Building a Flower: The Influence of Cell Wall Composition on Flower Development and Reproduction
Abstract
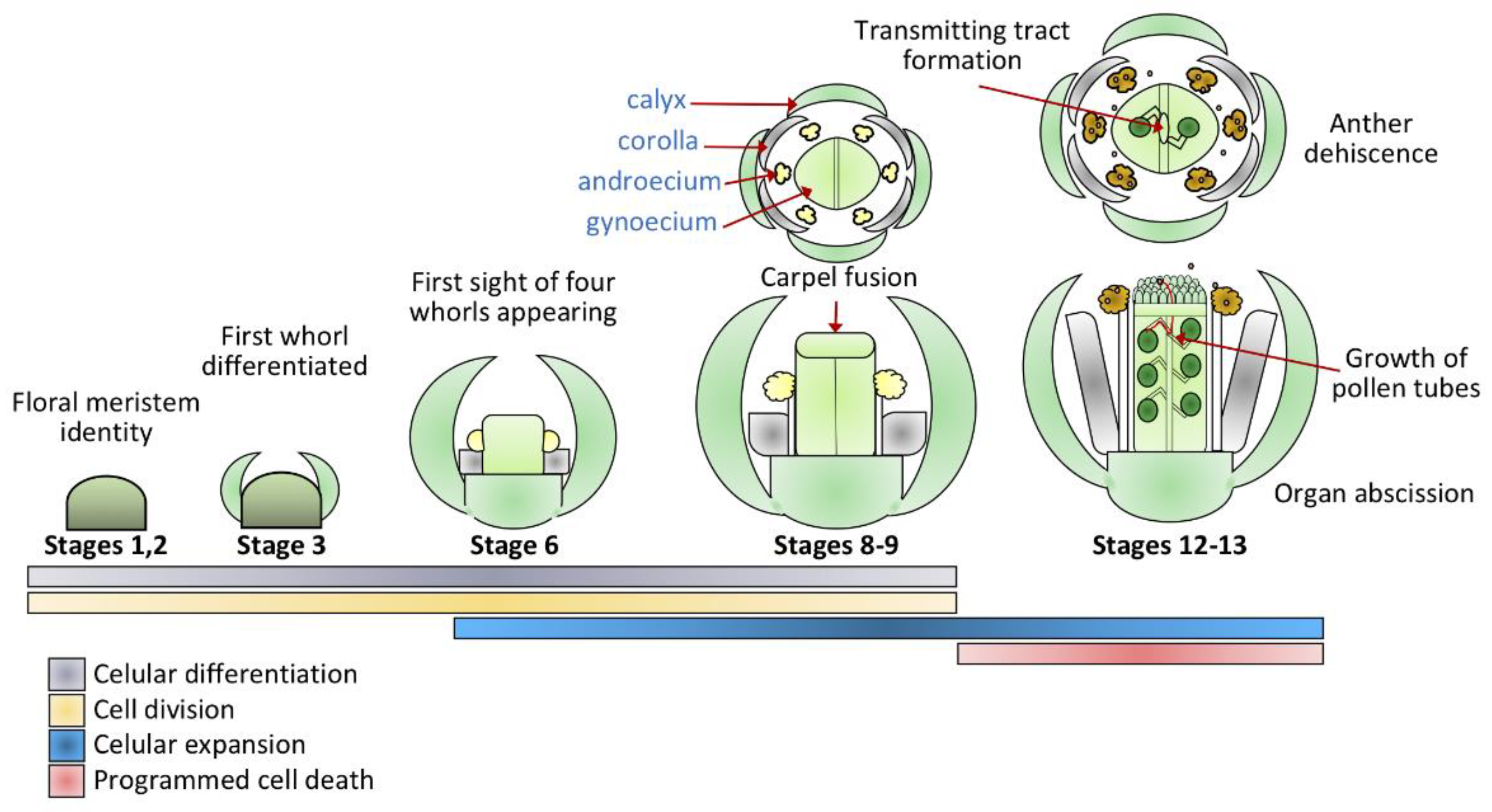
1. Introduction
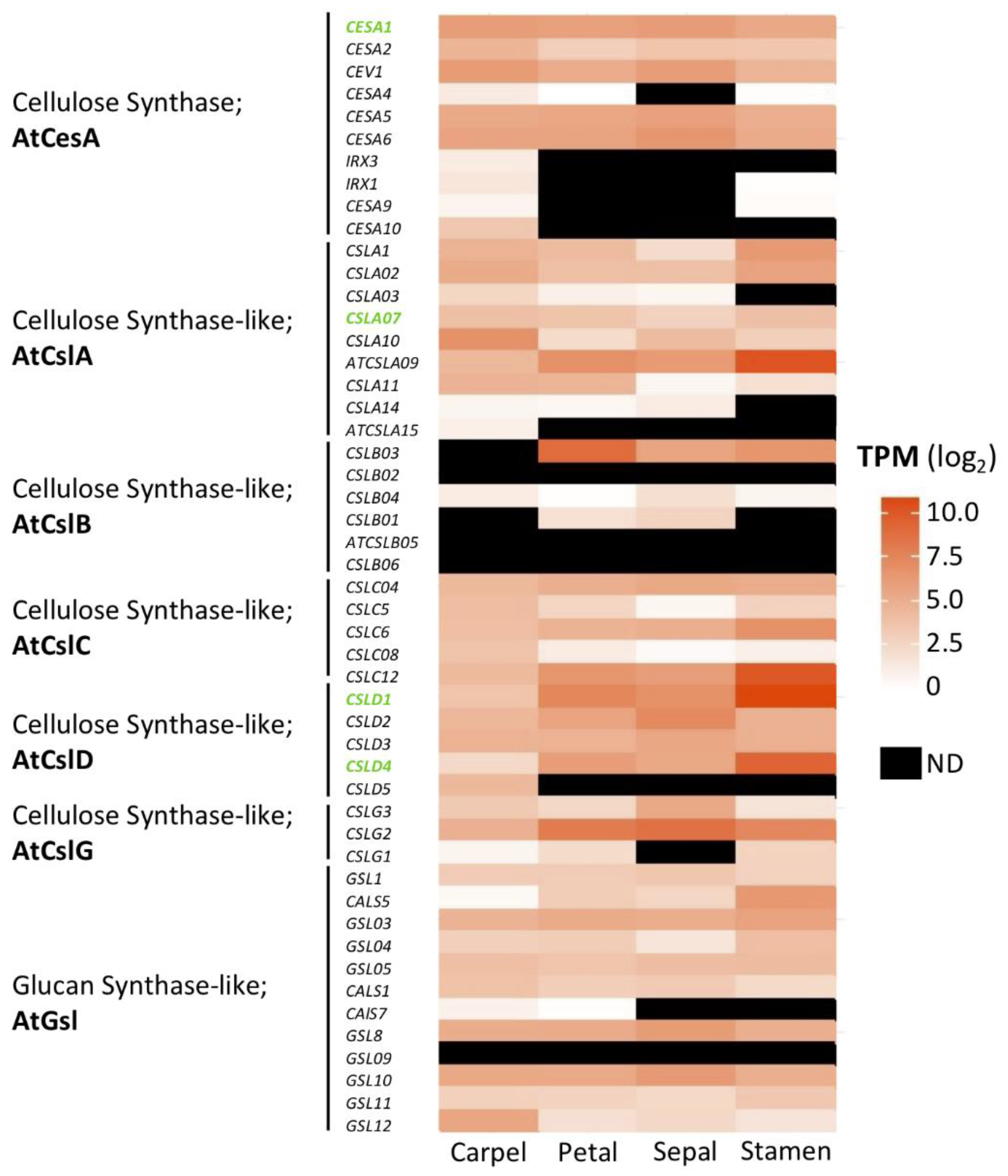
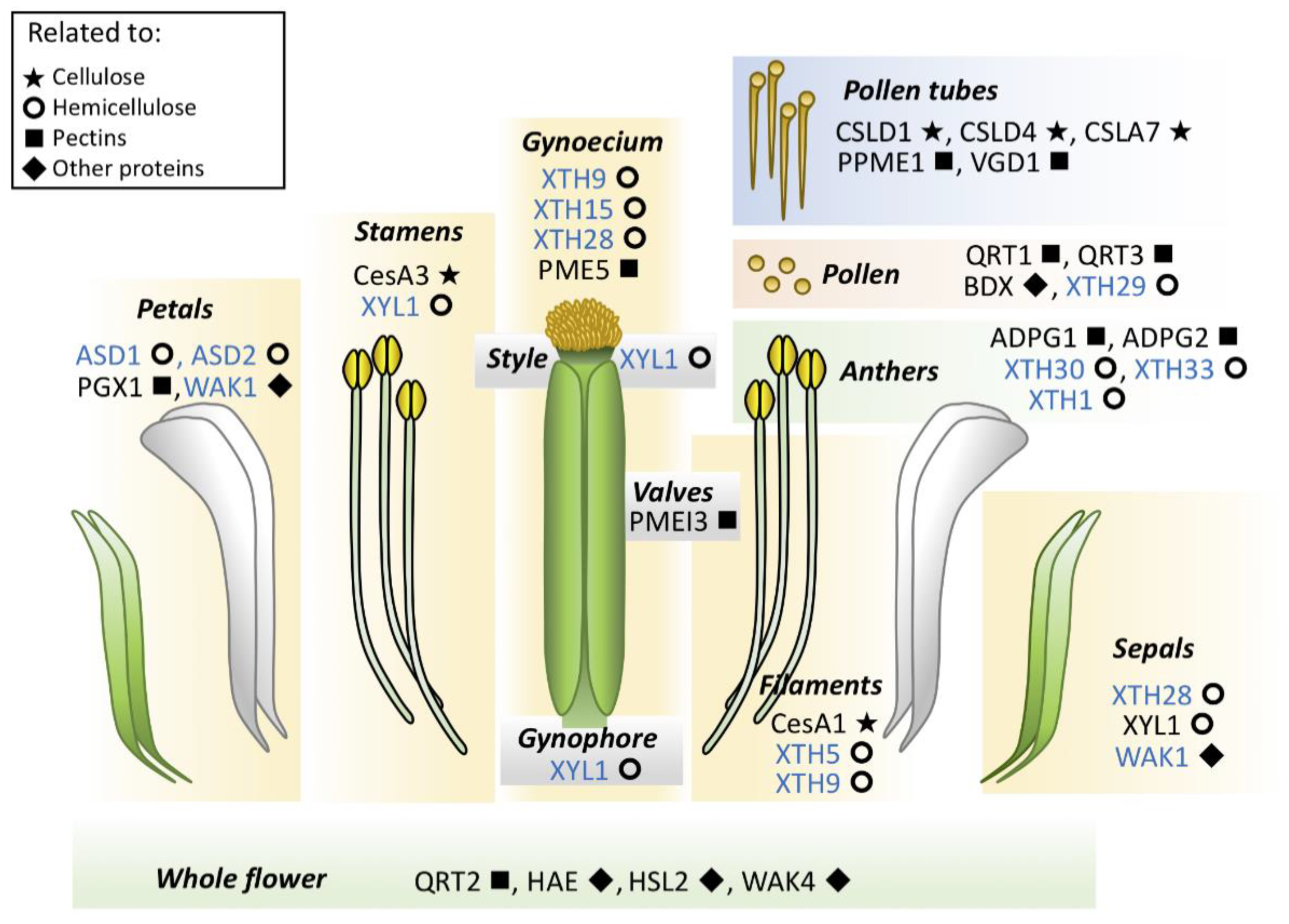
2. Cellulose
3. Hemicelluloses
4. Pectins
5. Other Cell Wall Proteins
6. Transcriptional Control and Hormonal Modulation
7. Concluding Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Alvarez-Buylla, E.R.; Benítez, M.; Corvera-Poiré, A.; Chaos Cador, A.; de Folter, S.; Gamboa de Buen, A.; Garay-Arroyo, A.; García-Ponce, B.; Jaimes-Miranda, F.; Pérez-Ruiz, R.V.; et al. Flower Development. Arab. Book 2010, 8, e0127. [Google Scholar] [CrossRef] [PubMed]
- Coen, E.S.; Meyerowitz, E.M. The War of the Whorls: Genetic Interactions Controlling Flower Development. Nature 1991, 353, 31–37. [Google Scholar] [CrossRef] [PubMed]
- Ferrándiz, C.; Fourquin, C.; Prunet, N.; Scutt, C.P.; Sundberg, E.; Trehin, C.; Vialette-Guiraud, A.C.M. Carpel Development. In Advances in Botanical Research; Academic Press: London, UK, 2010; Volume 55, pp. 1–73. ISBN 9780123808684. [Google Scholar]
- Smyth, D.R.; Bowman, J.L.; Meyerowitz, E.M. Early Flower Development in Arabidopsis. Plant. Cell 1990, 2, 755–767. [Google Scholar] [CrossRef]
- Irish, V. The ABC Model of Floral Development. Curr. Biol. 2017, 27, R887–R890. [Google Scholar] [CrossRef]
- Tucker, M.R.; Lou, H.; Aubert, M.K.; Wilkinson, L.G.; Little, A.; Houston, K.; Pinto, S.C.; Shirley, N.J. Exploring the Role of Cell Wall-Related Genes and Polysaccharides During Plant Development. Plants 2018, 7, 42. [Google Scholar] [CrossRef]
- Herrera-Ubaldo, H.; de Folter, S. Exploring Cell Wall Composition and Modifications During the Development of the Gynoecium Medial Domain in Arabidopsis. Front. Plant. Sci. 2018, 9, 454. [Google Scholar] [CrossRef]
- Somerville, C.; Bauer, S.; Brininstool, G.; Facette, M.; Hamann, T.; Milne, J.; Osborne, E.; Paredez, A.; Persson, S.; Raab, T.; et al. Toward a Systems Approach to Understanding Plant Cell Walls. Science 2004, 306, 2206–2211. [Google Scholar] [CrossRef]
- Wolf, S.; Hématy, K.; Höfte, H. Growth Control and Cell Wall Signaling in Plants. Annu. Rev. Plant. Biol. 2012, 63, 381–407. [Google Scholar] [CrossRef]
- Armezzani, A.; Abad, U.; Ali, O.; Andres Robin, A.; Vachez, L.; Larrieu, A.; Mellerowicz, E.J.; Taconnat, L.; Battu, V.; Stanislas, T.; et al. Transcriptional Induction of Cell Wall Remodelling Genes Is Coupled to Microtubule-Driven Growth Isotropy at the Shoot Apex in Arabidopsis. Development 2018, 145. [Google Scholar] [CrossRef]
- Yang, W.; Schuster, C.; Beahan, C.T.; Charoensawan, V.; Peaucelle, A.; Bacic, A.; Doblin, M.S.; Wightman, R.; Meyerowitz, E.M. Regulation of Meristem Morphogenesis by Cell Wall Synthases in Arabidopsis. Curr. Biol. 2016, 26, 1404–1415. [Google Scholar] [CrossRef] [PubMed]
- Sampathkumar, A.; Peaucelle, A.; Fujita, M.; Schuster, C.; Persson, S.; Wasteneys, G.O.; Meyerowitz, E.M. Primary Wall Cellulose Synthase Regulates Shoot Apical Meristem Mechanics and Growth. Development 2019, 146. [Google Scholar] [CrossRef]
- Zhao, F.; Chen, W.; Sechet, J.; Martin, M.; Bovio, S.; Lionnet, C.; Long, Y.; Battu, V.; Mouille, G.; Monéger, F.; et al. Xyloglucans and Microtubules Synergistically Maintain Meristem Geometry and Phyllotaxis. Plant. Physiol. 2019, 181, 1191–1206. [Google Scholar] [CrossRef] [PubMed]
- Peaucelle, A.; Braybrook, S.A.; Le Guillou, L.; Bron, E.; Kuhlemeier, C.; Höfte, H. Pectin-Induced Changes in Cell Wall Mechanics Underlie Organ Initiation in Arabidopsis. Curr. Biol. 2011, 21, 1720–1726. [Google Scholar] [CrossRef]
