, F. Assessment of clonal evolution at Ig/TCR loci in acute lymphoblastic leukaemia by single-strand conformation polymorphism studies and highly resolutive PCR derived methods: Implication for a general strategy of minimal residual disease detection. Br. J. Haematol. 1995, 90, 85–93. [Google Scholar] [CrossRef]
- Szczeparński, T.; Willemse, M.J.; Brinkhof, B.; Van Wering, E.R.; Van Der Burg, M.; Van Dongen, J.J.M. Comparative analysis of Ig and TCR gene rearrangements at diagnosis and at relapse of childhood precursor-B-ALL provides improved strategies for selection of stable PCR targets for monitoring of minimal residual disease. Blood 2002, 99, 2315–2323. [Google Scholar] [CrossRef]
- Van der Velden, V.H.J.; Willemse, M.J.; van der Schoot, C.E.; Hählen, K.; van Wering, E.R.; van Dongen, J.J.M. Immunoglobulin kappa deleting element rearrangements in precursor-B acute lymphoblastic leukemia are stable targets for detection of minimal residual disease by real-time quantitative PCR. Leukemia 2002, 16, 928–936. [Google Scholar] [CrossRef]
- Theunissen, P.M.J.; de Bie, M.; van Zessen, D.; de Haas, V.; Stubbs, A.P.; van der Velden, V.H.J. Next-generation antigen receptor sequencing of paired diagnosis and relapse samples of B-cell acute lymphoblastic leukemia: Clonal evolution and implications for minimal residual disease target selection. Leuk. Res. 2019, 76, 98–104. [Google Scholar] [CrossRef]
- Szczepański, T.; van der Velden, V.H.J.; Raff, T.; Jacobs, D.C.H.; van Wering, E.R.; Brüggemann, M.; Kneba, M.; van Dongen, J.J.M. Comparative analysis of T-cell receptor gene rearrangements at diagnosis and relapse of T-cell acute lymphoblastic leukemia (T-ALL) shows high stability of clonal markers for monitoring of minimal residual disease and reveals the occurence of second T-ALL. Leukemia 2003, 17, 2149–2156. [Google Scholar] [CrossRef]
- Van der Velden, V.H.J.; Wijkhuijs, J.M.; Jacobs, D.C.H.; van Wering, E.R.; van Dongen, J.J.M. T cell receptor gamma gene rearrangements as targets for detection of minimal residual disease in acute lymphoblastic leukemia by real-time quantitative PCR analysis. Leukemia 2002, 16, 1372–1380. [Google Scholar] [CrossRef]
- Reiter, A.; Schrappe, M.; Ludwig, W.D.; Hiddemann, W.; Sauter, S.; Henze, G.; Zimmermann, M.; Lampert, F.; Havers, W.; Niethammer, D. Chemotherapy in 998 unselected childhood acute lymphoblastic leukemia patients. Results and conclusions of the multicenter trial ALL-BFM 86. Blood 1994, 84, 3122–3133. [Google Scholar] [CrossRef]
- Gökbuget, N.; Kneba, M.; Raff, T.; Trautmann, H.; Bartram, C.R.; Arnold, R.; Fietkau, R.; Freund, M.; Ganser, A.; Ludwig, W.D.; et al. Adult patients with acute lymphoblastic leukemia and molecular failure display a poor prognosis and are candidates for stem cell transplantation and targeted therapies. Blood 2012, 120, 1868–1876. [Google Scholar] [CrossRef] [PubMed]
- Bassan, R.; Brüggemann, M.; Radcliffe, H.S.; Hartfield, E.; Kreuzbauer, G.; Wetten, S. A systematic literature review and metaanalysis of minimal residual disease as a prognostic indicator in adult B-cell acute lymphoblastic leukemia. Haematologica 2019, 104, 2028–2039. [Google Scholar] [CrossRef] [PubMed]
