Genome Features of Asaia sp. W12 Isolated from the Mosquito Anopheles stephensi Reveal Symbiotic Traits
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Growth Conditions
2.2. Genome Sequencing, Assembly, and Annotation
2.3. Bioinformatics
2.4. Accession of the Genome Sequences
2.5. Statistical Analyses
3. Results
3.1. Genome Features
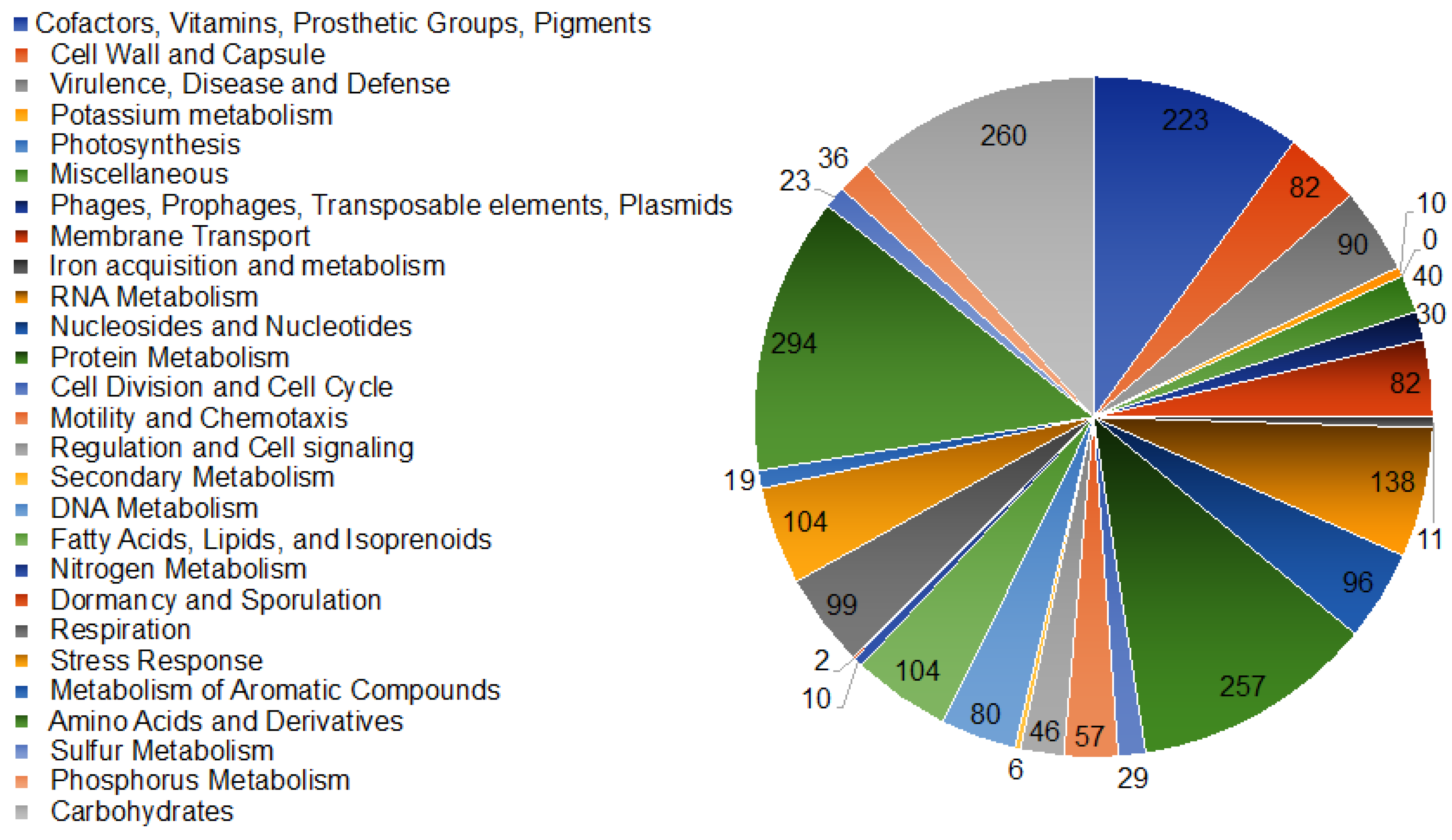
3.2. Gene Repertoire of Asaia spp.
3.3. Carbohydrate-Active EnZymes (CAZymes) in Asaia spp.
3.4. Regulatory Systems in Asaia spp.
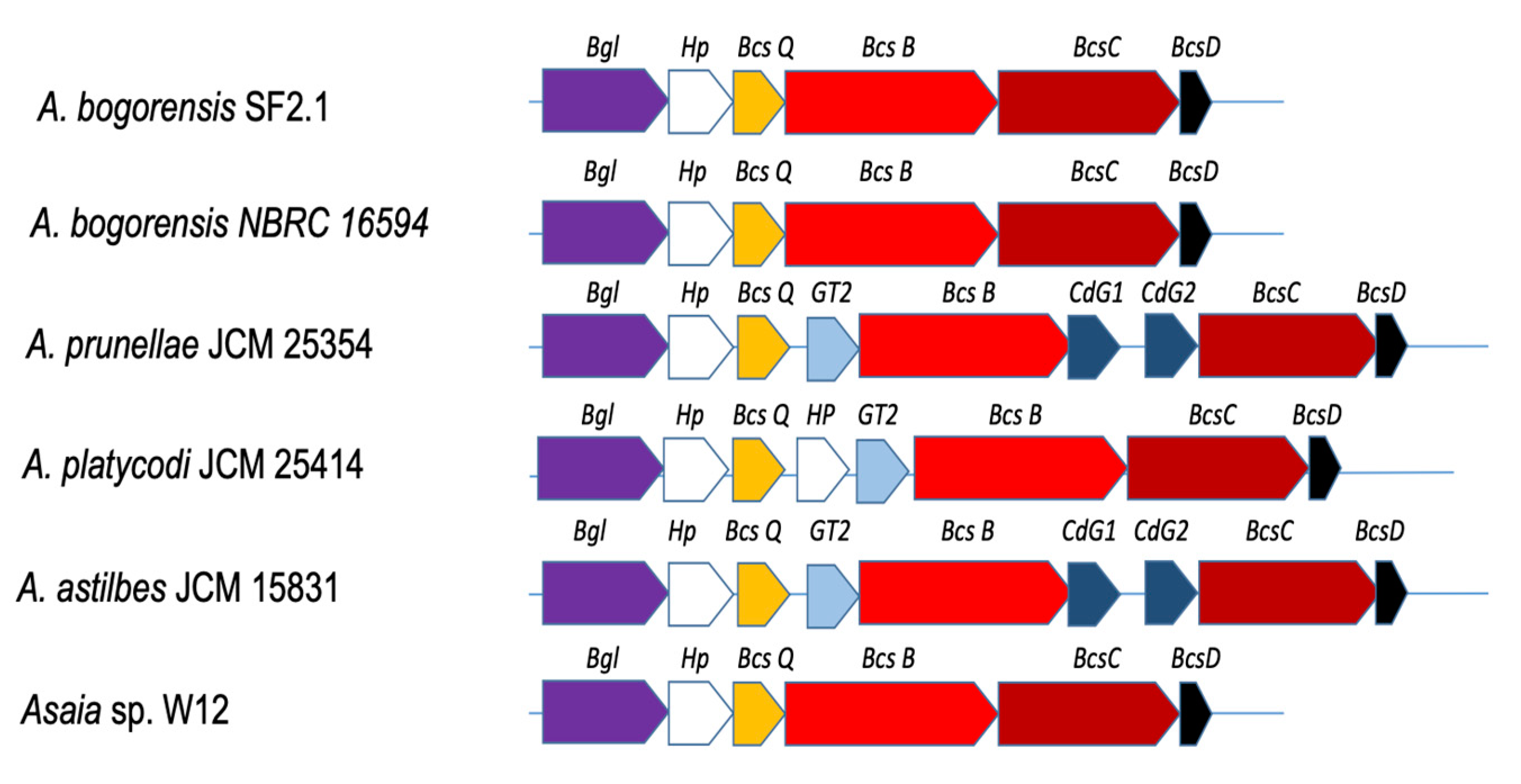
3.5. Genes Involved in other Symbiosis Traits
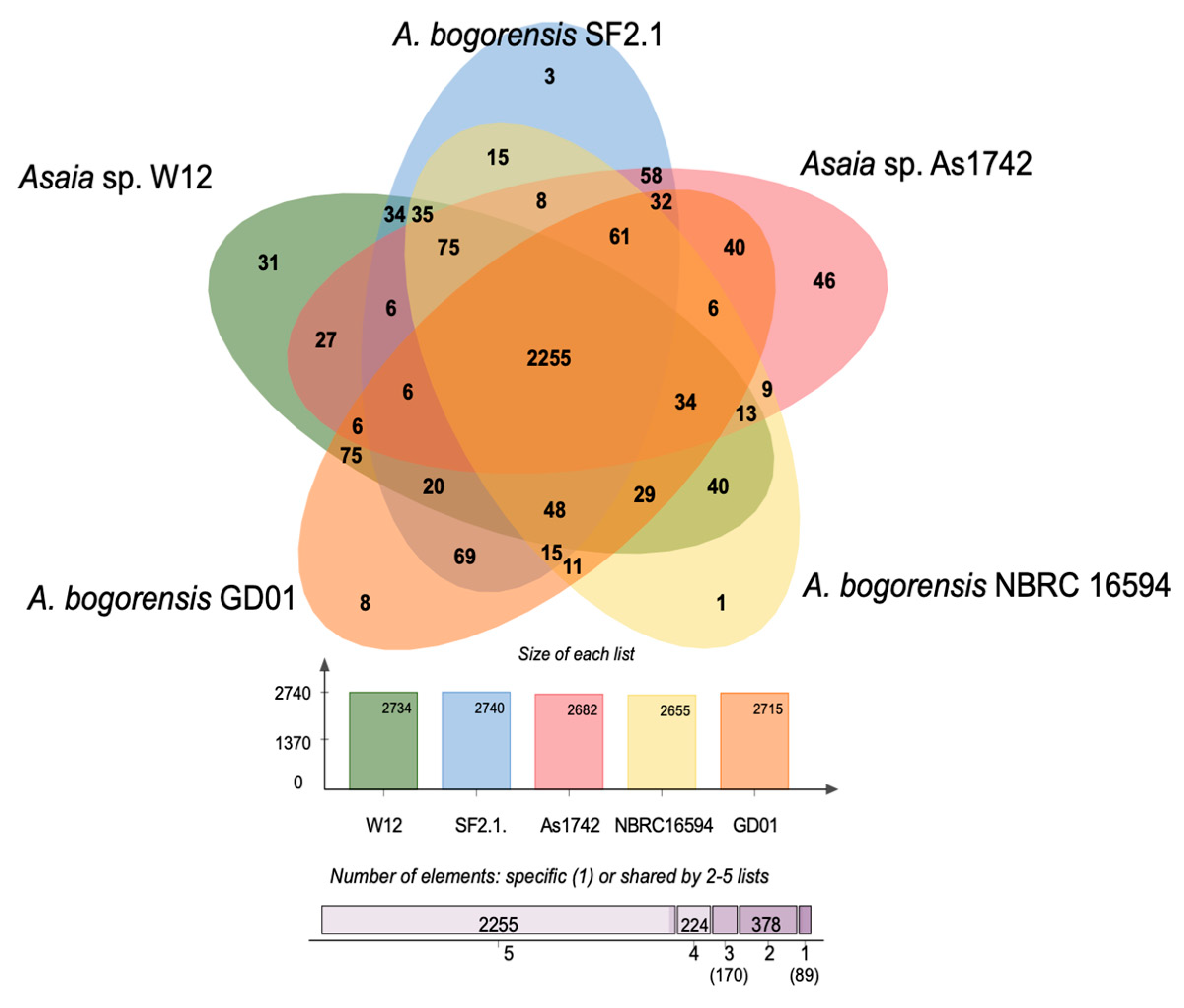
3.6. Comparative Genome Plasticity
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Favia, G.; Ricci, I.; Damiani, C.; Raddadi, N.; Crotti, E.; Marzorati, M.; Rizzi, A.; Urso, R.; Brusetti, L.; Borin, S.; et al. Bacteria of the genus Asaia stably associate with Anopheles stephensi, an Asian malarial mosquito vector. Proc. Natl. Acad. Sci. USA 2007, 104, 9047–9051. [Google Scholar] [CrossRef]
- Bassene, H.; Niang, E.H.A.; Fenollar, F.; Doucoure, S.; Faye, O.; Raoult, D.; Sokhna, C.; Mediannikov, O. Role of plants in the transmission of Asaia sp., which potentially inhibit the Plasmodium sporogenic cycle in Anopheles mosquitoes. Sci. Rep. 2020, 10, 7144. [Google Scholar] [CrossRef] [PubMed]
- Crotti, E.; Damiani, C.; Pajoro, M.; Gonella, E.; Rizzi, A.; Ricci, I.; Negri, I.; Scuppa, P.; Rossi, P.; Ballarini, P.; et al. Asaia, a versatile acetic acid bacterial symbiont, capable of cross-colonizing insects of phylogenetically distant genera and orders. Environ. Microbiol. 2009, 11, 3252–3264. [Google Scholar] [CrossRef]
- Chouaia, B.; Rossi, P.; Montagna, M.; Ricci, I.; Crotti, E.; Damiani, C.; Epis, S.; Faye, I.; Sagnon, N.; Alma, A.; et al. Molecular evidence for multiple infections as revealed by typing of Asaia bacterial symbionts of four mosquito species. Appl. Environ. Microbiol. 2010, 76, 7444–7450. [Google Scholar] [CrossRef]
- Minard, G.; Tran, F.; Raharimalala, F.; Hellard, E.; Ravelonandro, P.; Mavingui, P.; Valiente, M. Prevalence, genomic and metabolic profiles of Acinetobacter and Asaia associated with field-caught Aedes albopictus from Madagascar. FEMS Microbiol. Ecol. 2013, 83, 63–73. [Google Scholar] [CrossRef]
- Chen, S.; Zhao, J.; Joshi, D.; Xi, Z.; Norman, B.; Walker, E.D. Persistent infection by Wolbachia wAlbB has no effect on composition of the gut microbiota in adult female Anopheles stephensi. Front. Microbiol. 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Damiani, C.; Ricci, I.; Crotti, E.; Rossi, P.; Rizzi, A.; Scuppa, P.; Capone, A.; Ulissi, U.; Epis, S.; Genchi, M.; et al. Mosquito-bacteria symbiosis: The case of Anopheles gambiae and Asaia. Microb. Ecol. 2010, 60, 644–654. [Google Scholar] [CrossRef] [PubMed]