- McFarlane, H.E.; Döring, A.; Persson, S. The Cell Biology of Cellulose Synthesis. Annu. Rev. Plant. Biol. 2014, 65, 69–94. [Google Scholar] [CrossRef] [PubMed]
- Williamson, R.E.; Burn, J.E.; Birch, R.; Baskin, T.I.; Arioli, T.; Betzner, A.S.; Cork, A. Morphology of Rsw1, a Cellulose-Deficient Mutant of Arabidopsis Thaliana. Protoplasma 2001, 215, 116–127. [Google Scholar] [CrossRef] [PubMed]
- Persson, S.; Paredez, A.; Carroll, A.; Palsdottir, H.; Doblin, M.; Poindexter, P.; Khitrov, N.; Auer, M.; Somerville, C.R. Genetic Evidence for Three Unique Components in Primary Cell-Wall Cellulose Synthase Complexes in Arabidopsis. Proc. Natl. Acad. Sci. USA 2007, 104, 15566–15571. [Google Scholar] [CrossRef]
- Burn, J.E.; Hocart, C.H.; Birch, R.J.; Cork, A.C.; Williamson, R.E. Functional Analysis of the Cellulose Synthase Genes CesA1, CesA2, and CesA3 in Arabidopsis. Plant. Physiol. 2002, 129, 797–807. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Wang, L.; Chen, C.; Xiong, G.; Tan, X.-Y.; Yang, K.-Z.; Wang, Z.-C.; Zhou, Y.; Ye, D.; Chen, L.-Q. Arabidopsis CSLD1 and CSLD4 Are Required for Cellulose Deposition and Normal Growth of Pollen Tubes. J. Exp. Bot. 2011, 62, 5161–5177. [Google Scholar] [CrossRef]
- Goubet, F.; Misrahi, A.; Park, S.K.; Zhang, Z.; Twell, D.; Dupree, P. AtCSLA7, a Cellulose Synthase-Like Putative Glycosyltransferase, Is Important for Pollen Tube Growth and Embryogenesis in Arabidopsis. Plant. Physiol. 2003, 131, 547–557. [Google Scholar] [CrossRef] [PubMed]
- Coleman, H.D.; Beamish, L.; Reid, A.; Park, J.-Y.; Mansfield, S.D. Altered Sucrose Metabolism Impacts Plant Biomass Production and Flower Development. Transgenic Res. 2010, 19, 269–283. [Google Scholar] [CrossRef]
- Yoneda, A.; Ito, T.; Higaki, T.; Kutsuna, N.; Saito, T.; Ishimizu, T.; Osada, H.; Hasezawa, S.; Matsui, M.; Demura, T. Cobtorin Target Analysis Reveals that Pectin Functions in the Deposition of Cellulose Microfibrils in Parallel with Cortical Microtubules. Plant. J. 2010, 64, 657–667. [Google Scholar] [CrossRef] [PubMed]
- Mergner, J.; Frejno, M.; List, M.; Papacek, M.; Chen, X.; Chaudhary, A.; Samaras, P.; Richter, S.; Shikata, H.; Messerer, M.; et al. Mass-Spectrometry-Based Draft of the Arabidopsis Proteome. Nature 2020, 579, 409–414. [Google Scholar] [CrossRef]
- Pauly, M.; Gille, S.; Liu, L.; Mansoori, N.; de Souza, A.; Schultink, A.; Xiong, G. Hemicellulose Biosynthesis. Planta 2013, 238, 627–642. [Google Scholar] [CrossRef] [PubMed]
- Scheller, H.V.; Ulvskov, P. Hemicelluloses. Annu. Rev. Plant. Biol. 2010, 61, 263–289. [Google Scholar] [CrossRef] [PubMed]
- de Vetten, N.C.; Huber, D.J.; Gross, K.C. Endoglycanase-Catalyzed Degradation of Hemicelluloses during Development of Carnation (Dianthus caryophyllus L.) Petals. Plant. Physiol. 1991, 95, 853–860. [Google Scholar] [CrossRef] [PubMed]
- Becnel, J.; Natarajan, M.; Kipp, A.; Braam, J. Developmental Expression Patterns of Arabidopsis XTH Genes Reported by Transgenes and Genevestigator. Plant. Mol. Biol. 2006, 61, 451–467. [Google Scholar] [CrossRef] [PubMed]
- Tsuchiya, M.; Satoh, S.; Iwai, H. Distribution of XTH, Expansin, and Secondary-Wall-Related Cesa in Floral and Fruit Abscission Zones during Fruit Development in Tomato (Solanum lycopersicum). Front. Plant. Sci. 2015, 6, 323. [Google Scholar] [CrossRef] [PubMed]
- Harada, T.; Torii, Y.; Morita, S.; Onodera, R.; Hara, Y.; Yokoyama, R.; Nishitani, K.; Satoh, S. Cloning, Characterization, and Expression of Xyloglucan Endotransglucosylase/Hydrolase and Expansin Genes Associated with Petal Growth and Development during Carnation Flower Opening. J. Exp. Bot. 2011, 62, 815–823. [Google Scholar] [CrossRef] [PubMed]
