Genomic Investigation of Carbapenem-Resistant Klebsiella pneumonia Colonization in an Intensive Care Unit in South Africa
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Sample Collection and Screening for CPEs
2.3. Antimicrobial Susceptibility Testing (AST)
2.4. Genomic Characterization
2.4.1. DNA Extraction of CPKP Isolates
2.4.2. Genomic Fingerprinting
2.4.3. Whole Genome Sequencing (WGS) of CPKP Isolates
2.4.4. Genomic Analyses and Annotation
2.4.5. Phylogenomics and Epidemiological Insights
3. Results
3.1. Population Demographics
3.2. Antimicrobial Resistance Profiles
3.3. Genomic Fingerprinting
3.4. Genome Characteristics of the Isolates
3.5. In Silico ARGs Analysis
3.6. Sequence Types Analysis (MLST), In Silico Mobile Genetic Elements (Mobilome), Virulome and Capsular Serotypes
3.7. Phylogenomic Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Klein, E.Y.; Van Boeckel, T.P.; Martinez, E.M.; Pant, S.; Gandra, S.; Levin, S.A.; Goossens, H.; Laxminarayan, R. Global increase and geographic convergence in antibiotic consumption between 2000 and 2015. Proc. Natl. Acad. Sci. USA 2018, 115, E3463–E3470. [Google Scholar] [CrossRef]
- Bush, K.; Bradford, P.A. β-lactams and β-lactamase inhibitors: An overview. Cold Spring Harb. Perspect. Med. 2016, 6, a025247. [Google Scholar] [CrossRef]
- Ramsamy, Y.; Mlisana, K.P.; Allam, M.; Amoako, D.G.; Abia, A.L.K.; Ismail, A.; Singh, R.; Kisten, T.; Han, K.S.; Muckart, D.J.J.; et al. Genomic analysis of carbapenemase-producing extensively drug-resistant Klebsiella pneumonia isolates reveals the horizontal spread of p18-43_01 plasmid encoding blandm-1 in South Africa. Microorganisms 2020, 8, 137. [Google Scholar] [CrossRef]
- Pitout, J.D.D.; Nordmann, P.; Poirel, L. Carbapenemase-producing klebsiella pneumoniae, a key pathogen set for global nosocomial dominance. Antimicrob. Agents Chemother. 2015, 59, 5873–5884. [Google Scholar] [CrossRef]
- Bush, K.; Bradford, P.A. Epidemiology of β-lactamase-producing pathogens. Clin. Microbiol. Rev. 2020, 33, e00047-19. [Google Scholar] [CrossRef]
- Cui, X.; Zhang, H.; Du, H. Carbapenemases in Enterobacteriaceae: Detection and antimicrobial therapy. Front. Microbiol. 2019, 10, 1823. [Google Scholar] [CrossRef]
- Somboro, A.M.; Osei Sekyere, J.; Amoako, D.G.; Essack, S.Y.; Bester, L.A. Diversity and proliferation of metallo-β-lactamases: A clarion call for clinically effective metallo-β-lactamase inhibitors. Appl. Environ. Microbiol. 2018, 84, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Lomovskaya, O.; Sun, D.; Rubio-Aparicio, D.; Nelson, K.; Tsivkovski, R.; Griffith, D.C.; Dudley, M.N. Vaborbactam: Spectrum of beta-lactamase inhibition and impact of resistance mechanisms on activity in enterobacteriaceae. Antimicrob. Agents Chemother. 2017, 61, 61. [Google Scholar] [CrossRef] [PubMed]
- Brink, A.J.; Coetzee, J.; Corcoran, C.; Clay, C.G.; Hari-Makkan, D.; Jacobson, R.K.; Richards, G.A.; Feldman, C.; Nutt, L.; van Greune, J. Emergence of OXA-48 and OXA-181 carbapenemases among Enterobacteriaceae in South Africa and evidence of in vivo selection of colistin resistance as a consequence of selective decontamination of the gastrointestinal tract. J. Clin. Microbiol. 2013, 51, 369–372. [Google Scholar] [CrossRef] [PubMed]
- Poirel, L.; Potron, A.; Nordmann, P. OXA-48-like carbapenemases: The phantom menace. J. Antimicrob. Chemother. 2012, 67, 1597–1606. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. WHO Priority Pathogens List for R&D of New Antibiotics. Available online: http://www.who.int/bulletin/volumes/94/9/16-020916.pdf (accessed on 3 September 2020).