- Damiani, C.; Ricci, I.; Crotti, E.; Rossi, P.; Rizzi, A.; Scuppa, P. Paternal transmission of symbiotic bacteria in malaria vectors. Curr. Biol. 2008, 18, R1087–R1088. [Google Scholar] [CrossRef] [PubMed]
- Capone, A.; Ricci, I.; Damiani, C.; Mosca, M.; Rossi, P.; Scuppa, P.; Crotti, E.; Epis, S.; Angeletti, M.; Valzano, M.; et al. Interactions between Asaia, Plasmodium and Anopheles: New insights into mosquito symbiosis and implications in malaria symbiotic control. Parasite Vector 2013, 6. [Google Scholar] [CrossRef]
- Ricci, I.; Damiani, C.; Rossi, P.; Capone, A.; Scuppa, P.; Cappelli, A.; Ulissi, U.; Mosca, M.; Valzano, M.; Epis, S.; et al. Mosquito symbioses: From basic research to the paratransgenic control of mosquito-borne diseases. J. Appl. Entomol. 2011, 135, 487–493. [Google Scholar] [CrossRef]
- Mancini, M.V.; Damiani, C.; Short, S.M.; Cappelli, A.; Ulissi, U.; Capone, A.; Serrao, A.; Rossi, P.; Amici, A.; Kalogris, C.; et al. Inhibition of Asaia in adult mosquitoes causes male-specific mortality and diverse transcriptome changes. Pathogens 2020, 9, 380. [Google Scholar] [CrossRef]
- Rossi, P.; Ricci, I.; Cappelli, A.; Damiani, C.; Ulissi, U.; Mancini, M.V.; Valzano, M.; Capone, A.; Epis, S.; Crotti, E.; et al. Mutual exclusion of Asaia and Wolbachia in the reproductive organs of mosquito vectors. Parasite Vector 2015, 8, 278. [Google Scholar] [CrossRef] [PubMed]
- Favia, G.; Ricci, I.; Marzorati, M.; Negri, I.; Alma, A.; Sacchi, L.; Bandi, C.; Daffonchio, D. Bacteria of the genus Asaia: A potential paratransgenic weapon against malaria. In Transgenesis and the Management of Vector-Borne Disease; Aksoy, S., Ed.; Springer: New York, NY, USA, 2008; pp. 49–59. [Google Scholar] [CrossRef]
- Favia, G.; Ricci, I.; Marzorati, M.; Negri, I.; Alma, A.; Sacchi, L. Bacteria of the genus Asaia: A potential weapon against malaria. Adv. Exp. Med. Biol. 2008, 627, 49–59. [Google Scholar]
- Epis, S.; Varotto-Boccazzi, I.; Crotti, E.; Damiani, C.; Giovati, L.; Mandrioli, M.; Biggiogera, M.; Gabrieli, P.; Genchi, M.; Polonelli, L.; et al. Chimeric symbionts expressing a Wolbachia protein stimulate mosquito immunity and inhibit filarial parasite development. Commun. Biol. 2020, 3, 105. [Google Scholar] [CrossRef] [PubMed]
- Bongio, N.J.; Lampe, D.J. Inhibition of Plasmodium berghei development in mosquitoes by effector proteins secreted from Asaia sp. bacteria using a novel native secretion signal. PLoS ONE 2015, 10, e0143541. [Google Scholar] [CrossRef]