- Raghothama, K.G.; Lawton, K.A.; Goldsbrough, P.B.; Woodson, W.R. Characterization of an Ethylene-Regulated Flower Senescence-Related Gene from Carnation. Plant. Mol. Biol. 1991, 17, 61–71. [Google Scholar] [CrossRef] [PubMed]
- O’Donoghue, E.M.; Eason, J.R.; Somerfield, S.D.; Ryan, D.A. Galactosidases in Opening, Senescing and Water-Stressed Sandersonia Aurantiaca Flowers. Funct. Plant. Biol. 2005, 32, 911–922. [Google Scholar] [CrossRef] [PubMed]
- Fulton, L.M.; Cobbett, C.S. Two Alpha-L-Arabinofuranosidase Genes in Arabidopsis Thaliana Are Differentially Expressed during Vegetative Growth and Flower Development. J. Exp. Bot. 2003, 54, 2467–2477. [Google Scholar] [CrossRef]
- Sampedro, J.; Pardo, B.; Gianzo, C.; Guitián, E.; Revilla, G.; Zarra, I. Lack of α-xylosidase Activity in Arabidopsis Alters Xyloglucan Composition and Results in Growth Defects. Plant. Physiol. 2010, 154, 1105–1115. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Yuan, D.; Gao, W.; Li, Y.; Tan, J.; Zhang, X. A Comparative Genome Analysis of PME and PMEI Families Reveals the Evolution of Pectin Metabolism in Plant Cell Walls. PLoS ONE 2013, 8, e72082. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, P.; Bosneaga, E.; Auer, M. Plant Cell Walls throughout Evolution: Towards a Molecular Understanding of Their Design Principles. J. Exp. Bot. 2009, 60, 3615–3635. [Google Scholar] [CrossRef]
- Atmodjo, M.A.; Hao, Z.; Mohnen, D. Evolving Views of Pectin Biosynthesis. Annu. Rev. Plant. Biol. 2013, 64, 747–779. [Google Scholar] [CrossRef]
- Ridley, B.L.; O’Neill, M.A.; Mohnen, D. Pectins: Structure, Biosynthesis, and Oligogalacturonide-Related Signaling. Phytochemistry 2001, 57, 929–967. [Google Scholar] [CrossRef]
- Gou, J.-Y.; Miller, L.M.; Hou, G.; Yu, X.-H.; Chen, X.-Y.; Liu, C.-J. Acetylesterase-Mediated Deacetylation of Pectin Impairs Cell Elongation, Pollen Germination, and Plant Reproduction. Plant. Cell 2012, 24, 50–65. [Google Scholar] [CrossRef]
- Hasegawa, K.; Kamada, S.; Takehara, S.; Takeuchi, H.; Nakamura, A.; Satoh, S.; Iwai, H. Rice Putative Methyltransferase Gene Ospmt16 Is Required for Pistil Development Involving Pectin Modification. Front. Plant. Sci. 2020, 11, 475. [Google Scholar] [CrossRef] [PubMed]
- Francis, K.E.; Lam, S.Y.; Copenhaver, G.P. Separation of Arabidopsis Pollen Tetrads Is Regulated by QUARTET1, a Pectin Methylesterase Gene. Plant. Physiol. 2006, 142, 1004–1013. [Google Scholar] [CrossRef]
- Jiang, L.; Yang, S.-L.; Xie, L.-F.; Puah, C.S.; Zhang, X.-Q.; Yang, W.-C.; Sundaresan, V.; Ye, D. VANGUARD1 Encodes a Pectin Methylesterase that Enhances Pollen Tube Growth in the Arabidopsis Style and Transmitting Tract. Plant. Cell 2005, 17, 584–596. [Google Scholar] [CrossRef]
- Andres-Robin, A.; Reymond, M.C.; Dupire, A.; Battu, V.; Dubrulle, N.; Mouille, G.; Lefebvre, V.; Pelloux, J.; Boudaoud, A.; Traas, J.; et al. Evidence for the Regulation of Gynoecium Morphogenesis by ETTIN Via Cell Wall Dynamics. Plant. Physiol. 2018, 178, 1222–1232. [Google Scholar] [CrossRef]
- Tian, G.-W.; Chen, M.-H.; Zaltsman, A.; Citovsky, V. Pollen-Specific Pectin Methylesterase Involved in Pollen Tube Growth. Dev. Biol. 2006, 294, 83–91. [Google Scholar] [CrossRef]
- Bosch, M.; Hepler, P.K. Silencing of the Tobacco Pollen Pectin Methylesterase Ntppme1 Results in Retarded In Vivo Pollen Tube Growth. Planta 2006, 223, 736–745. [Google Scholar] [CrossRef]
- Wormit, A.; Usadel, B. The Multifaceted Role of Pectin Methylesterase Inhibitors (PMEIs). Int. J. Mol. Sci. 2018, 19, 2878. [Google Scholar] [CrossRef] [PubMed]