- Berry, D.A.; Zhou, S.; Higley, H.; Mukundan, L.; Fu, S.; Reaman, G.H.; Wood, B.L.; Kelloff, G.J.; Jessup, J.M.; Radich, J.P. Association of minimal residual disease with clinical outcome in pediatric and adult acute lymphoblastic leukemia: A meta-analysis. JAMA Oncol. 2017, 3, e170580. [Google Scholar] [CrossRef]
- Beldjord, K.; Chevret, S.; Asnafi, V.; Huguet, F.; Boulland, M.L.; Leguay, T.; Thomas, X.; Cayuela, J.M.; Grardel, N.; Chalandon, Y.; et al. Oncogenetics and minimal residual disease are independent outcome predictors in adult patients with acute lymphoblastic leukemia. Blood 2014, 123, 3739–3749. [Google Scholar] [CrossRef] [PubMed]
- Bassan, R.; Spinelli, O.; Oldani, E.; Intermesoli, T.; Tosi, M.; Peruta, B.; Rossi, G.; Borlenghi, E.; Pogliani, E.M.; Terruzzi, E.; et al. Improved risk classification for risk-specific therapy based on the molecular study of minimal residual disease (MRD) in adult acute lymphoblastic leukemia (ALL). Blood 2009, 113, 4153–4162. [Google Scholar] [CrossRef] [PubMed]
- Ribera, J.M.; Oriol, A.; Morgades, M.; Montesinos, P.; Sarrà, J.; González-Campos, J.; Brunet, S.; Tormo, M.; Fernández-Abellán, P.; Guàrdia, R.; et al. Treatment of high-risk Philadelphia chromosome-negative acute lymphoblastic leukemia in adolescents and adults according to early cytologic response and minimal residual disease after consolidation assessed by flow cytometry: Final results of the PETHEMA ALL-AR-03 trial. J. Clin. Oncol. 2014, 32, 1595–1604. [Google Scholar] [CrossRef] [PubMed]
- Van der Velden, V.H.J.; Corral, L.; Valsecchi, M.G.; Jansen, M.W.J.C.; De Lorenzo, P.; Cazzaniga, G.; Panzer-Grümayer, E.R.; Schrappe, M.; Schrauder, A.; Meyer, C.; et al. Prognostic significance of minimal residual disease in infants with acute lymphoblastic leukemia treated within the Interfant-99 protocol. Leukemia 2009, 23, 1073–1079. [Google Scholar] [CrossRef]
- Fronkova, E.; Mejstrikova, E.; Avigad, S.; Chik, K.W.; Castillo, L.; Manor, S.; Reznickova, L.; Valova, T.; Zdrahalova, K.; Hrusak, O.; et al. Minimal residual disease (MRD) analysis in the non-MRD-based ALL IC-BFM 2002 protocol for childhood ALL: Is it possible to avoid MRD testing? Leukemia 2008, 22, 989–997. [Google Scholar] [CrossRef]
- Schultz, K.R.; Pullen, D.J.; Sather, H.N.; Shuster, J.J.; Devidas, M.; Borowitz, M.J.; Carroll, A.J.; Heerema, N.A.; Rubnitz, J.E.; Loh, M.L.; et al. Risk- and response-based classification of childhood B-precursor acute lymphoblastic leukemia: A combined analysis of prognostic markers from the Pediatric Oncology Group (POG) and Children’s Cancer Group (CCG). Blood 2007, 109, 926–935. [Google Scholar] [CrossRef] [PubMed]
- Van Dongen, J.J.M.; Seriu, T.; Panzer-Grümayer, E.R.; Biondi, A.; Pongers-Willemse, M.J.; Corral, L.; Stolz, F.; Schrappe, M.; Masera, G.; Kamps, W.A.; et al. Prognostic value of minimal residual disease in acute lymphoblastic leukaemia in childhood. Lancet 1998, 352, 1731–1738. [Google Scholar] [CrossRef]
