Integrative Modelling of Gene Expression and Digital Phenotypes to Describe Senescence in Wheat
Abstract
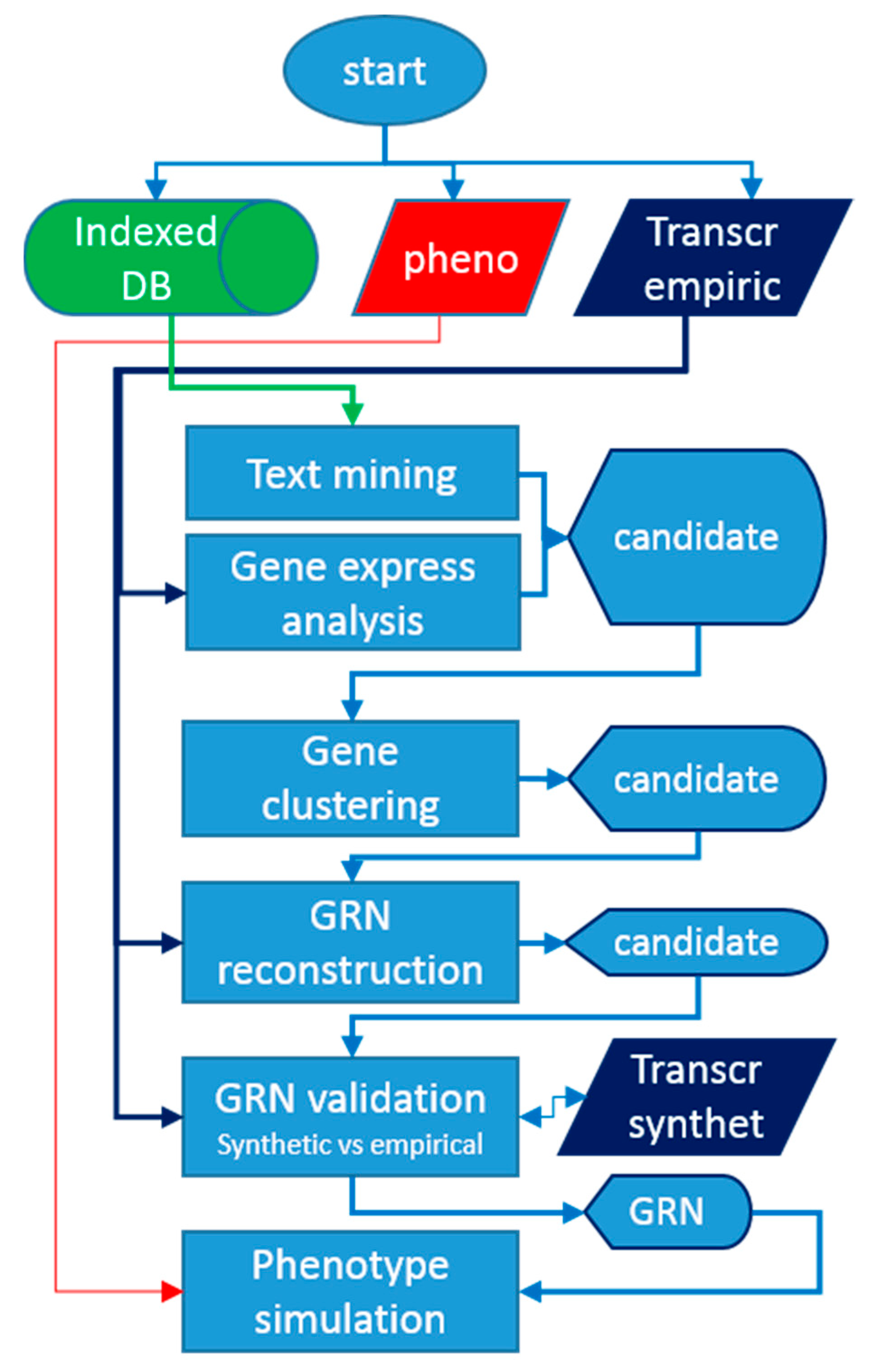
1. Introduction
2. Results
2.1. Gene Expression Profiles Vary across Genes and Time Points
2.2. Colour Distribution
2.3. GRN Reconstruction Reveals Interactions across Developmental Time Points
2.4. In Silico Reconstruction of GRN Modelling Senescence
2.5. Modelling the Senescence Phenotype
3. Discussion
4. Materials and Methods
4.1. Plant Material and Imaging
4.2. Gene Expression Analysis and Gene Selection
4.3. Selection of Senescence Clusters
4.4. Gene Regulatory Network Inference
4.5. GRN Simulation Transsys
4.6. Network Parameter Optimization and Dissection of GRN Topology
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wojciechowska, N.; Sobieszczuk-Nowicka, E.; Bagniewska-Zadworna, A. Plant organ senescence–regulation by manifold pathways. Plant Biol. 2018, 20, 167–181. [Google Scholar] [CrossRef]
- Woo, H.R.; Masclaux-Daubresse, C.; Lim, P.O. Plant senescence: How plants know when and how to die. J. Exp. Bot. 2018, 69, 715–718. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.; Gao, J.; Zhu, X.; Song, Y.; Li, Z.; Ren, G.; Zhou, X.; Kuai, B. ABF2, ABF3, and ABF4 promote ABA-mediated chlorophyll degradation and leaf senescence by transcriptional activation of chlorophyll catabolic genes and senescence-associated genes in Arabidopsis. Mol. Plant 2016, 9, 1272–1285. [Google Scholar] [CrossRef] [PubMed]
- Tamary, E.; Nevo, R.; Naveh, L.; Levin-Zaidman, S.; Kiss, V.; Savidor, A.; Levin, Y.; Eyal, Y.; Reich, Z.; Adam, Z. Chlorophyll catabolism precedes changes in chloroplast structure and proteome during leaf senescence. Plant Direct. 2019, 3, e00127. [Google Scholar] [CrossRef] [PubMed]
- Del Duca, S.; Serafini-Fracassini, D.; Cai, G. Senescence and programmed cell death in plants: Polyamine action mediated by transglutaminase. Front. Plant Sci. 2014, 5, 120. [Google Scholar] [CrossRef]
- Woo, H.R.; Kim, H.J.; Nam, H.G.; Lim, P.O. Plant leaf senescence and death–regulation by multiple layers of control and implications for aging in general. J. Cell Sci. 2013, 126, 4823–4833. [Google Scholar] [CrossRef] [PubMed]
- Woo, H.R.; Kim, J.H.; Kim, J.; Kim, J.; Lee, U.; Song, I.-J.; Kim, J.-H.; Lee, H.-Y.; Nam, H.G.; Lim, P.O. The rav1 transcription factor positively regulates leaf senescence in Arabidopsis. J. Exp. Bot. 2010, 61, 3947–3957. [Google Scholar] [CrossRef]
- Zhang, B.; Hu, Z.; Zhang, Y.; Li, Y.; Zhou, S.; Chen, G. A putative functional MYB transcription factor induced by low temperature regulates anthocyanin biosynthesis in purple kale (Brassica Oleracea var. acephala f. tricolor). Plant Cell Rep. 2012, 31, 281–289. [Google Scholar] [CrossRef]
- Sharabi-Schwager, M.; Samach, A.; Porat, R. Overexpression of the CBF2 transcriptional activator in Arabidopsis suppresses the responsiveness of leaf tissue to the stress hormone ethylene. Plant Biol. 2010, 12, 630–638. [Google Scholar]