- Röckel, N.; Wolf, S.; Kost, B.; Rausch, T.; Greiner, S. Elaborate Spatial Patterning of Cell-Wall PME and PMEI at the Pollen Tube Tip Involves PMEI Endocytosis, and Reflects the Distribution of Esterified and De-Esterified Pectins. Plant. J. 2008, 53, 133–143. [Google Scholar] [CrossRef]
- Zhang, G.Y.; Feng, J.; Wu, J.; Wang, X.W. BoPMEI1, a Pollen-Specific Pectin Methylesterase Inhibitor, Has an Essential Role in Pollen Tube Growth. Planta 2010, 231, 1323–1334. [Google Scholar] [CrossRef] [PubMed]
- Tan, C.; Liu, Z.; Huang, S.; Li, C.; Ren, J.; Tang, X.; Liu, W.; Peng, S.; Feng, H. Pectin Methylesterase Inhibitor (PMEI) Family Can Be Related to Male Sterility in Chinese Cabbage (Brassica Rapa Ssp. Pekinensis). Mol. Genet. Genom. 2018, 293, 343–357. [Google Scholar] [CrossRef]
- Cankar, K.; Kortstee, A.; Toonen, M.A.J.; Wolters-Arts, M.; Houbein, R.; Mariani, C.; Ulvskov, P.; Jorgensen, B.; Schols, H.A.; Visser, R.G.F.; et al. Pectic Arabinan Side Chains Are Essential for Pollen Cell Wall Integrity During Pollen Development. Plant. Biotechnol. J. 2014, 12, 492–502. [Google Scholar] [CrossRef] [PubMed]
- de Souza, A.J.; Pauly, M. Comparative Genomics of Pectinacetylesterases: Insight on Function and Biology. Plant. Signal. Behav. 2015, 10, e1055434. [Google Scholar] [CrossRef]
- Philippe, F.; Pelloux, J.; Rayon, C. Plant Pectin Acetylesterase Structure and Function: New Insights from Bioinformatic Analysis. BMC Genom. 2017, 18, 456. [Google Scholar] [CrossRef] [PubMed]
- Rhee, S.Y.; Osborne, E.; Poindexter, P.D.; Somerville, C.R. Microspore Separation in the Quartet 3 Mutants of Arabidopsis Is Impaired by a Defect in a Developmentally Regulated Polygalacturonase Required for Pollen Mother Cell Wall Degradation. Plant. Physiol. 2003, 133, 1170–1180. [Google Scholar] [CrossRef]
- Ogawa, M.; Kay, P.; Wilson, S.; Swain, S.M. ARABIDOPSIS DEHISCENCE ZONE POLYGALACTURONASE1 (ADPG1), ADPG2, and QUARTET2 are Polygalacturonases Required for Cell Separation during Reproductive Development in Arabidopsis. Plant. Cell 2009, 21, 216–233. [Google Scholar] [CrossRef]
- Huang, L.; Ye, Y.; Zhang, Y.; Zhang, A.; Liu, T.; Cao, J. BcMF9, a Novel Polygalacturonase Gene, Is Required for Both Brassica Campestris Intine and Exine Formation. Ann. Bot. 2009, 104, 1339–1351. [Google Scholar] [CrossRef]
- Huang, L.; Cao, J.; Zhang, A.; Ye, Y.; Zhang, Y.; Liu, T. The Polygalacturonase Gene BcMF2 from Brassica Campestris Is Associated with Intine Development. J. Exp. Bot. 2009, 60, 301–313. [Google Scholar] [CrossRef]
- Liao, J.; Chen, Z.; Wei, X.; Tao, K.; Zhang, J.; Qin, X.; Pan, Z.; Ma, W.; Pan, L.; Yang, S.; et al. Identification of Pollen and Pistil Polygalacturonases in Nicotiana Tabacum and Their Function in Interspecific Stigma Compatibility. Plant. Reprod. 2020, 33, 173–190. [Google Scholar] [CrossRef] [PubMed]
- Sander, L.; Child, R.; Ulvskov, P.; Albrechtsen, M.; Borkhardt, B. Analysis of a Dehiscence Zone Endo-Polygalacturonase in Oilseed Rape (Brassica napus) and Arabidopsis Thaliana: Evidence for Roles in Cell Separation in Dehiscence and Abscission Zones, and in Stylar Tissues during Pollen Tube Growth. Plant. Mol. Biol. 2001, 46, 469–479. [Google Scholar] [CrossRef] [PubMed]
- González-Carranza, Z.H.; Elliott, K.A.; Roberts, J.A. Expression of Polygalacturonases and Evidence to Support Their Role during Cell Separation Processes in Arabidopsis thaliana. J. Exp. Bot. 2007, 58, 3719–3730. [Google Scholar] [CrossRef]
- Zhang, A.; Qiu, L.; Huang, L.; Yu, X.; Lu, G.; Cao, J. Isolation and Characterization of an Anther-Specific Polygalacturonase Gene, BcMF16, in Brassica campestris ssp. chinensis. Plant. Mol. Biol. Rep. 2012, 30, 330–338. [Google Scholar] [CrossRef]