- Martin, A.; Fahrbach, K.; Zhao, Q.; Lodise, T. Association between carbapenem resistance and mortality among adult, hospitalized patients with serious infections due to enterobacteriaceae: Results of a systematic literature review and meta-analysis. Open Forum Infect. Dis. 2018, 5, ofy150. [Google Scholar] [CrossRef]
- Tamma, P.D.; Goodman, K.E.; Harris, A.D.; Tekle, T.; Roberts, A.; Taiwo, A.; Simner, P.J. Comparing the outcomes of patients with carbapenemase-producing and non-carbapenemase- producing carbapenem-resistant enterobacteriaceae bacteremia. Clin. Infect. Dis. 2017, 64, 257–264. [Google Scholar] [CrossRef]
- Lynch, T.; Petkau, A.; Knox, N.; Graham, M.; Domselaar, V. A primer on infectious disease bacterial genomics. Clin. Microbiol. Rev. 2016, 29, 881–913. [Google Scholar] [CrossRef] [PubMed]
- Rozwandowicz, M.; Brouwer, M.S.M.; Fischer, J.; Wagenaar, J.A.; Gonzalez-Zorn, B.; Guerra, B.; Mevius, D.J.; Hordijk, J. Plasmids carrying antimicrobial resistance genes in Enterobacteriaceae. J. Antimicrob. Chemother. 2018, 73, 1121–1137. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.U.; Maryam, L.; Zarrilli, R. Structure, genetics and worldwide spread of New Delhi metallo-β-lactamase (NDM): A threat to public health. BMC Microbiol. 2017, 17, 101. [Google Scholar] [CrossRef] [PubMed]
- Pitout, J.D.D.; Peirano, G.; Kock, M.M.; Strydom, K.-A.; Matsumura, Y. The global ascendency of OXA-48-type carbapenemases. Clin. Microbiol. Rev. 2019, 33. [Google Scholar] [CrossRef] [PubMed]
- Partridge, S.R. Analysis of antibiotic resistance regions in Gram-negative bacteria. FEMS Microbiol. Rev. 2011, 35, 820–855. [Google Scholar] [CrossRef] [PubMed]
- Mathers, A.J.; Peirano, G.; Pitout, J.D.D. The role of epidemic resistance plasmids and international high-risk clones in the spread of multidrug-resistant enterobacteriaceae. Clin. Microbiol. Rev. 2015, 28, 565–591. [Google Scholar] [CrossRef]
- Xu, L.; Sun, X.; Ma, X. Systematic review and meta-analysis of mortality of patients infected with carbapenem-resistant Klebsiella pneumoniae. Ann. Clin. Microbiol. Antimicrob. 2017, 16, 18. [Google Scholar] [CrossRef]
- Tatusova, T.; Dicuccio, M.; Badretdin, A.; Chetvernin, V.; Nawrocki, P.; Zaslavsky, L.; Lomsadze, A.; Pruitt, K.D.; Borodovsky, M.; Ostell, J. NCBI prokaryotic genome annotation pipeline. Microbiol. Resour. Announc. 2016, 44, 6614–6624. [Google Scholar] [CrossRef]
- Zankari, E.; Hasman, H.; Cosentino, S.; Vestergaard, M.; Rasmussen, S.; Lund, O.; Aarestrup, F.M.; Larsen, M.V. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 2012, 67, 2640–2644. [Google Scholar] [CrossRef]
- Carattoli, A.; Zankari, E.; García-Fernández, A.; Voldby Larsen, M.; Lund, O.; Villa, L.; Møller Aarestrup, F.; Hasman, H. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef]
- Overbeek, R.; Olson, R.; Pusch, G.D.; Olsen, G.J.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Parrello, B.; Shukla, M.; et al. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res. 2014, 42, D206–D214. [Google Scholar] [CrossRef] [PubMed]