- Conter, V.; Bartram, C.R.; Valsecchi, M.G.; Schrauder, A.; Panzer-Grümayer, R.; Möricke, A.; Aricò, M.; Zimmermann, M.; Mann, G.; De Rossi, G.; et al. Molecular response to treatment redefines all prognostic factors in children and adolescents with B-cell precursor acute lymphoblastic leukemia: Results in 3184 patients of the AIEOP-BFMALL 2000 study. Blood 2010, 115, 3206–3214. [Google Scholar] [CrossRef]
- Strati, P.; Keating, M.J.; O’Brien, S.M.; Burger, J.; Ferrajoli, A.; Jain, N.; Tambaro, F.P.; Estrov, Z.; Jorgensen, J.; Challagundla, P.; et al. Eradication of bone marrow minimal residual disease may prompt early treatment discontinuation in CLL. Blood 2014, 123, 3727–3732. [Google Scholar] [CrossRef] [PubMed]
- Santacruz, R.; Villamor, N.; Aymerich, M.; Martínez-Trillos, A.; López, C.; Navarro, A.; Rozman, M.; Beà, S.; Royo, C.; Cazorla, M.; et al. The prognostic impact of minimal residual disease in patients with chronic lymphocytic leukemia requiring first-line therapy. Haematologica 2014, 99, 873–880. [Google Scholar] [CrossRef]
- Logan, A.C.; Zhang, B.; Narasimhan, B.; Carlton, V.; Zheng, J.; Moorhead, M.; Krampf, M.R.; Jones, C.D.; Waqar, A.N.; Faham, M.; et al. Minimal residual disease quantification using consensus primers and high-throughput IGH sequencing predicts post-transplant relapse in chronic lymphocytic leukemia. Leukemia 2013, 27, 1659–1665. [Google Scholar] [CrossRef] [PubMed]
- Kwok, M.; Rawstron, A.C.; Varghese, A.; Evans, P.A.S.; O’Connor, S.J.M.; Doughty, C.; Newton, D.J.; Moreton, P.; Hillmen, P. Minimal residual disease is an independent predictor for 10-year survival in CLL. Blood 2016, 128, 2770–2773. [Google Scholar] [CrossRef]
- Kovacs, G.; Robrecht, S.; Fink, A.M.; Bahlo, J.; Cramer, P.; Von Tresckow, J.; Maurer, C.; Langerbeins, P.; Fingerle-Rowson, G.; Ritgen, M.; et al. Minimal residual disease assessment improves prediction of outcome in patients with chronic lymphocytic Leukemia (CLL) who achieve partial response: Comprehensive analysis of two phase III Studies of the German CLL Study Group. J. Clin. Oncol. 2016, 34, 3758–3765. [Google Scholar] [CrossRef]
- Dimier, N.; Delmar, P.; Ward, C.; Morariu-Zamfir, R.; Fingerle-Rowson, G.; Bahlo, J.; Fischer, K.; Eichhorst, B.; Goede, V.; Van Dongen, J.J.M.; et al. A model for predicting effect of treatment on progression-free survival using MRD as a surrogate end point in CLL. Blood 2018, 131, 955–962. [Google Scholar] [CrossRef] [PubMed]
- Böttcher, S.; Ritgen, M.; Fischer, K.; Stilgenbauer, S.; Busch, R.M.; Fingerle-Rowson, G.; Fink, A.M.; Buḧler, A.; Zenz, T.; Wenger, M.K.; et al. Minimal residual disease quantification is an independent predictor of progression-free and overall survival in chronic lymphocytic leukemia: A Multivariate analysis from the randomized GCLLSG CLL8 trial. J. Clin. Oncol. 2012, 30, 980–988. [Google Scholar] [CrossRef]
- Paiva, B.; Puig, N.; Cedena, M.T.; Rosiñol, L.; Cordón, L.; Vidriales, M.B.; Burgos, L.; Flores-Montero, J.; Sanoja-Flores, L.; Lopez-Anglada, L.; et al. Measurable residual disease by next-generation flow cytometry in multiple myeloma. J. Clin. Oncol. 2020, 38, 784–792. [Google Scholar] [CrossRef]
- Lahuerta, J.J.; Paiva, B.; Vidriales, M.B.; Cordón, L.; Cedena, M.T.; Puig, N.; Martinez-Lopez, J.; Rosiñol, L.; Gutierrez, N.C.; Martín-Ramos, M.L.; et al. Depth of response in multiple myeloma: A pooled analysis of three PETHEMA/GEM clinical trials. J. Clin. Oncol. 2017, 35, 2900–2910. [Google Scholar] [CrossRef]