- Cappelli, A.; Damiani, C.; Mancini, M.V.; Valzano, M.; Rossi, P.; Serrao, A.; Ricci, I.; Favia, G. Asaia activates immune genes in mosquito eliciting an anti-Plasmodium response: Implications in malaria control. Front. Genet. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Crotti, E.; Rizzi, A.; Chouaia, B.; Ricci, I.; Favia, G.; Alma, A.; Sacchi, L.; Bourtzis, K.; Mandrioli, M.; Cherif, A.; et al. Acetic acid bacteria, newly emerging symbionts of insects. Appl. Environ. Microbiol. 2010, 76, 6963–6970. [Google Scholar] [CrossRef]
- Chen, S.; Bagdasarian, M.; Walker, E.D. Elizabethkingia anophelis: Molecular manipulation and interactions with mosquito hosts. Appl. Environ. Microbiol. 2015. [Google Scholar] [CrossRef]
- Overbeek, R.; Olson, R.; Pusch, G.D.; Olsen, G.J.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Parrello, B.; Shukla, M.; et al. The SEED and the rapid annotation of microbial genomes using subsystems technology (RAST). Nucleic Acids Res. 2014, 42, D206–D214. [Google Scholar] [CrossRef]
- Blom, J.; Kreis, J.; Spänig, S.; Juhre, T.; Bertelli, C.; Ernst, C.; Goesmann, A. EDGAR 2.0: An enhanced software platform for comparative gene content analyses. Nucleic Acids Res. 2016, 44, W22–W28. [Google Scholar] [CrossRef]
- Chaumeil, P.-A.; Mussig, A.J.; Hugenholtz, P.; Parks, D.H. GTDB-Tk: A toolkit to classify genomes with the Genome Taxonomy Database. Bioinformatics 2019, 36, 1925–1927. [Google Scholar] [CrossRef]
- Bertelli, C.; Laird, M.R.; Williams, K.P.; Simon Fraser University Research Computing Group; Lau, B.Y.; Hoad, G.; Winsor, G.L.; Brinkman, F.S. IslandViewer 4: Expanded prediction of genomic islands for larger-scale datasets. Nucleic Acids Res. 2017, 45, W30–W35. [Google Scholar] [CrossRef]
- Lombard, V.; Golaconda Ramulu, H.; Drula, E.; Coutinho, P.M.; Henrissat, B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014, 42, D490–D495. [Google Scholar] [CrossRef]
- Terrapon, N.; Lombard, V.; Drula, É.; Lapébie, P.; Al-Masaudi, S.; Gilbert, H.J.; Henrissat, B. PULDB: The expanded database of polysaccharide utilization loci. Nucleic Acids Res. 2018, 46, D677–D683. [Google Scholar] [CrossRef]
- Cantarel, B.L.; Coutinho, P.M.; Rancurel, C.; Bernard, T.; Lombard, V.; Henrissat, B. The Carbohydrate-Active EnZymes database (CAZy): An expert resource for glycogenomics. Nucleic Acids Res. 2008, 37, D233–D238. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Steinke, K.; Villebro, R.; Ziemert, N.; Lee, S.Y.; Medema, M.H.; Weber, T. antiSMASH 5.0: Updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 2019, 47, W81–W87. [Google Scholar] [CrossRef] [PubMed]
- Sharma, M.P.; Grover, M.; Chourasiya, D.; Bharti, A.; Agnihotri, R.; Maheshwari, H.S.; Pareek, A.; Buyer, J.S.; Sharma, S.K.; Schütz, L.; et al. Deciphering the role of trehalose in tripartite symbiosis among rhizobia, arbuscular mycorrhizal fungi, and legumes for enhancing abiotic stress tolerance in crop plants. Front. Microbiol. 2020, 11. [Google Scholar] [CrossRef] [PubMed]