- Johansson, M.H.K.; Bortolaia, V.; Tansirichaiya, S.; Aarestrup, F.M.; Roberts, A.P.; Petersen, T.N. Detection of mobile genetic elements associated with antibiotic resistance in Salmonella enterica using a newly developed web tool: MobileElementFinder. J. Antimicrob. Chemother. 2021, 76, 101–109. [Google Scholar] [CrossRef] [PubMed]
- Wyres, K.L.; Wick, R.R.; Gorrie, C.; Jenney, A.; Follador, R.; Thomson, N.R.; Holt, K.E. Identification of Klebsiella capsule synthesis loci from whole genome data. Microb. Genom. 2016, 2, e000102. [Google Scholar] [CrossRef] [PubMed]
- Kaas, R.S.; Leekitcharoenphon, P.; Aarestrup, F.M.; Lund, O. Solving the problem of comparing whole bacterial genomes across different sequencing platforms. PLoS ONE 2014, 9, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Sheu, C.-C.; Chang, Y.-T.; Lin, S.-Y.; Chen, Y.-H.; Hsueh, P.-R. Infections Caused by Carbapenem-Resistant Enterobacteriaceae: An Update on Therapeutic Options. Front. Microbiol. 2019, 10, 80. [Google Scholar] [CrossRef] [PubMed]
- Navon-Venezia, S.; Kondratyeva, K.; Carattoli, A. Klebsiella pneumoniae: A major worldwide source and shuttle for antibiotic resistance. FEMS Microbiol. Rev. 2017, 41, 252–275. [Google Scholar] [CrossRef]
- Jacobson, R.K.; Manesen, M.R.; Moodley, C.; Smith, M.; Williams, S.; Nicol, M.; Bamford, C. Molecular characterisation and epidemiological investigation of an outbreak of blaOXA-181 carbapenemase-producing isolates of Klebsiella pneumoniae in South Africa. S. Afr. Med. J. 2015, 105, 1030. [Google Scholar] [CrossRef]
- Haidar, G.; Alkroud, A.; Cheng, S.; Churilla, T.M.; Churilla, B.M.; Shields, R.K.; Doi, Y.; Clancy, C.J.; Nguyen, M.H. Association between the presence of aminoglycoside-modifying enzymes and in vitro activity of gentamicin, tobramycin, amikacin, and plazomicin against klebsiella pneumoniae carbapenemase- and extended-spectrum-β-lactamase-producing enterobacter species. Antimicrob. Agents Chemother. 2016, 60, 5208–5214. [Google Scholar] [CrossRef]
- Almaghrabi, R.; Clancy, C.J.; Doi, Y.; Hao, B.; Chen, L.; Shields, R.K.; Press, E.G.; Iovine, N.M.; Townsend, B.M.; Wagener, M.M.; et al. Carbapenem-resistant klebsiella pneumoniae strains exhibit diversity in aminoglycoside-modifying enzymes, which exert differing effects on plazomicin and other agents. Antimicrob. Agents Chemother. 2014, 58, 4443–4451. [Google Scholar] [CrossRef]
- Butler, D.A.; Rana, A.P.; Krapp, F.; Patel, S.R.; Huang, Y.; Ozer, E.A.; Hauser, A.R.; Bulman, Z.P. Optimizing aminoglycoside selection for KPC-producing Klebsiella pneumoniae with the aminoglycoside-modifying enzyme (AME) gene aac(6′)-Ib. J. Antimicrob. Chemother. 2021, 76, 671–679. [Google Scholar] [CrossRef]
- Kopotsa, K.; Mbelle, N.M.; Osei Sekyere, J. Epigenomics, genomics, resistome, mobilome, virulome and evolutionary phylogenomics of carbapenem-resistant Klebsiella pneumoniae clinical strains. Microb. Genom. 2020, 6, e000474. [Google Scholar] [CrossRef]
- Kayama, S.; Koba, Y.; Shigemoto, N.; Kuwahara, R.; Kakuhama, T.; Kimura, K.; Hisatsune, J.; Onodera, M.; Yokozaki, M.; Ohge, H.; et al. Imipenem-susceptible, meropenem-resistant Klebsiella pneumoniae producing OXA-181 in Japan. Antimicrob. Agents Chemother. 2015, 59, 1379–1380. [Google Scholar] [CrossRef][Green Version]