- Rawstron, A.C.; Child, J.A.; De Tute, R.M.; Davies, F.E.; Gregory, W.M.; Bell, S.E.; Szubert, A.J.; Navarro-Coy, N.; Drayson, M.T.; Feyler, S.; et al. Minimal residual disease assessed by multiparameter flow cytometry in multiple myeloma: Impact on outcome in the Medical Research Council Myeloma IX study. J. Clin. Oncol. 2013, 31, 2540–2547. [Google Scholar] [CrossRef] [PubMed]
- Munshi, N.C.; Avet-Loiseau, H.; Rawstron, A.C.; Owen, R.G.; Child, J.A.; Thakurta, A.; Sherrington, P.; Samur, M.K.; Georgieva, A.; Anderson, K.C.; et al. Association of Minimal Residual Disease With Superior Survival Outcomes in Patients With Multiple Myeloma: A Meta-analysis. JAMA Oncol. 2017, 3, 28. [Google Scholar] [CrossRef] [PubMed]
- Szczepański, T. Why and how to quantify minimal residual disease in acute lymphoblastic leukemia? Leukemia 2007, 21, 622–626. [Google Scholar] [CrossRef]
- Rawstron, A.C.; Kennedy, B.; Evans, P.A.S.; Davies, F.E.; Richards, S.J.; Haynes, A.P.; Russell, N.H.; Hale, G.; Morgan, G.J.; Jack, A.S.; et al. Quantitation of minimal disease levels in chronic lymphocytic leukemia using a sensitive flow cytometric assay improves the prediction of outcome and can be used to optimize therapy. Blood 2001, 98, 29–35. [Google Scholar] [CrossRef] [PubMed]
- Theunissen, P.; Mejstrikova, E.; Sedek, L.; Van Der Sluijs-Gelling, A.J.; Gaipa, G.; Bartels, M.; Sobral da Costa, E.; Kotrová, M.; Novakova, M.; Sonneveld, E.; et al. Standardized flow cytometry for highly sensitive MRD measurements in B-cell acute lymphoblastic leukemia. Blood 2017, 129, 347–357. [Google Scholar] [CrossRef]
- Van Dongen, J.J.M.; Langerak, A.W.; Brüggemann, M.; Evans, P.A.S.; Hummel, M.; Lavender, F.L.; Delabesse, E.; Davi, F.; Schuuring, E.; García-Sanz, R.; et al. Design and standardization of PCR primers and protocols for detection of clonal immunoglobulin and T-cell receptor gene recombinations in suspect lymphoproliferations: Report of the BIOMED-2 concerted action BMH4-CT98-3936. Leukemia 2003, 17, 2257–2317. [Google Scholar] [CrossRef]
- Brüggemann, M.; Kotrová, M.; Knecht, H.; Bartram, J.; Boudjogrha, M.; Bystry, V.; Fazio, G.; Froňková, E.; Giraud, M.; Grioni, A.; et al. Standardized next-generation sequencing of immunoglobulin and T-cell receptor gene recombinations for MRD marker identification in acute lymphoblastic leukaemia; a EuroClonality-NGS validation study. Leukemia 2019, 33, 2241–2253. [Google Scholar] [CrossRef]
- Van der Velden, V.H.J.; Cazzaniga, G.; Schrauder, A.; Hancock, J.; Bader, P.; Panzer-Grumayer, E.R.; Flohr, T.; Sutton, R.; Cave, H.; Madsen, H.O.; et al. Analysis of minimal residual disease by Ig/TCR gene rearrangements: Guidelines for interpretation of real-time quantitative PCR data. Leukemia 2007, 21, 604–611. [Google Scholar] [CrossRef]
- Kotrova, M.; Van Der Velden, V.H.J.; Van Dongen, J.J.M.; Formankova, R.; Sedlacek, P.; Brüggemann, M.; Zuna, J.; Stary, J.; Trka, J.; Fronkova, E. Next-generation sequencing indicates false-positive MRD results and better predicts prognosis after SCT in patients with childhood ALL. Bone Marrow Transplant. 2017, 52, 962–968. [Google Scholar] [CrossRef]