- MacIntyre, A.M.; Barth, J.X.; Pellitteri Hahn, M.C.; Scarlett, C.O.; Genin, S.; Allen, C. Trehalose synthesis contributes to osmotic stress tolerance and virulence of the bacterial wilt pathogen Ralstonia solanacearum. Mol. Plant-Microbe Interact. 2020, 33, 462–473. [Google Scholar] [CrossRef] [PubMed]
- Kawai, M.; Higashiura, N.; Hayasaki, K.; Okamoto, N.; Takami, A.; Hirakawa, H.; Matsushita, K.; Azuma, Y. Complete genome and gene expression analyses of Asaia bogorensis reveal unique responses to culture with mammalian cells as a potential opportunistic human pathogen. DNA Res. 2015, 22, 357–366. [Google Scholar] [CrossRef]
- Oleńska, E.; Małek, W.; Wójcik, M.; Swiecicka, I.; Thijs, S.; Vangronsveld, J. Beneficial features of plant growth-promoting rhizobacteria for improving plant growth and health in challenging conditions: A methodical review. Sci. Total Environ. 2020, 743, 140682. [Google Scholar] [CrossRef] [PubMed]
- Farag, M.A.; Song, G.C.; Park, Y.-S.; Audrain, B.; Lee, S.; Ghigo, J.-M.; Kloepper, J.W.; Ryu, C.-M. Biological and chemical strategies for exploring inter- and intra-kingdom communication mediated via bacterial volatile signals. Nat. Protoc. 2017, 12, 1359–1377. [Google Scholar] [CrossRef]
- Michell, R.H. Inositol derivatives: Evolution and functions. Nat. Rev. Mol. Cell Biol. 2008, 9, 151–161. [Google Scholar] [CrossRef]
- Enriquez, T.; Colinet, H. Cold acclimation triggers major transcriptional changes in Drosophila suzukii. BMC Genom. 2019, 20, 413. [Google Scholar] [CrossRef]
- Rojas, C.M.; Ham, J.H.; Deng, W.-L.; Doyle, J.J.; Collmer, A. HecA, a member of a class of adhesins produced by diverse pathogenic bacteria, contributes to the attachment, aggregation, epidermal cell killing, and virulence phenotypes of Erwinia chrysanthemi EC16 on Nicotiana clevelandii seedlings. Proc. Natl. Acad. Sci. USA 2002, 99, 13142–13147. [Google Scholar] [CrossRef]
- Comandatore, F.; Damiani, C.; Cappelli, A.; Ribolla, P.E.M.; Gasperi, G.; Gradoni, F.; Capelli, G.; Piazza, A.; Montarsi, F.; Mancini, M.V.; et al. Phylogenomics reveals that Asaia symbionts from insects underwent convergent genome reduction, preserving an insecticide-degrading gene. mBio 2021, 12, e00106–e00121. [Google Scholar] [CrossRef]
- Alonso, D.P.; Mancini, M.V.; Damiani, C.; Cappelli, A.; Ricci, I.; Alvarez, M.V.N.; Bandi, C.; Ribolla, P.E.M.; Favia, G. Genome reduction in the mosquito symbiont Asaia. Genome Biol. Evol. 2018, 11, 1–10. [Google Scholar] [CrossRef]
- Chouaia, B.; Gaiarsa, S.; Crotti, E.; Comandatore, F.; Degli Esposti, M.; Ricci, I.; Alma, A.; Favia, G.; Bandi, C.; Daffonchio, D. Acetic acid bacteria genomes reveal functional traits for adaptation to life in insect guts. Genome Biol. Evol. 2014, 6, 912–920. [Google Scholar] [CrossRef]