- Castanheira, M.; Farrell, S.E.; Wanger, A.; Rolston, K.V.; Jones, R.N.; Mendes, R.E. Rapid expansion of KPC-2-producing klebsiella pneumoniae isolates in two texas hospitals due to clonal Spread of ST258 and ST307 Lineages. Microb. Drug Resist. 2013, 19, 295–297. [Google Scholar] [CrossRef]
- Habeeb, M.A.; Haque, A.; Nematzadeh, S.; Iversen, A.; Giske, C.G. High prevalence of 16S rRNA methylase RmtB among CTX-M extended-spectrum β-lactamase-producing Klebsiella pneumoniae from Islamabad, Pakistan. Int. J. Antimicrob. Agents 2013, 41, 524–526. [Google Scholar] [CrossRef]
- Lazareva, I.V.; Ageevets, V.A.; Ershova, T.A.; Zueva, L.P.; Goncharov, A.E.; Darina, M.G.; Svetlichnaya, Y.S.; Uskov, A.N.; Sidorenko, S.V. Prevalence and antibiotic resistance of carbapenemase-producing gram-negative bacteria in Saint Petersburg and some other regions of the Russian Federation. Antibiot. Chemoterapy 2016, 61, 28–38. [Google Scholar]
- Ruiz-Garbajosa, P.; Hernández-García, M.; Beatobe, L.; Tato, M.; Méndez, M.I.; Grandal, M.; Aranzábal, L.; Alonso, S.; Lópaz, M.Á.; Astray, J.; et al. A single-day point-prevalence study of faecal carriers in long-term care hospitals in Madrid (Spain) depicts a complex clonal and polyclonal dissemination of carbapenemase-producing Enterobacteriaceae. J. Antimicrob. Chemother. 2016, 71, 348–352. [Google Scholar] [CrossRef]
- Dropa, M.; Lincopan, N.; Balsalobre, L.C.; Oliveira, D.E.; Moura, R.A.; Fernandes, M.R.; da Silva, Q.M.; Matté, G.R.; Sato, M.I.Z.; Matté, M.H. Genetic background of novel sequence types of CTX-M-8- and CTX-M-15-producing Escherichia coli and Klebsiella pneumoniae from public wastewater treatment plants in São Paulo, Brazil. Environ. Sci. Pollut. Res. 2016, 23, 4953–4958. [Google Scholar] [CrossRef]
- Harada, K.; Shimizu, T.; Mukai, Y.; Kuwajima, K.; Sato, T.; Usui, M.; Tamura, Y.; Kimura, Y.; Miyamoto, T.; Tsuyuki, Y.; et al. Phenotypic and molecular characterization of antimicrobial resistance in Klebsiella spp. isolates from companion animals in Japan: Clonal dissemination of multidrug-resistant extended-spectrum β-lactamase-producing klebsiella pneumoniae. Front. Microbiol. 2016, 7, 1021. [Google Scholar] [CrossRef]
- Baek, E.-H.; Kim, S.-E.; Kim, S.; Lee, S.; Cho, O.-H.; In Hong, S.; Shin, J.H.; Hwang, I. Successful control of an extended-spectrum beta-lactamase-producing Klebsiella pneumoniae ST307 outbreak in a neonatal intensive care unit. BMC Infect. Dis. 2020, 20, 166. [Google Scholar] [CrossRef] [PubMed]
- Ocampo, A.M.; Chen, L.; Cienfuegos, A.V.; Roncancio, G.; Chavda, K.D.; Kreiswirth, B.N.; Jiménez, J.N. A two-year surveillance in five Colombian tertiary care hospitals reveals high frequency of non-CG258 Clones of carbapenem-resistant klebsiella pneumoniae with distinct clinical characteristics. Antimicrob. Agents Chemother. 2016, 60, 332–342. [Google Scholar] [CrossRef]
- Long, S.W.; Olsen, R.J.; Eagar, T.N.; Beres, S.B.; Zhao, P.; Davis, J.J.; Brettin, T.; Xia, F.; Musser, J.M. Population Genomic Analysis of 1,777 Extended-spectrum beta-lactamase-producing klebsiella pneumoniae isolates, Houston, Texas: Unexpected abundance of clonal group 307. MBio 2017, 8, e00489-17. [Google Scholar] [CrossRef] [PubMed]