- Lim, P.O.; Lee, I.C.; Kim, J.; Kim, H.J.; Ryu, J.S.; Woo, H.R.; Nam, H.G. Auxin response factor 2 (ARF2) plays a major role in regulating auxin-mediated leaf longevity. J. Exp. Bot. 2010, 61, 1419–1430. [Google Scholar] [CrossRef]
- Nie, H.; Zhao, C.; Wu, G.; Wu, Y.; Chen, Y.; Tang, D. SR1, a calmodulin-binding transcription factor, modulates plant defense and ethylene-induced senescence by directly regulating NDR1 and EIN3. Plant Physiol. 2012, 158, 1847–1859. [Google Scholar] [CrossRef]
- Thomas, H.; Howarth, C.J. Five ways to stay green. J. Exp. Bot. 2000, 51, 329–337. [Google Scholar] [CrossRef]
- Jan, S.; Abbas, N.; Ashraf, M.; Ahmad, P. Roles of potential plant hormones and transcription factors in controlling leaf senescence and drought tolerance. Protoplasma 2019, 256, 313–329. [Google Scholar] [CrossRef]
- Sakuraba, Y.; Bülbül, S.; Piao, W.; Choi, G.; Paek, N.C. Arabidopsis early FLOWERING3 increases salt tolerance by suppressing salt stress response pathways. Plant J. Cell Mol. Biol. 2017, 92, 1106–1120. [Google Scholar] [CrossRef]
- Brouwer, B.; Ziolkowska, A.; Bagard, M.; Keech, O.; Gardeström, P. The impact of light intensity on shade-induced leaf senescence. Plant Cell Environ. 2012, 35, 1084–1098. [Google Scholar] [CrossRef]
- Wingler, A.; Masclaux-Daubresse, C.; Fischer, A.M. Sugars, senescence, and ageing in plants and heterotrophic organisms. J. Exp. Bot. 2009, 60, 1063–1066. [Google Scholar] [CrossRef]
- Abdelrahman, M.; El-Sayed, M.; Jogaiah, S.; Burritt, D.J.; Tran, L.-S.P. The “stay-green” trait and phytohormone signaling networks in plants under heat stress. Plant Cell Rep. 2017, 36, 1009–1025. [Google Scholar] [CrossRef] [PubMed]
- Van Doorn, W.G. Is the onset of senescence in leaf cells of intact plants due to low or high sugar levels? J. Exp. Bot. 2008, 59, 1963–1972. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Zhang, D.; Hazbun, T.R.; Zhang, M. Inferring gene regulatory networks from a population of yeast segregants. Sci. Rep. 2019, 9, 1–9. [Google Scholar] [CrossRef] [PubMed]
- William, R.L. Spatial and temporal variation in plant hydraulic traits and their relevance for climate change impacts on vegetation. New Phytol. 2015, 205, 1008–1014. [Google Scholar]
- Friedman, N.; Linial, M.; Nachman, I.; Pe’er, D. Using Bayesian networks to analyze expression data. J. Comput. Biol. 2000, 7, 601–620. [Google Scholar] [CrossRef]
- Li, P.; Zhang, C.; Perkins, E.J.; Gong, P.; Deng, Y. Comparison of probabilistic Boolean network and dynamic Bayesian network approaches for inferring gene regulatory networks. BMC Bioinform. 2007, 8, S13. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; He, H.L.; Church, G.M. Modeling gene expression with differential equations. In Pacific Symposium on Biocomputing; World Scientific: Singapore, 1999; pp. 29–40. [Google Scholar]