- Shane, J.L.; Bongio, N.J.; Favia, G.; Lampe, D.J. Draft genome sequence of Asaia sp. strain SF2.1, an important member of the microbiome of Anopheles mosquitoes. Genome Announc. 2014, 2, e01202–e01213. [Google Scholar] [CrossRef]
- Perez, J.C.; Groisman, E.A. Evolution of transcriptional regulatory circuits in bacteria. Cell 2009, 138, 233–244. [Google Scholar] [CrossRef]
- Lacombe-Harvey, M.-È.; Brzezinski, R.; Beaulieu, C. Chitinolytic functions in actinobacteria: Ecology, enzymes, and evolution. Appl. Microbiol. Biotechnol. 2018, 102, 7219–7230. [Google Scholar] [CrossRef]
- Augimeri, R.V.; Varley, A.J.; Strap, J.L. Establishing a role for bacterial cellulose in environmental interactions: Lessons learned from diverse biofilm-producing proteobacteria. Front. Microbiol. 2015, 6. [Google Scholar] [CrossRef]
- Penttilä, P.A.; Imai, T.; Capron, M.; Mizuno, M.; Amano, Y.; Schweins, R.; Sugiyama, J. Multimethod approach to understand the assembly of cellulose fibrils in the biosynthesis of bacterial cellulose. Cellulose 2018, 25, 2771–2783. [Google Scholar] [CrossRef]
- Liu, M.; Liu, L.; Jia, S.; Li, S.; Zou, Y.; Zhong, C. Complete genome analysis of Gluconacetobacter xylinus CGMCC 2955 for elucidating bacterial cellulose biosynthesis and metabolic regulation. Sci. Rep. 2018, 8, 6266. [Google Scholar] [CrossRef] [PubMed]
- Kumagai, A.; Mizuno, M.; Nozaki, K.; MSaxena, I.; Amano, Y. Comparative study on the ability to produce gentiobiose in cellulose-producing bacteria Asaia bogorensis and Gluconacetobacter xylinus. J. Appl. Glycosci. 2011, 58, 147–150. [Google Scholar] [CrossRef]
- Krasteva, P.V.; Bernal-Bayard, J.; Travier, L.; Martin, F.A.; Kaminski, P.-A.; Karimova, G.; Fronzes, R.; Ghigo, J.-M. Insights into the structure and assembly of a bacterial cellulose secretion system. Nat. Commun. 2017, 8, 2065. [Google Scholar] [CrossRef]
- Wong, H.C.; Fear, A.L.; Calhoon, R.D.; Eichinger, G.H.; Mayer, R.; Amikam, D.; Benziman, M.; Gelfand, D.H.; Meade, J.H.; Emerick, A.W. Genetic organization of the cellulose synthase operon in Acetobacter xylinum. Proc. Natl. Acad. Sci. USA 1990, 87, 8130. [Google Scholar] [CrossRef]
- Díez-Vives, C.; Moitinho-Silva, L.; Nielsen, S.; Reynolds, D.; Thomas, T. Expression of eukaryotic-like protein in the microbiome of sponges. Mol. Ecol. 2017, 26, 1432–1451. [Google Scholar] [CrossRef]
- Wybouw, N.; Pauchet, Y.; Heckel, D.G.; Van Leeuwen, T. Horizontal gene transfer contributes to the evolution of arthropod herbivory. Genome Biol. Evol. 2016, 8, 1785–1801. [Google Scholar] [CrossRef]