- Cejas, D.; Elena, A.; Guevara Nuñez, D.; Sevillano Platero, P.; De Paulis, A.; Magariños, F.; Alfonso, C.; Berger, M.A.; Fernández-Canigia, L.; Gutkind, G.; et al. Changing epidemiology of KPC-producing Klebsiella pneumoniae in Argentina: Emergence of hypermucoviscous ST25 and high-risk clone ST307. J. Glob. Antimicrob. Resist. 2019, 18, 238–242. [Google Scholar] [CrossRef]
- Bonura, C.; Giuffrè, M.; Aleo, A.; Fasciana, T.; Di Bernardo, F.; Stampone, T.; Giammanco, A.; MDR-GN Working Group; Palma, D.M.; Mammina, C. An update of the evolving epidemic of blaKPC carrying klebsiella pneumoniae in Sicily, Italy, 2014: Emergence of multiple non-ST258 clones. PLoS ONE 2015, 10, e0132936. [Google Scholar] [CrossRef]
- Lowe, M.; Kock, M.M.; Coetzee, J.; Hoosien, E.; Peirano, G.; Strydom, K.-A.; Ehlers, M.M.; Mbelle, N.M.; Shashkina, E.; Haslam, D.B.; et al. Klebsiella pneumoniae ST307 with blaOXA-181, South Africa, 2014–2016. Emerg. Infect. Dis. 2019, 25, 739–747. [Google Scholar] [CrossRef]
- Perovic, O.; Ismail, H.; Van Schalkwyk, E.; Lowman, W.; Prentice, E.; Senekal, M.; Govind, C.N. Antimicrobial resistance surveillance in the South African private sector report for 2016. South. Afr. J. Infect. Dis. 2018, 33, 114–117. [Google Scholar] [CrossRef]
- Osei Sekyere, J. Current state of resistance to antibiotics of last-resort in South Africa: A review from a public health perspective. Front. Public Health 2016, 4, 209. [Google Scholar] [CrossRef]
- Asante, J.; Osei Sekyere, J. Understanding antimicrobial discovery and resistance from a metagenomic and metatranscriptomic perspective: Advances and applications. Environ. Microbiol. Rep. 2019, 11, 62–86. [Google Scholar] [CrossRef] [PubMed]
- Partridge, S.R.; Kwong, S.M.; Firth, N.; Jensen, S.O. Mobile genetic elements associated with antimicrobial resistance. Clin. Microbiol. Rev. 2018, 31, 1–61. [Google Scholar] [CrossRef]
- Kopotsa, K.; Osei Sekyere, J.; Mbelle, N.M. Plasmid evolution in carbapenemase-producing Enterobacteriaceae: A review. Ann. N. Y. Acad. Sci. 2019, 1457, 61–91. [Google Scholar] [CrossRef]
- Mbelle, N.M.; Feldman, C.; Osei Sekyere, J.; Maningi, N.E.; Modipane, L.; Essack, S.Y. The resistome, mobilome, virulome and phylogenomics of multidrug-resistant escherichia coli clinical isolates from Pretoria, South Africa. Sci. Rep. 2019, 9, 16457. [Google Scholar] [CrossRef]
- Pedersen, T.; Sekyere, J.O.; Govinden, U.; Moodley, K.; Sivertsen, A.; Samuelsen, Ø.; Essack, S.Y.; Sundsfjord, A. Spread of plasmid-encoded NDM-1 and GES-5 carbapenemases among extensively drug-resistant and pandrug-resistant clinical enterobacteriaceae in Durban, South Africa. Antimicrob. Agents Chemother. 2018, 62, e02178–17. [Google Scholar] [CrossRef] [PubMed]
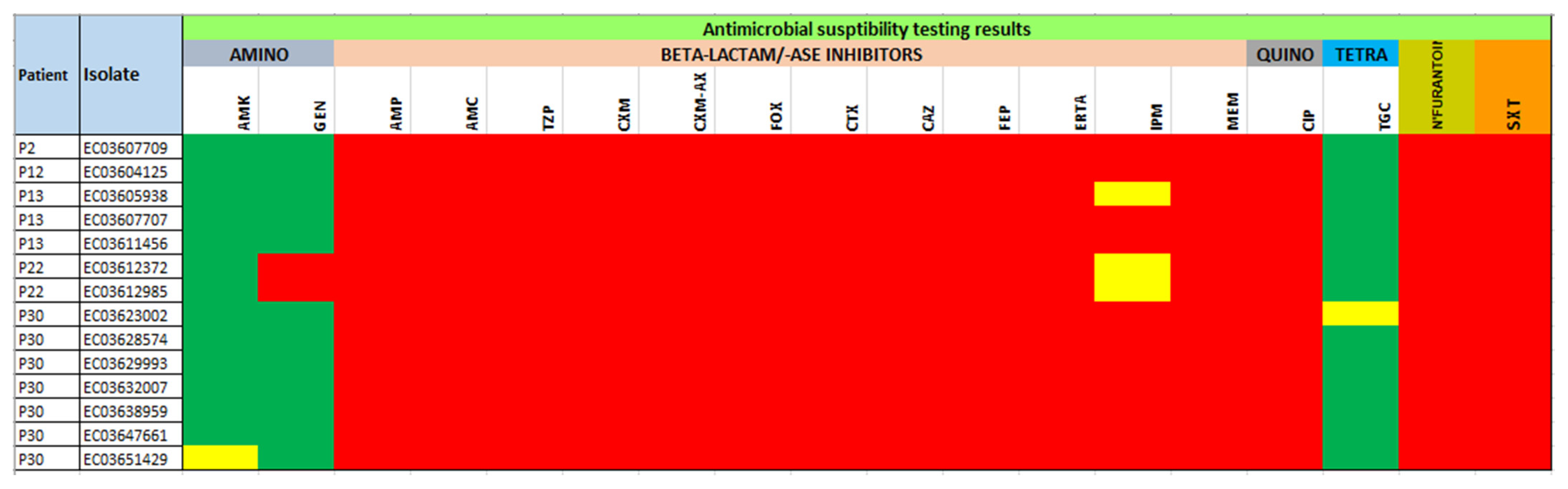
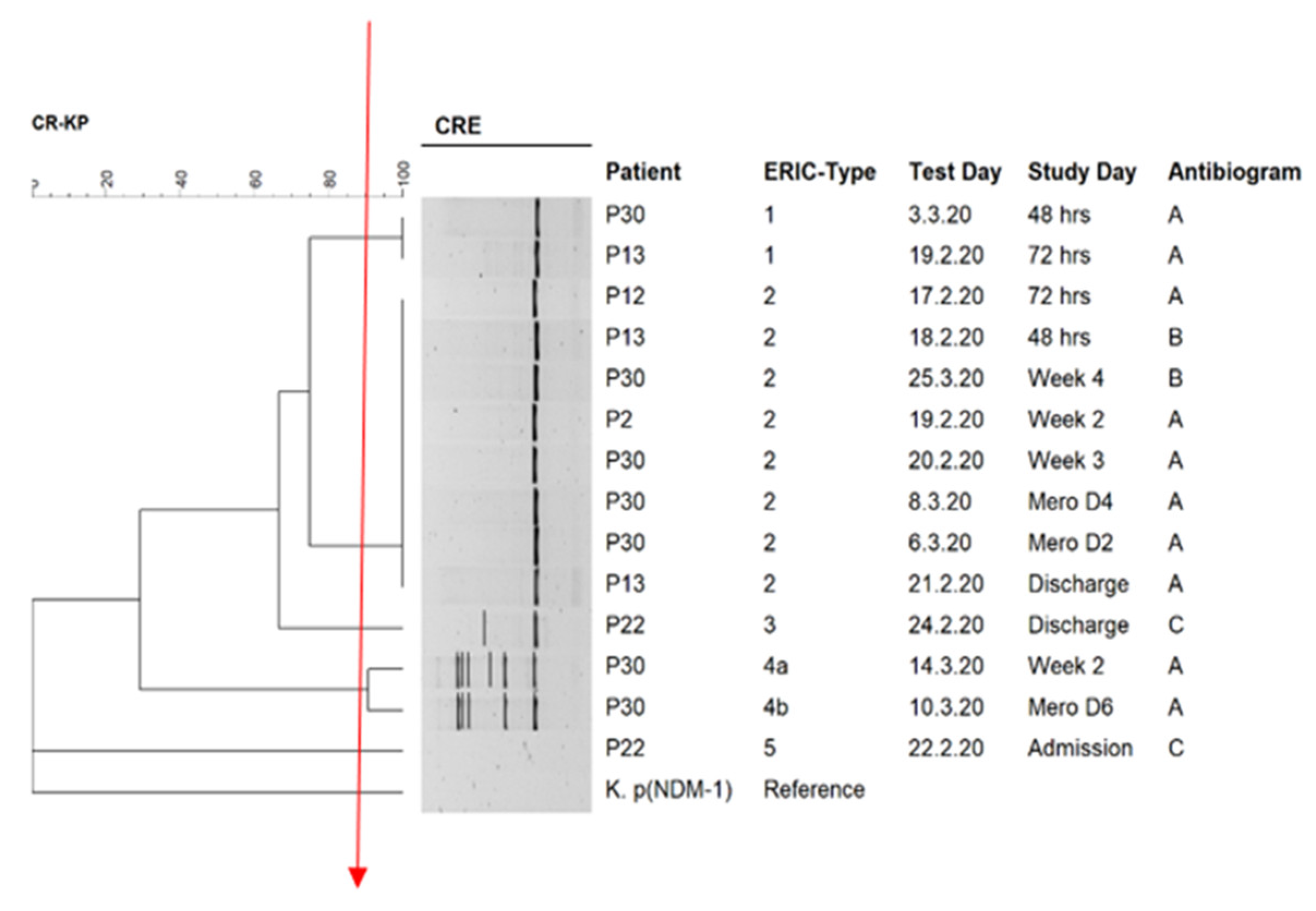

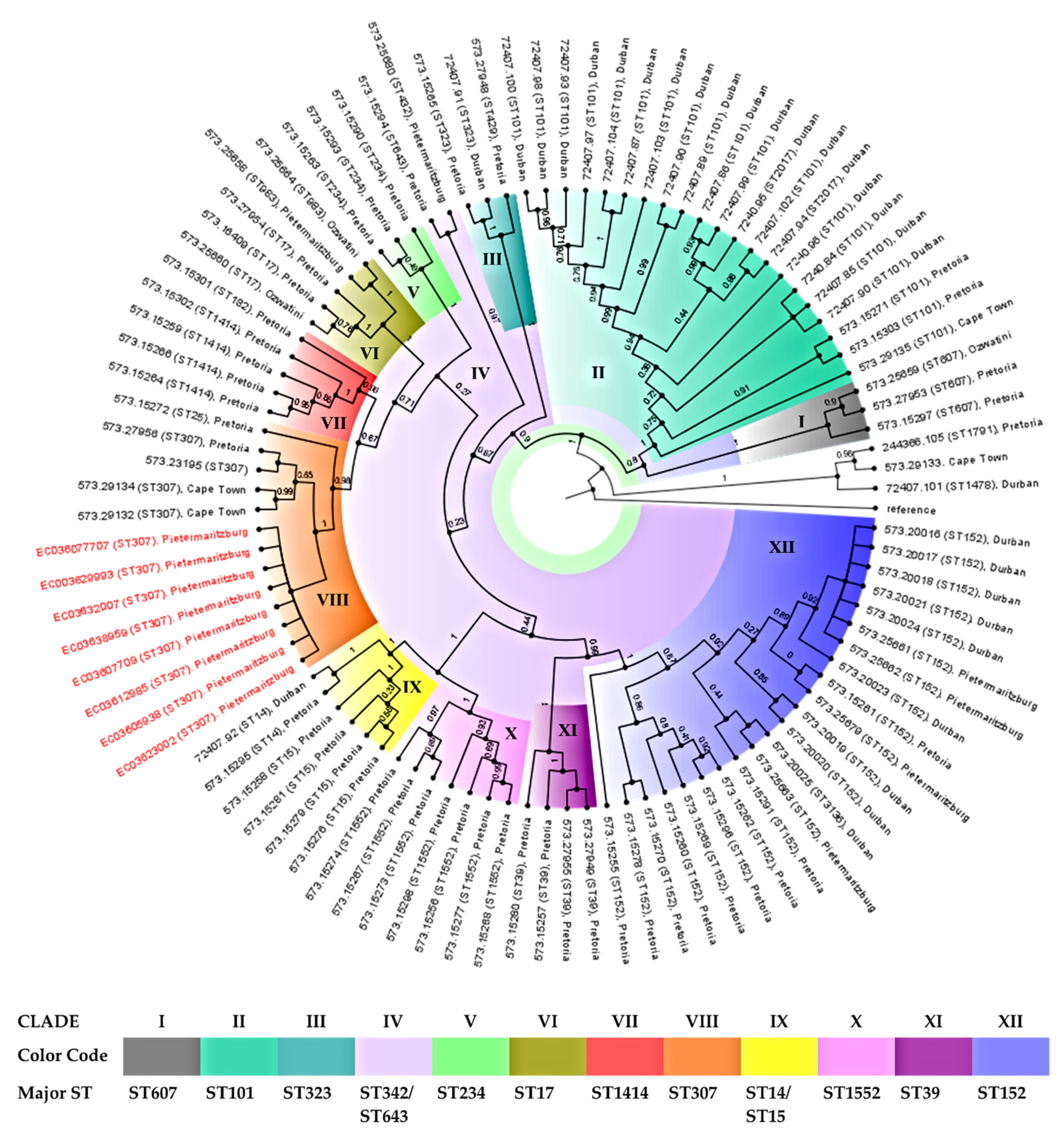
| Isolate | Demographic Information | Resistome | Plasmid Incompatibility Group | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Patient | Gender | Date | Study Day | Antibiogram | ERIC Type | Other β-Lactamases | Other ARGs | ||