- Petralia, F.; Wang, P.; Yang, J.; Tu, Z. Integrative random forest for gene regulatory network inference. Bioinformatics 2015, 31, i197–i205. [Google Scholar] [CrossRef] [PubMed]
- Bonneau, R.; Reiss, D.J.; Shannon, P.; Facciotti, M.; Hood, L.; Baliga, N.S.; Thorsson, V. The Inferelator: An algorithm for learning parsimonious regulatory networks from systems-biology data sets de novo. Genome Biol. 2006, 7, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Park, J.-H.; Kim, J.; Kim, J.J.; Hong, S.; Kim, J.; Kim, J.H.; Woo, H.R.; Hyeon, C.; Lim, P.O.; et al. Time-evolving genetic networks reveal a NAC troika that negatively regulates leaf senescence in Arabidopsis. Proc. Natl. Acad. Sci. USA 2018, 115, E4930–E4939. [Google Scholar] [CrossRef] [PubMed]
- Band, L.R.; Wells, D.M.; Larrieu, A.; Sun, J.; Middleton, A.M.; French, A.P.; Brunoud, G.; Sato, E.M.; Wilson, M.H.; Péret, B.; et al. Root gravitropism is regulated by a transient lateral auxin gradient controlled by a tipping-point mechanism. Proc. Natl. Acad. Sci. USA 2012, 109, 4668–4673. [Google Scholar] [CrossRef]
- Hill, K.; Porco, S.; Lobet, G.; Zappala, S.; Mooney, S.; Draye, X.; Bennett, M.J. Root systems biology: Integrative modeling across scales, from gene regulatory networks to the rhizosphere. Plant Physiol. 2013, 163, 1487–1503. [Google Scholar] [CrossRef] [PubMed]
- Chew, Y.H.; Wenden, B.; Flis, A.; Mengin, V.; Taylor, J.; Davey, C.L.; Tindal, C.; Thomas, H.; Ougham, H.J.; De Reffye, P. Multiscale digital Arabidopsis predicts individual organ and whole-organism growth. Proc. Natl. Acad. Sci. USA 2014, 111, E4127–E4136. [Google Scholar] [CrossRef]
- Zardilis, A.; Hume, A.; Millar, A.J. A multi-model framework for the Arabidopsis life cycle. J. Exp. Bot. 2019, 70, 2463–2477. [Google Scholar] [CrossRef]
- Kannan, K.; Wang, Y.; Lang, M.; Challa, G.S.; Long, S.P.; Marshall-Colon, A. Combining gene network, metabolic and leaf-level models shows means to future-proof soybean photosynthesis under rising CO2. In Silico Plants 2019, 1, 1–18. [Google Scholar] [CrossRef]
- Matthews, M.L.; Wang, J.P.; Sederoff, R.; Chiang, V.L.; Williams, C.M. A multiscale model of lignin biosynthesis for predicting bioenergy traits in Populus trichocarpa. Comput. Struct. Biotechnol. J. 2021, 19, 168–182. [Google Scholar] [CrossRef]
- Gregersen, P.L.; Culetic, A.; Boschian, L.; Krupinska, K. Plant senescence and crop productivity. Plant Mol. Biol. 2013, 82, 603–622. [Google Scholar] [CrossRef]
- Chen, L.; Xiang, S.; Chen, Y.; Li, D.; Yu, D. Arabidopsis WRKY45 interacts with the DELLA protein RGL1 to positively regulate age-triggered leaf senescence. Mol. Plant 2017, 10, 1174–1189. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Lv, X.; Li, Y.; Li, F.; Geng, M.; Mi, Y.; Ni, Z.; Wang, X.; Xie, C.; Sun, Q. Haynaldia villosa NAM-V1 is linked with the powdery mildew resistance gene Pm21 and contributes to increasing grain protein content in wheat. BMC Genet. 2016, 17, 82. [Google Scholar] [CrossRef] [PubMed]