- Siozios, S.; Ioannidis, P.; Klasson, L.; Andersson, S.G.E.; Braig, H.R.; Bourtzis, K. The diversity and evolution of Wolbachia ankyrin repeat domain genes. PLoS ONE 2013, 8, e55390. [Google Scholar] [CrossRef] [PubMed]
- Walker, T.; Klasson, L.; Sebaihia, M.; Sanders, M.J.; Thomson, N.R.; Parkhill, J.; Sinkins, S.P. Ankyrin repeat domain-encoding genes in the wPip strain of Wolbachia from the Culex pipiens group. BMC Biol. 2007, 5, 39. [Google Scholar] [CrossRef]
- Wu, M.; Sun, L.V.; Vamathevan, J.; Riegler, M.; Deboy, R.; Brownlie, J.C.; McGraw, E.A.; Martin, W.; Esser, C.; Ahmadinejad, N.; et al. Phylogenomics of the reproductive parasite Wolbachia pipientis wMel: A streamlined genome overrun by mobile genetic elements. PLoS Biol. 2004, 2, e69. [Google Scholar] [CrossRef]
- Klasson, L.; Walker, T.; Sebaihia, M.; Sanders, M.J.; Quail, M.A.; Lord, A.; Sanders, S.; Earl, J.; O′Neill, S.L.; Thomson, N.; et al. Genome evolution of Wolbachia strain wPip from the Culex pipiens group. Mol. Biol. Evol. 2008, 25, 1877–1887. [Google Scholar] [CrossRef] [PubMed]
- Penz, T.; Schmitz-Esser, S.; Kelly, S.E.; Cass, B.N.; Müller, A.; Woyke, T.; Malfatti, S.A.; Hunter, M.S.; Horn, M. Comparative genomics suggests an independent origin of cytoplasmic incompatibility in Cardinium hertigii. PLoS Genet. 2012, 8, e1003012. [Google Scholar] [CrossRef]
- Coon, K.L.; Valzania, L.; McKinney, D.A.; Vogel, K.J.; Brown, M.R.; Strand, M.R. Bacteria-mediated hypoxia functions as a signal for mosquito development. Proc. Natl. Acad. Sci. USA 2017, 114, E5362. [Google Scholar] [CrossRef]
- Zhang, L.; Sun, J.; Hao, Y.; Zhu, J.; Chu, J.; Wei, D.; Shen, Y. Microbial production of 2, 3-butanediol by a surfactant (serrawettin)-deficient mutant of Serratia marcescens H30. J. Ind. Microbiol. Biotechnol. 2010, 37, 857–862. [Google Scholar] [CrossRef] [PubMed]
- Köpke, M.; Mihalcea, C.; Liew, F.; Tizard, J.H.; Ali, M.S.; Conolly, J.J.; Al-Sinawi, B.; Simpson, S.D. 2,3-butanediol production by acetogenic bacteria, an alternative route to chemical synthesis, using industrial waste gas. Appl. Environ. Microbiol. 2011, 77, 5467–5475. [Google Scholar] [CrossRef]
- Nguyen, M.; Sharma, A.; Wu, W.; Gomi, R.; Sung, B.; Hospodsky, D.; Angenent, L.T.; Worgall, S. The fermentation product 2,3-butanediol alters P. aeruginosa clearance, cytokine response and the lung microbiome. ISME J. 2016, 10, 2978–2983. [Google Scholar] [CrossRef]
- Venkataraman, A.; Rosenbaum, M.A.; Werner, J.J.; Winans, S.C.; Angenent, L.T. Metabolite transfer with the fermentation product 2,3-butanediol enhances virulence by Pseudomonas aeruginosa. ISME J. 2014, 8, 1210–1220. [Google Scholar] [CrossRef]
- Reynolds, T.B. Strategies for acquiring the phospholipid metabolite inositol in pathogenic bacteria, fungi and protozoa: Making it and taking it. Microbiology 2009, 155, 1386–1396. [Google Scholar] [CrossRef] [PubMed]