| EC03607709 | P2 | Male | 19 February 2020 | Week 2 | A | 2 | blaCTX-M-15, blaOXA-1, blaSHV-106, blaTEM-1B | aac(6′)-Ib-cr, aph(3″)-Ib, aph(6)-Id, qnrS1, qnrB1, oqxB, oqxA, dfrA14, sul2, fosA, catB3, tet(A) | ColKP3, IncFIB(K), IncFII(K), IncX3 |
| EC03605938 | P13 | Male | 18 February 2020 | 48 h | B | 2 | blaCTX-M-15, blaOXA-1, blaSHV-106, blaTEM-1B | aac(6′)-Ib-cr, aph(3′)-VIa, armA, qnrS1, oqxB, oqxA, dfrA14, fosA, msr(E), mph(E), catB3, tet(A), cmlA1, ARR-2 | ColKP3, IncFIB(K), IncFII(K), IncX3 |
| EC03607707 | P13 | Male | 19 February 2020 | 72 h | A | 1 | blaCTX-M-15, blaOXA-1, blaSHV-106, blaTEM-1B | aac(6′)-Ib-cr, aph(3′)-VIa, armA, qnrS1, oqxB, oqxA, dfrA14, fosA, msr(E), mph(E), catB3, tet(A), cmlA1 | ColKP3, IncFIB(K), IncFII(K), IncX3 |
| EC03612985 | P22 | Female | 24 February 2020 | Discharge | C | 5 | blaCTX-M-15, blaOXA-1, blaSHV-106, blaTEM-1C | aac(6′)-Ib-cr, aph(3″)-Ib, aph(6)-Id, qnrS1, qnrB1, oqxB, oqxA, dfrA14, sul2, fosA, catB3, tet(A) | ColKP3, IncFIB(pNDM-Mar), IncX3 |
| EC03623002 | P30 | Male | 3 March 2020 | 48 h | A | 1 | blaCTX-M-15, blaOXA-1, blaSHV-106, blaTEM-1B | aac(6′)-Ib-cr, aph(3″)-Ib, aph(6)- Id, qnrS1, qnrB1, oqxB, oqxA, dfrA14, sul2, fosA, catB3, tet(A) | ColKP3, IncFIB(K), IncFII(K), IncX3 |
| EC03629993 | P30 | Male | 8 March 2020 | Mero D4 | A | 2 | blaCTX-M-15, blaOXA-1, blaSHV-106, blaTEM-1B | aac(6′)-Ib-cr, aph(3″)-Ib, aph(6)-Id, qnrS1, qnrB1, oqxB, oqxA, dfrA14, sul2, fosA, catB3, tet(A) | ColKP3, IncFIB(K), IncFII(K), IncX3 |
| EC03632007 | P30 | Male | 10 March 2020 | Mero D6 | A | 4b | blaCTX-M-15, blaOXA-1, blaSHV-106, blaTEM-1B | aac(6′)-Ib-cr, aph(3″)-Ib, aph(6)-Id, qnrS1, qnrB1, oqxB, oqxA, dfrA14, sul2, fosA, catB3, tet(A) | ColKP3, IncFIB(K), IncFII(K) |
| EC03638959 | P30 | Male | 14 March 2020 | Week 2 | A | 4a | blaCTX-M-15, blaOXA-1, blaSHV-106, blaTEM-1B | aac(6′)-Ib-cr, aph(3″)-Ib, aph(6)-Id, qnrS1, qnrB1, oqxB, oqxA, dfrA14, sul2, fosA, catB3, tet(A) | ColKP3, IncFII(K) |
| Isolate | Plasmids Replicons | pMLST | Integron Class | Integron | Cassette Arrays | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| GC1 a | GC2 | GC3 | GC4 | GC5 | GC6 | |||||
| EC03607709 | ColKP3, IncFIB(K), IncFII(K), IncX3 | [K7:A-:B-] | Integron integrase IntI1 | In191 | dfrA14 | — b | — | — | — | — |
| EC03638959 | ColKP3, IncFII(K) | [K7:A-:B-] | Integron integrase IntI1 | In191 | dfrA14 | — | — | — | — | — |
| EC03632007 | ColKP3, IncFIB(K), IncFII(K) | [K7:A-:B-] | ND | ND | dfrA14 | — | — | — | — | — |
| EC03629993 | ColKP3, IncFIB(K), IncFII(K), IncX3 | [K7:A-:B-] | Integron integrase IntI1 | In191 | dfrA14 | — | — | — | — | — |
| EC03607707 | ColKP3, IncFIB(K), IncFII(K), IncX3 | [K7:A-:B-] | Integron integrase IntI1 | In191 | dfrA14 | — | — | — | — | — |
| EC03623002 | ColKP3, IncFIB(K), IncFII(K), IncX3 | [K7:A-:B-] | Integron integrase IntI1 | In191 | dfrA14 | — | — | — | — | — |
| EC03605938 | ColKP3, IncFIB(K), IncFII(K), IncX3 | [K7:A-:B-] | Integron integrase IntI1 | In191 | dfrA14 | — | — | — | — | — |
| EC03612985 | ColKP3, IncFIB(pNDM-Mar), IncX3 | [F-:A-:B-] | Integron integrase IntI1 | In27 | dfrA12 | gcuF | aadA2 | — | — | — |