- Adamski, N.M.; Borrill, P.; Brinton, J.; Harrington, S.A.; Marchal, C.; Bentley, A.R.; Bovill, W.D.; Cattivelli, L.; Cockram, J.; Contreras-Moreira, B.; et al. A roadmap for gene functional characterisation in crops with large genomes: Lessons from polyploid wheat. ELife 2020, 9, e55646. [Google Scholar] [CrossRef] [PubMed]
- Appels, R.; Eversole, K.; Stein, N.; Feuillet, C.; Keller, B.; Rogers, J.; Pozniak, C.J.; Choulet, F.; Distelfeld, A.; Poland, J.; et al. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 2018, 361. [Google Scholar] [CrossRef]
- Krasileva, K.V.; Vasquez-Gross, H.A.; Howell, T.; Bailey, P.; Paraiso, F.; Clissold, L.; Simmonds, J.; Ramirez-Gonzalez, R.H.; Wang, X.; Borrill, P. Uncovering hidden variation in polyploid wheat. Proc. Natl. Acad. Sci. USA 2017, 114, E913–E921. [Google Scholar] [CrossRef] [PubMed]
- Borrill, P.; Ramirez-Gonzalez, R.; Uauy, C. expVIP: A customizable RNA-seq data analysis and visualization platform. Plant Physiol. 2016, 170, 2172–2186. [Google Scholar] [CrossRef]
- Camargo, A.V.; Mackay, I.; Mott, R.; Han, J.; Doonan, J.H.; Askew, K.; Corke, F.; Williams, K.; Bentley, A.R. Functional mapping of quantitative trait loci (QTLs) associated with plant performance in a wheat MAGIC mapping population. Front. Plant Sci. 2018, 9, 887. [Google Scholar] [CrossRef]
- Miryeganeh, M.; Yamaguchi, M.; Kudoh, H. Synchronisation of Arabidopsis flowering time and whole-plant senescence in seasonal environments. Sci. Rep. 2018, 8, 10282. [Google Scholar] [CrossRef] [PubMed]
- Xie, Q.; Mayes, S.; Sparkes, D.L. Early anthesis and delayed but fast leaf senescence contribute to individual grain dry matter and water accumulation in wheat. Field Crops Res. 2016, 187, 24–34. [Google Scholar] [CrossRef]
- Borrill, P.; Harrington, S.A.; Simmonds, J.; Uauy, C. Identification of transcription factors regulating senescence in wheat through gene regulatory network modelling. Plant Physiol. 2019, 180, 1740–1755. [Google Scholar] [CrossRef]
- Uauy, C.; Distelfeld, A.; Fahima, T.; Blechl, A.; Dubcovsky, J. A NAC gene regulating senescence improves grain protein, zinc, and iron content in wheat. Science 2006, 314, 1298–1301. [Google Scholar] [CrossRef]
- Camargo, A.V.; Mott, R.; Gardner, K.A.; Mackay, I.J.; Corke, F.; Doonan, J.H.; Kim, J.T.; Bentley, A.R. Determining phenological patterns associated with the onset of senescence in a wheat MAGIC mapping population. Front. Plant Sci. 2016, 7, 1540. [Google Scholar] [CrossRef]
- Mackay, I.J.; Bansept-Basler, P.; Barber, T.; Bentley, A.R.; Cockram, J.; Gosman, N.; Greenland, A.J.; Horsnell, R.; Howells, R.; O’Sullivan, D.M.; et al. An eight-parent multiparent advanced generation inter-cross population for winter-sown wheat: Creation, properties, and validation. G3 Genes Genomes Genet. 2014, 4, 1603–1610. [Google Scholar] [CrossRef] [PubMed]