- Chhetri, D.R. Myo-Inositol and its derivatives: Their emerging role in the treatment of human diseases. Front. Pharmacol. 2019, 10, 1172. [Google Scholar] [CrossRef]
- Vesala, L.; Salminen, T.S.; Koštál, V.; Zahradníčková, H.; Hoikkala, A. Myo-inositol as a main metabolite in overwintering flies: Seasonal metabolomic profiles and cold stress tolerance in a northern drosophilid fly. J. Exp. Biol. 2012, 215, 2891–2897. [Google Scholar] [CrossRef][Green Version]
- Dutta, C.; Sarkar, M. Horizontal Gene Transfer and Bacterial Diversity. In Encyclopedia of Metagenomics: Genes, Genomes and Metagenomes: Basics, Methods, Databases and Tools; Nelson, K.E., Ed.; Springer: Boston, MA, USA, 2015; pp. 251–257. [Google Scholar] [CrossRef]
- Burrus, V.; Waldor, M.K. Shaping bacterial genomes with integrative and conjugative elements. Res. Microbiol. 2004, 155, 376–386. [Google Scholar] [CrossRef]
- Ramisetty, B.C.M.; Sudhakari, P.A. Bacterial ‘grounded’ prophages: Hotspots for genetic renovation and innovation. Front. Genet. 2019, 10, 65. [Google Scholar] [CrossRef]
- Bobay, L.-M.; Rocha, E.P.C.; Touchon, M. The adaptation of temperate bacteriophages to their host genomes. Mol. Biol. Evol. 2012, 30, 737–751. [Google Scholar] [CrossRef]

| Species | Sources | Size (Mb) | GC% | Total RNA Number * | CDS | GenBank Accession Number |
|---|---|---|---|---|---|---|
| A. bogorensis NBRC 16594 | Bauhinia purpurea | 3.20 | 59.8 | 59 | 2896 | GCA_001547995.1 |
| A. astilbis JCM 15831 | Astilbe thunbergii | 3.15 | 58.0 | 49 | 3482 | GCA_000613845.1 |
| A. prunellae JCM 25354 | Prunella vulgaris | 3.18 | 55.8 | 48 | 3583 | GCA_900465315.1 |
| A. platycodi JCM 25414 | Platycodon grandiflorum | 3.15 | 59.3 | 49 | 3774 | GCA_000614545.1 |
| A. bogorensis IPC-01 | Flower | 3.17 | 59.7 | 54 | 2880 | GCA_900465315.1 |
| A. bogorensis GD01 | Anopheles gambiae | 3.34 | 59.8 | 60 | 3226 | GCA_900465345.1 |
| A. bogorensis SF2.1 | Anopheles stephensi | 3.52 | 59.8 | 45 | 3005 | GCA_000724025.1 |
| Asaia sp. As-1742 | Atta sexdens | 3.74 | 59.6 | 54 | 3308 | GCA_011753235.1 |
| Asaia sp. W12 | A. stephensi | 3.94 | 60.1 | 59 | 3580 | GCA_003994335.1 |
| Glycoside Hydrolases (GHs) | Glycosyltransferases (GTs) | Carbohydrate Esterases (CEs) | |
|---|---|---|---|
| A. astilbis JCM 15831 | 17 | 37 | 1 |
| A. platycodi JCM 25414 | 15 | 34 | 1 |
| A. prunellae JCM 25354 | 22 | 55 | 1 |
| A. bogorensis NBRC 16594 | 28 | 54 | 3 |
| A. bogorensis IPC-01 | 27 | 54 | 3 |
| A. bogorensis GD-01 | 28 | 55 | 3 |
| A. bogorensis SF2.1 | 32 | 53 | 3 |
| Asaia sp. As-1742 | 32 | 53 | 3 |
| Asaia sp. W12 | 32 | 52 | 3 |
| Predicted Regulatory Proteins | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Two Component Systems | Transcription Factors | Other DNA-Binding Proteins | Total | ||||||