- Xiao, C.; Somerville, C.; Anderson, C.T. POLYGALACTURONASE INVOLVED IN EXPANSION1 Functions in Cell Elongation and Flower Development in Arabidopsis. Plant. Cell 2014, 26, 1018–1035. [Google Scholar] [CrossRef]
- Albenne, C.; Canut, H.; Jamet, E. Plant Cell Wall Proteomics: The Leadership of Arabidopsis thaliana. Front. Plant. Sci. 2013, 4, 111. [Google Scholar] [CrossRef] [PubMed]
- San Clemente, H.; Jamet, E. WallProtDB, a Database Resource for Plant Cell Wall Proteomics. Plant. Methods 2015, 11, 2. [Google Scholar] [CrossRef]
- Stenvik, G.-E.; Butenko, M.A.; Urbanowicz, B.R.; Rose, J.K.C.; Aalen, R.B. Overexpression of INFLORESCENCE DEFICIENT IN ABSCISSION Activates Cell Separation in Vestigial Abscission Zones in Arabidopsis. Plant. Cell 2006, 18, 1467–1476. [Google Scholar] [CrossRef] [PubMed]
- Ventimilla, D.; Velázquez, K.; Ruiz-Ruiz, S.; Terol, J.; Pérez-Amador, M.A.; Vives, M.C.; Guerri, J.; Talon, M.; Tadeo, F.R. IDA (INFLORESCENCE DEFICIENT IN ABSCISSION)-Like Peptides and HAE (HAESA)-Like Receptors Regulate Corolla Abscission in Nicotiana Benthamiana Flowers. BMC Plant. Biol. 2021, 21, 226. [Google Scholar] [CrossRef]
- Cho, S.K.; Larue, C.T.; Chevalier, D.; Wang, H.; Jinn, T.-L.; Zhang, S.; Walker, J.C. Regulation of Floral Organ Abscission in Arabidopsis Thaliana. Proc. Natl. Acad. Sci. USA 2008, 105, 15629–15634. [Google Scholar] [CrossRef]
- Lally, D.; Ingmire, P.; Tong, H.Y.; He, Z.H. Antisense Expression of a Cell Wall-Associated Protein Kinase, WAK4, Inhibits Cell Elongation and Alters Morphology. Plant. Cell 2001, 13, 1317–1331. [Google Scholar] [CrossRef]
- Cruz-Valderrama, J.E.; Gómez-Maqueo, X.; Salazar-Iribe, A.; Zúñiga-Sánchez, E.; Hernández-Barrera, A.; Quezada-Rodríguez, E.; Gamboa-deBuen, A. Overview of the Role of Cell Wall DUF642 Proteins in Plant Development. Int. J. Mol. Sci. 2019, 20, 3333. [Google Scholar] [CrossRef] [PubMed]
- Zúñiga-Sánchez, E.; Soriano, D.; Martínez-Barajas, E.; Orozco-Segovia, A.; Gamboa-deBuen, A. BIIDXI, the At4g32460 DUF642 gene, is Involved in Pectin Methyl Esterase Regulation during Arabidopsis Thaliana Seed Germination and Plant Development. BMC Plant. Biol. 2014, 14, 338. [Google Scholar] [CrossRef]
- Cosgrove, D.J. Loosening of Plant Cell Walls by Expansins. Nature 2000, 407, 321–326. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Xu, L.; Lin, H.; Cao, J. Two Expansin Genes, AtEXPA4 and AtEXPB5, Are Redundantly Required for Pollen Tube Growth and AtEXPA4 Is Involved in Primary Root Elongation in Arabidopsis thaliana. Genes 2021, 12, 249. [Google Scholar] [CrossRef]
- Zenoni, S.; Reale, L.; Tornielli, G.B.; Lanfaloni, L.; Porceddu, A.; Ferrarini, A.; Moretti, C.; Zamboni, A.; Speghini, A.; Ferranti, F.; et al. Downregulation of the Petunia Hybrida Alpha-Expansin Gene Phexp1 Reduces the Amount of Crystalline Cellulose in Cell Walls and Leads to Phenotypic Changes in Petal Limbs. Plant. Cell 2004, 16, 295–308. [Google Scholar] [CrossRef] [PubMed]
- Michael, A.J. A cDNA from Pea Petals with Sequence Similarity to Pollen Allergen, Cytokinin-Induced and Genetic Tumour-Specific Genes: Identification of a New Family of Related Sequences. Plant. Mol. Biol. 1996, 30, 219–224. [Google Scholar] [CrossRef] [PubMed]
- Brummell, D.A.; Harpster, M.H.; Dunsmuir, P. Differential Expression of Expansin Gene Family Members during Growth and Ripening of Tomato Fruit. Plant. Mol. Biol. 1999, 39, 161–169. [Google Scholar] [CrossRef] [PubMed]
- Ochiai, M.; Matsumoto, S.; Maesaka, M.; Yamada, K. Expression of mRNAs and Proteins Associated with Cell-wall-loosening during Eustoma Flower Opening. J. Jpn. Soc. Hort. Sci. 2013, 82, 154–160. [Google Scholar] [CrossRef][Green Version]