- Cavalli, M.; De Novi, L.A.; Della Starza, I.; Cappelli, L.V.; Nunes, V.; Pulsoni, A.; Del Giudice, I.; Guarini, A.; Foà, R. Comparative analysis between RQ-PCR and digital droplet PCR of BCL2/IGH gene rearrangement in the peripheral blood and bone marrow of early stage follicular lymphoma. Br. J. Haematol. 2017, 177, 588–596. [Google Scholar] [CrossRef]
- Drandi, D.; Kubiczkova-Besse, L.; Ferrero, S.; Dani, N.; Passera, R.; Mantoan, B.; Gambella, M.; Monitillo, L.; Saraci, E.; Ghione, P.; et al. Minimal Residual Disease Detection by Droplet Digital PCR in Multiple Myeloma, Mantle Cell Lymphoma, and Follicular Lymphoma: A Comparison with Real-Time PCR. J. Mol. Diagn. 2015, 17, 652–660. [Google Scholar] [CrossRef]
- Della Starza, I.; Nunes, V.; Cavalli, M.; De Novi, L.A.; Ilari, C.; Apicella, V.; Vitale, A.; Testi, A.M.; Del Giudice, I.; Chiaretti, S.; et al. Comparative analysis between RQ-PCR and digital-droplet-PCR of immunoglobulin/T-cell receptor gene rearrangements to monitor minimal residual disease in acute lymphoblastic leukaemia. Br. J. Haematol. 2016, 174, 541–549. [Google Scholar] [CrossRef] [PubMed]
- Della Starza, I.; Nunes, V.; Lovisa, F.; Silvestri, D.; Cavalli, M.; Garofalo, A.; Campeggio, M.; De Novi, L.A.; Soscia, R.; Oggioni, C.; et al. Droplet Digital PCR Improves IG-/TR-based MRD Risk Definition in Childhood B-cell Precursor Acute Lymphoblastic Leukemia. HemaSphere 2021, 5, e543. [Google Scholar] [CrossRef]
- Hansen, M.H.; Cédile, O.; Larsen, T.S.; Abildgaard, N.; Nyvold, C.G. Perspective: Sensitive detection of residual lymphoproliferative disease by clonal gene rearrangements and NGS–How low can you go? Exp. Hematol. 2021, 98, 14–24. [Google Scholar] [CrossRef] [PubMed]
- Knecht, H.; Reigl, T.; Kotrová, M.; Appelt, F.; Stewart, P.; Bystry, V.; Krejci, A.; Grioni, A.; Pal, K.; Stranska, K.; et al. Quality control and quantification in IG/TR next-generation sequencing marker identification: Protocols and bioinformatic functionalities by EuroClonality-NGS. Leukemia 2019, 33, 2254–2265. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kotrova, M.; Darzentas, N.; Pott, C.; Baldus, C.D.; Brüggemann, M. Immune Gene Rearrangements: Unique Signatures for Tracing Physiological Lymphocytes and Leukemic Cells. Genes 2021, 12, 979. https://doi.org/10.3390/genes12070979
Kotrova M, Darzentas N, Pott C, Baldus CD, Brüggemann M. Immune Gene Rearrangements: Unique Signatures for Tracing Physiological Lymphocytes and Leukemic Cells. Genes. 2021; 12(7):979. https://doi.org/10.3390/genes12070979
Chicago/Turabian StyleKotrova, Michaela, Nikos Darzentas, Christiane Pott, Claudia D. Baldus, and Monika Brüggemann. 2021. "Immune Gene Rearrangements: Unique Signatures for Tracing Physiological Lymphocytes and Leukemic Cells" Genes 12, no. 7: 979. https://doi.org/10.3390/genes12070979
APA StyleKotrova, M., Darzentas, N., Pott, C., Baldus, C. D., & Brüggemann, M. (2021). Immune Gene Rearrangements: Unique Signatures for Tracing Physiological Lymphocytes and Leukemic Cells. Genes, 12(7), 979. https://doi.org/10.3390/genes12070979