| RR | PP | HK | OCS | RR | TR | SF | ODP | ||
| A. astilbis JCM 15831 | 19 | 1 | 15 | 27 | 10 | 65 | 5 | 8 | 150 |
| A. bogorensis NBRC 16594 | 25 | 0 | 22 | 42 | 15 | 62 | 7 | 11 | 184 |
| A. bogorensis IPC-01 | 24 | 0 | 20 | 35 | 14 | 64 | 7 | 13 | 177 |
| A. prunellae JCM 25354 | 23 | 2 | 14 | 30 | 13 | 69 | 7 | 12 | 170 |
| A. platycodi JCM 25414 | 16 | 2 | 16 | 33 | 13 | 59 | 8 | 10 | 157 |
| A. bogorensis GD-01 | 21 | 1 | 19 | 39 | 13 | 83 | 7 | 15 | 198 |
| A. bogorensis SF2.1 | 25 | 1 | 21 | 40 | 15 | 88 | 7 | 16 | 213 |
| Asaia sp. As-1742 | 24 | 1 | 21 | 57 | 14 | 96 | 7 | 15 | 235 |
| Asaia sp. W12 | 30 | 1 | 26 | 57 | 18 | 85 | 9 | 17 | 243 |
| Species | cyoABCD | Acetoin, Butanediol and Inositol | Flagella | Large Adhesin |
|---|---|---|---|---|
| A. astilbis JCM 15831 | 1 | + | + | + |
| A. bogorensis NBRC 16594 | 2 | + | + | + |
| A. bogorensis IPC-01 | 2 | + | + | + |
| A. prunellae JCM 25354 | 2 | + | + | + |
| A. platycodi JCM 25414 | 2 | + | + | + |
| A. bogorensis GD-01 | 2 | + | + | + |
| Asaia sp. As-1742 | 2 | + | + | + |
| A. bogorensis SF2.1 | 2 | + | + | - |
| Asaia sp. W12 | 2 | + | + | + |
| Tetratrico Peptide Repeats | Ankyrin Repeats | Sel1 Repeats | Fibronectin Type III | Total Motif | |
|---|---|---|---|---|---|
| A. astilbis JCM 15831 | 9 | 2 | 2 | 2 | 15 |
| A. bogorensis NBRC 16594 | 9 | 1 | 3 | 1 | 14 |
| A. bogorensis IPC 01 | 9 | 1 | 3 | 1 | 14 |
| A. platycodi JCM 25414 | 7 | 1 | 3 | 1 | 12 |
| A. prunellae JCM 25354 | 9 | 1 | 3 | 3 | 16 |
| A. bogorensis GD 01 | 8 | 1 | 3 | 1 | 13 |
| A. bogorensis SF2.1 | 9 | 1 | 3 | 2 | 15 |
| Asaia As-1742 | 8 | 1 | 3 | 1 | 13 |
| Asaia sp. W12 | 10 | 1 | 2 | 1 | 14 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, S.; Yu, T.; Terrapon, N.; Henrissat, B.; Walker, E.D. Genome Features of Asaia sp. W12 Isolated from the Mosquito Anopheles stephensi Reveal Symbiotic Traits. Genes 2021, 12, 752. https://doi.org/10.3390/genes12050752
Chen S, Yu T, Terrapon N, Henrissat B, Walker ED. Genome Features of Asaia sp. W12 Isolated from the Mosquito Anopheles stephensi Reveal Symbiotic Traits. Genes. 2021; 12(5):752. https://doi.org/10.3390/genes12050752
Chicago/Turabian StyleChen, Shicheng, Ting Yu, Nicolas Terrapon, Bernard Henrissat, and Edward D. Walker. 2021. "Genome Features of Asaia sp. W12 Isolated from the Mosquito Anopheles stephensi Reveal Symbiotic Traits" Genes 12, no. 5: 752. https://doi.org/10.3390/genes12050752
APA StyleChen, S., Yu, T., Terrapon, N., Henrissat, B., & Walker, E. D. (2021). Genome Features of Asaia sp. W12 Isolated from the Mosquito Anopheles stephensi Reveal Symbiotic Traits. Genes, 12(5), 752. https://doi.org/10.3390/genes12050752