- Gookin, T.E.; Hunter, D.A.; Reid, M.S. Temporal Analysis of Alpha and Beta-Expansin Expression during Floral Opening and Senescence. Plant. Sci. 2003, 164, 769–781. [Google Scholar] [CrossRef]
- Showalter, A.M. Arabinogalactan-Proteins: Structure, Expression and Function. Cell Mol. Life Sci. 2001, 58, 1399–1417. [Google Scholar] [CrossRef]
- Coimbra, S.; Almeida, J.; Junqueira, V.; Costa, M.L.; Pereira, L.G. Arabinogalactan Proteins as Molecular Markers in Arabidopsis Thaliana Sexual Reproduction. J. Exp. Bot. 2007, 58, 4027–4035. [Google Scholar] [CrossRef] [PubMed]
- Silva, J.; Ferraz, R.; Dupree, P.; Showalter, A.M.; Coimbra, S. Three Decades of Advances in Arabinogalactan-Protein Biosynthesis. Front. Plant. Sci. 2020, 11, 610377. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Kieliszewski, M.J.; Showalter, A.M. Overexpression of Tomato LeAGP-1 Arabinogalactan-Protein Promotes Lateral Branching and Hampers Reproductive Development. Plant. J. 2004, 40, 870–881. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Sardar, H.S.; McGovern, K.R.; Zhang, Y.; Showalter, A.M. A Lysine-Rich Arabinogalactan Protein in Arabidopsis Is Essential for Plant Growth and Development, Including Cell Division and Expansion. Plant. J. 2007, 49, 629–640. [Google Scholar] [CrossRef]
- Lin, S.; Dong, H.; Zhang, F.; Qiu, L.; Wang, F.; Cao, J.; Huang, L. BcMF8, a Putative Arabinogalactan Protein-Encoding Gene, Contributes to Pollen Wall Development, Aperture Formation and Pollen Tube Growth in Brassica Campestris. Ann. Bot. 2014, 113, 777–788. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Long, T.; Wang, Y.; Tong, X.; Tang, J.; Li, J.; Wang, H.; Tang, L.; Li, Z.; Shu, Y.; et al. RMS2 Encoding a GDSL Lipase Mediates Lipid Homeostasis in Anthers to Determine Rice Male Fertility. Plant. Physiol. 2020, 182, 2047–2064. [Google Scholar] [CrossRef]
- Lombard, V.; Golaconda Ramulu, H.; Drula, E.; Coutinho, P.M.; Henrissat, B. The Carbohydrate-Active Enzymes Database (CAZy) in 2013. Nucleic Acids Res. 2014, 42, D490–D495. [Google Scholar] [CrossRef]
- Yadav, R.K.; Girke, T.; Pasala, S.; Xie, M.; Reddy, G.V. Gene Expression Map of the Arabidopsis Shoot Apical Meristem Stem Cell Niche. Proc. Natl. Acad. Sci. USA 2009, 106, 4941–4946. [Google Scholar] [CrossRef] [PubMed]
- Tian, C.; Wang, Y.; Yu, H.; He, J.; Wang, J.; Shi, B.; Du, Q.; Provart, N.J.; Meyerowitz, E.M.; Jiao, Y. A Gene Expression Map of Shoot Domains Reveals Regulatory Mechanisms. Nat. Commun. 2019, 10, 141. [Google Scholar] [CrossRef] [PubMed]
- Brady, S.M.; Orlando, D.A.; Lee, J.-Y.; Wang, J.Y.; Koch, J.; Dinneny, J.R.; Mace, D.; Ohler, U.; Benfey, P.N. A High-Resolution Root Spatiotemporal Map Reveals Dominant Expression Patterns. Science 2007, 318, 801–806. [Google Scholar] [CrossRef]
- Crawford, B.C.W.; Ditta, G.; Yanofsky, M.F. The NTT Gene Is Required for Transmitting-Tract Development in Carpels of Arabidopsis Thaliana. Curr. Biol. 2007, 17, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
- Crawford, B.C.W.; Yanofsky, M.F. The formation and function of the female reproductive tract in flowering plants. Curr. Biol. 2008, 18, R972–R978. [Google Scholar] [CrossRef]
- Herrera-Ubaldo, H.; Lozano-Sotomayor, P.; Ezquer, I.; Di Marzo, M.; Chávez Montes, R.A.; Gómez-Felipe, A.; Pablo-Villa, J.; Diaz-Ramirez, D.; Ballester, P.; Ferrándiz, C.; et al. New Roles of NO TRANSMITTING TRACT and SEEDSTICK During Medial Domain Development in Arabidopsis Fruits. Development 2019, 146. [Google Scholar] [CrossRef]
- Di Marzo, M.; Roig-Villanova, I.; Zanchetti, E.; Caselli, F.; Gregis, V.; Bardetti, P.; Chiara, M.; Guazzotti, A.; Caporali, E.; Mendes, M.A.; et al. MADS-Box and bHLH Transcription Factors Coordinate Transmitting Tract Development in Arabidopsis thaliana. Front. Plant. Sci. 2020, 11, 526. [Google Scholar] [CrossRef]