| Isolate | Contig | Synteny of Resistance Genes and MGEs | Plasmid/Chromosomal Sequence with the Closest Nucleotide Homology (Accession Number) |
|---|---|---|---|
| EC03607709 | 49 | blaOXA-181:EreA::ISKra4 (ISKpn19):recombinase::::recombinase:QnrS1:transposase | E. coli p010_B-OXA181 (99.98%), accession (CP048332.1) (plasmid) |
| 34 | IS5:IS5075 (IS110)::Sul2:aph(3″)-Ib:aph(6)-Id:IS91:TEM-1B:recombinase::IS1380 (ISEc9):blaCTX-M-15::Tn3 family:IS6 (IS6100) | p72_FIBkpn (100%), accession (CP034282.1) (plasmid) | |
| 54 | CatB3:blaOXA-1:aac(6′)-Ib-cr5 | E. coli pYJ3-a DNA (100%), accession (AP023228.1) (plasmid) | |
| 15 | ::::blaSHV-106:::: | K.p F16KP0053 chromosome (100%), accession (CP052727.1) | |
| 50 | IS6 (IS6100)::dfrA14:IntI1 | K.p pB16KP0177-1 (100%), accession (CP052525.1) (plasmid) | |
| EC03638959 | 95 | transposase:QnrS1:recombinase::::recombinase:ISKra4 (ISKpn19)::EreA:blaOXA-181 | E. coli p010_B-OXA181 (99.98%), accession (CP048332.1) (plasmid) |
| 98 | blaTEM-1B:recombinase::IS1380 (ISEc9):blaCTX-M-15::Tn3 family:IS6 (IS6100) | p72_FIBkpn (100%), accession (CP034282.1) (plasmid) | |
| 109 | aac(6′)-Ib-cr5:blaOXA-1:CatB3 | E. coli pYJ3-a DNA (100%), accession (AP023228.1) (plasmid) | |
| 24 | ::::blaSHV-106:::: | K.p F16KP0053 chromosome (100%), accession (CP052727.1) | |
| 102 | dfrA14::IS6 (IS6100) | K.p pB16KP0177-1 (100%), accession (CP052525.1) (plasmid) | |
| EC03632007 | 93 | blaOXA-181:EreA::ISKra4 (ISKpn19):recombinase::::recombinase:QnrS1:transposase | E. coli p010_B-OXA181 (99.98%), accession (CP048332.1) (plasmid) |
| 96 | Tn3 family::blaCTX-M-15:IS1380 (ISEc9)::recombinase:TEM-1B | p72_FIBkpn (100%), accession (CP034282.1) (plasmid) | |
| 112 | CatB3:blaOXA-1:aac(6′)-Ib-cr5 | E. coli pYJ3-a DNA (100%), accession (AP023228.1) (plasmid) | |
| 23 | ::::blaSHV-106:::: | K.p F16KP0053 chromosome (100%), accession (CP052727.1) | |
| 101 | dfrA14::IS6 (IS6100) | K.p pB16KP0177-1 (100%), accession (CP052525.1) (plasmid) | |
| EC03629993 | 47 | blaOXA-181:EreA::ISKra4 (ISKpn19):recombinase::::recombinase:QnrS1:transposase | E. coli p010_B-OXA181 (99.98%), accession (CP048332.1) (plasmid) |
| 35 | IS5:IS5075 (IS110)::Sul2:aph(3″)-Ib:aph(6)-Id:IS91:TEM-1B:recombinase::IS1380 (ISEc9):blaCTX-M-15::Tn3 family:IS6 (IS6100) | p72_FIBkpn (100%), accession (CP034282.1) (plasmid) | |
| 55 | aac(6′)-Ib-cr5:blaOXA-1:CatB3 | E. coli pYJ3-a DNA (100%), accession (AP023228.1) (plasmid) | |
| 16 | ::::blaSHV-106:::: | K.p F16KP0053 chromosome (100%), accession (CP052727.1) | |
| 49 | IntI1:dfrA14::IS6 (IS6100) | K.p pB16KP0177-1 (100%), accession (CP052525.1) (plasmid) | |
| EC03607707 | 50 | blaOXA-181:EreA::ISKra4 (ISKpn19):recombinase::::recombinase:QnrS1:transposase | E. coli p010_B-OXA181 (99.98%), accession (CP048332.1) (plasmid) |
| 36 | IS6 (IS6100):Tn3 family::blaCTX-M-15:IS1380 (ISEc9)::recombinase:TEM-1B:IS91:aph(6)-Id:aph(3″)-Ib:Sul2::IS5075 (IS110):IS5 | p72_FIBkpn (100%), accession (CP034282.1) (plasmid) | |
| 56 | CatB3:blaOXA-1:aac(6′)-Ib-cr5 | E. coli pYJ3-a DNA (100%), accession (AP023228.1) (plasmid) | |
| 13 | ::::blaSHV-106:::: | K.p F16KP0053 chromosome (100%), accession (CP052727.1) | |
| 52 | IS6(IS6100)::dfrA14:IntI1 | K.p pB16KP0177-1 (100%), accession (CP052525.1) (plasmid) | |