- Zadoks, J.C.; Chang, T.T.; Konzak, C.F. A decimal code for the growth stages of cereals. Weed Res. 1974, 14, 415–421. [Google Scholar] [CrossRef]
- The MathWorks. I MATLAB and Statistics Toolbox Release; The MathWorks: Natick, MA, USA, 2012. [Google Scholar]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Kondo, Y.; Salibian-Barrera, M.; Zamar, R. RSKC: An R package for a robust and sparse k-means clustering algorithm. J. Stat. Softw. 2016, 72, 1–26. [Google Scholar] [CrossRef]
- Tibshirani, R.; Suo, X. An ordered lasso and sparse time-lagged regression. Technometrics 2016, 58, 415–423. [Google Scholar] [CrossRef]
- Nguyen, P.; Braun, R. Time-lagged Ordered Lasso for network inference. BMC Bioinform. 2018, 19, 1–15. [Google Scholar] [CrossRef]
- Tibshirani, R. Regression shrinkage and selection via the lasso. J. R. Stat. Soc. Ser. B (Methodol.) 1996, 58, 267–288. [Google Scholar] [CrossRef]
- Csardi, G.; Nepusz, T. The IGRAPH software package for complex network research. Int. J. Complex Syst. 2006, 1695, 1–9. [Google Scholar]
- Kim, J. Transsys: A Generic Formalism for Modelling Regulatory Networks in Morphogenesis; Springer: Berlin, Germany, 2001. [Google Scholar]
- Camargo-Rodriguez, A.; Kim, J. DoGeNetS: Using optimisation to discriminate regulatory network topologies based on gene expression data. IET Syst. Biol. 2012, 6, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Artico, I.; Smolyarenko, I.; Vinciotti, V.; Wit, E. How rare are power-law networks really? Proc. R. Soc. A Math. Phys. Eng. Sci. 2020, 476, 20190742. [Google Scholar] [CrossRef] [PubMed]
- Camargo, A.; Kim, J.T. SimGenex: A System for Concisely Specifying Simulation of Biological Processes and Experimentation; BIOTECHNO: Venice, Italy, 2011. [Google Scholar]




| Stage | Name |
|---|---|
| GS10 | Seedling growth |
| GS20 | Tilling |
| GS30 | Stem elongation |
| GS40 | Booting |
| GS50 | Inflorescence emergence |
| GS60 | Anthesis |
| GS70 | Milk development |
| GS80 | Dough development |
| GS90 | Ripening |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Camargo Rodriguez, A.V. Integrative Modelling of Gene Expression and Digital Phenotypes to Describe Senescence in Wheat. Genes 2021, 12, 909. https://doi.org/10.3390/genes12060909
Camargo Rodriguez AV. Integrative Modelling of Gene Expression and Digital Phenotypes to Describe Senescence in Wheat. Genes. 2021; 12(6):909. https://doi.org/10.3390/genes12060909
Chicago/Turabian StyleCamargo Rodriguez, Anyela Valentina. 2021. "Integrative Modelling of Gene Expression and Digital Phenotypes to Describe Senescence in Wheat" Genes 12, no. 6: 909. https://doi.org/10.3390/genes12060909
APA StyleCamargo Rodriguez, A. V. (2021). Integrative Modelling of Gene Expression and Digital Phenotypes to Describe Senescence in Wheat. Genes, 12(6), 909. https://doi.org/10.3390/genes12060909