- Pereira, A.M.; Moreira, D.; Coimbra, S.; Masiero, S. Paving the Way for Fertilization: The Role of the Transmitting Tract. Int. J. Mol. Sci. 2021, 22, 2603. [Google Scholar] [CrossRef]
- Ezquer, I.; Mizzotti, C.; Nguema-Ona, E.; Gotté, M.; Beauzamy, L.; Viana, V.E.; Dubrulle, N.; Costa de Oliveira, A.; Caporali, E.; Koroney, A.-S.; et al. The Developmental Regulator SEEDSTICK Controls Structural and Mechanical Properties of the Arabidopsis Seed Coat. Plant. Cell 2016, 28, 2478–2492. [Google Scholar] [CrossRef]
- Mizzotti, C.; Ezquer, I.; Paolo, D.; Rueda-Romero, P.; Guerra, R.F.; Battaglia, R.; Rogachev, I.; Aharoni, A.; Kater, M.M.; Caporali, E.; et al. SEEDSTICK is a Master Regulator of Development and Metabolism in the Arabidopsis Seed Coat. PLoS Genet. 2014, 10, e1004856. [Google Scholar] [CrossRef]
- Balanzà, V.; Roig-Villanova, I.; Di Marzo, M.; Masiero, S.; Colombo, L. Seed Abscission and Fruit Dehiscence Required for Seed Dispersal Rely on Similar Genetic Networks. Development 2016, 143, 3372–3381. [Google Scholar] [CrossRef] [PubMed]
- Krizek, B.A.; Bequette, C.J.; Xu, K.; Blakley, I.C.; Fu, Z.Q.; Stratmann, J.W.; Loraine, A.E. RNA-Seq Links the Transcription Factors AINTEGUMENTA and AINTEGUMENTA-LIKE6 to Cell Wall Remodeling and Plant Defense Pathways. Plant. Physiol. 2016, 171, 2069–2084. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, N.; Huang, J.; Tatsumi, Y.; Abe, M.; Sugano, S.S.; Kojima, M.; Takebayashi, Y.; Kiba, T.; Yokoyama, R.; Nishitani, K.; et al. Chromatin-Mediated Feed-Forward Auxin Biosynthesis in Floral Meristem Determinacy. Nat. Commun. 2018, 9, 5290. [Google Scholar] [CrossRef] [PubMed]
- Majda, M.; Robert, S. The Role of Auxin in Cell Wall Expansion. Int. J. Mol. Sci. 2018, 19, 951. [Google Scholar] [CrossRef]
- Feraru, E.; Feraru, M.I.; Kleine-Vehn, J.; Martinière, A.; Mouille, G.; Vanneste, S.; Vernhettes, S.; Runions, J.; Friml, J. PIN Polarity Maintenance by the Cell Wall in Arabidopsis. Curr. Biol. 2011, 21, 338–343. [Google Scholar] [CrossRef]
- Velasquez, S.M.; Gallemi, M.; Aryal, B.; Venhuizen, P.; Barbez, E.; Dünser, K.; Kalyna, M.; Mouille, G.; Benkova, E.; Bhalerao, R.; et al. Auxin-Dependent Xyloglucan Remodelling Defines Differential Tissue Expansion in Arabidopsis thaliana. BioRxiv 2019. [Google Scholar] [CrossRef]
- Braybrook, S.A.; Peaucelle, A. Mechano-Chemical Aspects of Organ Formation in Arabidopsis Thaliana: The Relationship Between Auxin and Pectin. PLoS ONE 2013, 8, e57813. [Google Scholar] [CrossRef]
- Marsch-Martínez, N.; Ramos-Cruz, D.; Irepan Reyes-Olalde, J.; Lozano-Sotomayor, P.; Zúñiga-Mayo, V.M.; de Folter, S. The Role of Cytokinin during Arabidopsis Gynoecia and Fruit Morphogenesis and Patterning. Plant. J. 2012, 72, 222–234. [Google Scholar] [CrossRef]
- Reyes-Olalde, J.I.; Zúñiga-Mayo, V.M.; Serwatowska, J.; Chavez Montes, R.A.; Lozano-Sotomayor, P.; Herrera-Ubaldo, H.; Gonzalez-Aguilera, K.L.; Ballester, P.; Ripoll, J.J.; Ezquer, I.; et al. The bHLH Transcription Factor SPATULA Enables Cytokinin Signaling, and Both Activate Auxin Biosynthesis and Transport Genes at the Medial Domain of the Gynoecium. PLoS Genet. 2017, 13, e1006726. [Google Scholar] [CrossRef]
- Jung, K.W.; Oh, S.-I.; Kim, Y.Y.; Yoo, K.S.; Cui, M.H.; Shin, J.S. Arabidopsis Histidine-Containing Phosphotransfer Factor 4 (AHP4) Negatively Regulates Secondary Wall Thickening of the Anther Endothecium during Flowering. Mol. Cells 2008, 25, 294–300. [Google Scholar]