| EC03623002 | 45 | blaOXA-181:EreA::ISKra4 (ISKpn19):recombinase::::recombinase:QnrS1:transposase | E. coli p010_B-OXA181 (99.98%), accession (CP048332.1) (plasmid) |
| 32 | IS5:IS5075 (IS110)::Sul2:aph(3″)-Ib:aph(6)-Id:IS91:TEM-1B:recombinase::IS1380 (ISEc9):blaCTX-M-15::Tn3 family:IS6 (IS6100) | p72_FIBkpn (100%), accession (CP034282.1) (plasmid) | |
| 54 | aac(6′)-Ib-cr5:blaOXA-1:CatB3 | E. coli pYJ3-a DNA (100%), accession (AP023228.1) (plasmid) | |
| 16 | ::::blaSHV-106:::: | K.p F16KP0053 chromosome (100%), accession (CP052727.1) | |
| 47 | IntI1:dfrA14::IS6 (IS6100) | K.p pB16KP0177-1 (100%), accession (CP052525.1) (plasmid) | |
| EC03605938 | 48 | transposase:QnrS1:recombinase::::recombinase:ISKra4 (ISKpn19)::EreA:blaOXA-181 | E. coli p010_B-OXA181 (99.98%), accession (CP048332.1) (plasmid) |
| 34 | IS6 (IS6100):Tn3 family::blaCTX-M-15:IS1380 (ISEc9)::recombinase:TEM-1B:IS91:aph(6)-Id:aph(3″)-Ib:Sul2::IS5075 (IS110):IS5 | p72_FIBkpn (100%), accession (CP034282.1) (plasmid) | |
| 53 | aac(6′)-Ib-cr5:blaOXA-1:CatB3 | E. coli pYJ3-a DNA (100%), accession (AP023228.1) (plasmid) | |
| 14 | ::::blaSHV-106:::: | K.p F16KP0053 chromosome (100%), accession (CP052727.1) | |
| 50 | IntI1:dfrA14::IS6 (IS6100) | K.p pB16KP0177-1 (100%), accession (CP052525.1) (plasmid) | |
| EC03612985 | 48 | Tn3-like IS3000:blaOXA-181:EreA::ISKra4 (ISKpn19):recombinase::::recombinase:QnrS1:transposase | E. coli p010_B-OXA181 (99.98%), accession (CP048332.1) (plasmid) |
| 54 | ::blaCTX-M-15:IS1380 (ISEc9)::recombinase:blaTEM-1C:: | E. coli str. 473 pRCS52 (99.9%), accession (LO017736.1) (plasmid) | |
| 61 | aac(6′)-Ib-cr5:blaOXA-1:CatB3 | E. coli pYJ3-a DNA (100%), accession (AP023228.1) (plasmid) | |
| 2 | ::::blaSHV-106:::: | K.p F16KP0053 chromosome (100%), accession (CP052727.1) | |
| 49 | recombinase::IntI1:dfrA12:gcuF:aadA2::Sul1::::::::IS6 (IS6100)::::Mph(A) | K.p plasmid unnamed1 (99.99%), accession (CP060050.1) (plasmid) | |
| 59 | IS3:aac(3)-IIa | K.p pMS14393B (100%), accession (CP054305.1) (plasmid) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Madni, O.; Amoako, D.G.; Abia, A.L.K.; Rout, J.; Essack, S.Y. Genomic Investigation of Carbapenem-Resistant Klebsiella pneumonia Colonization in an Intensive Care Unit in South Africa. Genes 2021, 12, 951. https://doi.org/10.3390/genes12070951
Madni O, Amoako DG, Abia ALK, Rout J, Essack SY. Genomic Investigation of Carbapenem-Resistant Klebsiella pneumonia Colonization in an Intensive Care Unit in South Africa. Genes. 2021; 12(7):951. https://doi.org/10.3390/genes12070951
Chicago/Turabian StyleMadni, Osama, Daniel G. Amoako, Akebe Luther King Abia, Joan Rout, and Sabiha Yusuf Essack. 2021. "Genomic Investigation of Carbapenem-Resistant Klebsiella pneumonia Colonization in an Intensive Care Unit in South Africa" Genes 12, no. 7: 951. https://doi.org/10.3390/genes12070951
APA StyleMadni, O., Amoako, D. G., Abia, A. L. K., Rout, J., & Essack, S. Y. (2021). Genomic Investigation of Carbapenem-Resistant Klebsiella pneumonia Colonization in an Intensive Care Unit in South Africa. Genes, 12(7), 951. https://doi.org/10.3390/genes12070951