- Samalova, M.; Elsayad, K.; Melnikava, A.; Peaucelle, A.; Gahurova, E.; Gumulec, J.; Spyroglou, I.; Zemlyanskaya, E.V.; Ubogoeva, E.V.; Hejatko, J. Expansin-Controlled Cell Wall Stiffness Regulates Root Growth in Arabidopsis. BioRxiv 2020. [Google Scholar] [CrossRef]
- Pacifici, E.; Di Mambro, R.; Dello Ioio, R.; Costantino, P.; Sabatini, S. Acidic Cell Elongation Drives Cell Differentiation in the Arabidopsis Root. EMBO J. 2018, 37, e99134. [Google Scholar] [CrossRef] [PubMed]
- O’Donoghue, E.M. Flower Petal Cell Walls: Changes Associated with Flower Opening and Senescence. N. Z. J. For. Sci 2006, 36, 134–144. [Google Scholar]
- Sénéchal, F.; Wattier, C.; Rustérucci, C.; Pelloux, J. Homogalacturonan-Modifying Enzymes: Structure, Expression, and Roles in Plants. J. Exp. Bot. 2014, 65, 5125–5160. [Google Scholar] [CrossRef]
- Mollet, J.-C.; Leroux, C.; Dardelle, F.; Lehner, A. Cell Wall Composition, Biosynthesis and Remodeling during Pollen Tube Growth. Plants 2013, 2, 107–147. [Google Scholar] [CrossRef] [PubMed]
- Cascallares, M.; Setzes, N.; Marchetti, F.; López, G.A.; Distéfano, A.M.; Cainzos, M.; Zabaleta, E.; Pagnussat, G.C. A Complex Journey: Cell Wall Remodeling, Interactions, and Integrity During Pollen Tube Growth. Front. Plant. Sci. 2020, 11, 599247. [Google Scholar] [CrossRef]
- Ageez, A.; Kazama, Y.; Sugiyama, R.; Kawano, S. Male-Fertility Genes Expressed in Male Flower Buds of Silene Latifolia Include Homologs of Anther-Specific Genes. Genes Genet. Syst. 2005, 80, 403–413. [Google Scholar] [CrossRef] [PubMed]
- Kong, Y.; Zhou, G.; Yin, Y.; Xu, Y.; Pattathil, S.; Hahn, M.G. Molecular Analysis of a Family of Arabidopsis Genes Related to Galacturonosyltransferases. Plant. Physiol. 2011, 155, 1791–1805. [Google Scholar] [CrossRef]
- Li, Q.; Chen, J.; Xiao, Y.; Di, P.; Zhang, L.; Chen, W. The Dirigent Multigene Family in Isatis Indigotica: Gene Discovery and Differential Transcript Abundance. BMC Genom. 2014, 15, 388. [Google Scholar] [CrossRef] [PubMed]
- Xie, L.; Shang, Q. Genome-Wide Analysis of Purple Acid Phosphatase Structure and Expression in Ten Vegetable Species. BMC Genom. 2018, 19, 646. [Google Scholar] [CrossRef] [PubMed]
- Pereira, A.M.; Masiero, S.; Nobre, M.S.; Costa, M.L.; Solís, M.-T.; Testillano, P.S.; Sprunck, S.; Coimbra, S. Differential Expression Patterns of Arabinogalactan Proteins in Arabidopsis Thaliana Reproductive Tissues. J. Exp. Bot. 2014, 65, 5459–5471. [Google Scholar] [CrossRef]
- Zhu, H.; Qian, W.; Lu, X.; Li, D.; Liu, X.; Liu, K.; Wang, D. Expression Patterns of Purple Acid Phosphatase Genes in Arabidopsis Organs and Functional Analysis of Atpap23 Predominantly Transcribed in Flower. Plant. Mol. Biol. 2005, 59, 581–594. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cruz-Valderrama, J.E.; Bernal-Gallardo, J.J.; Herrera-Ubaldo, H.; de Folter, S. Building a Flower: The Influence of Cell Wall Composition on Flower Development and Reproduction. Genes 2021, 12, 978. https://doi.org/10.3390/genes12070978
Cruz-Valderrama JE, Bernal-Gallardo JJ, Herrera-Ubaldo H, de Folter S. Building a Flower: The Influence of Cell Wall Composition on Flower Development and Reproduction. Genes. 2021; 12(7):978. https://doi.org/10.3390/genes12070978
Chicago/Turabian StyleCruz-Valderrama, José Erik, Judith Jazmin Bernal-Gallardo, Humberto Herrera-Ubaldo, and Stefan de Folter. 2021. "Building a Flower: The Influence of Cell Wall Composition on Flower Development and Reproduction" Genes 12, no. 7: 978. https://doi.org/10.3390/genes12070978
APA StyleCruz-Valderrama, J. E., Bernal-Gallardo, J. J., Herrera-Ubaldo, H., & de Folter, S. (2021). Building a Flower: The Influence of Cell Wall Composition on Flower Development and Reproduction. Genes, 12(7), 978. https://doi.org/10.3390/genes12070978





