Identification and Validation of Reference Genes for Gene Expression Analysis in Schima superba
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Selection of Candidate Reference Genes
2.3. RT-qPCR Analysis
2.4. Validation of Identified Reference Genes
2.5. Statistical Data Analysis
3. Results
3.1. Candidate Reference Genes and PCR Amplification
3.2. Ct Values of Candidate Reference Genes
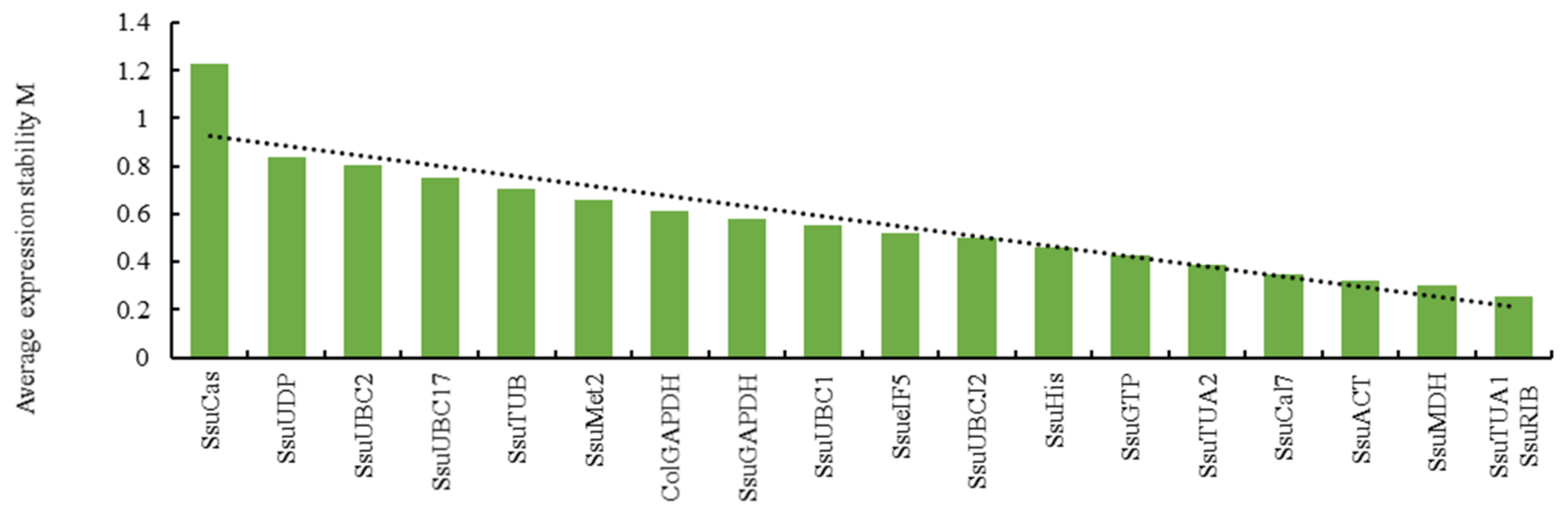
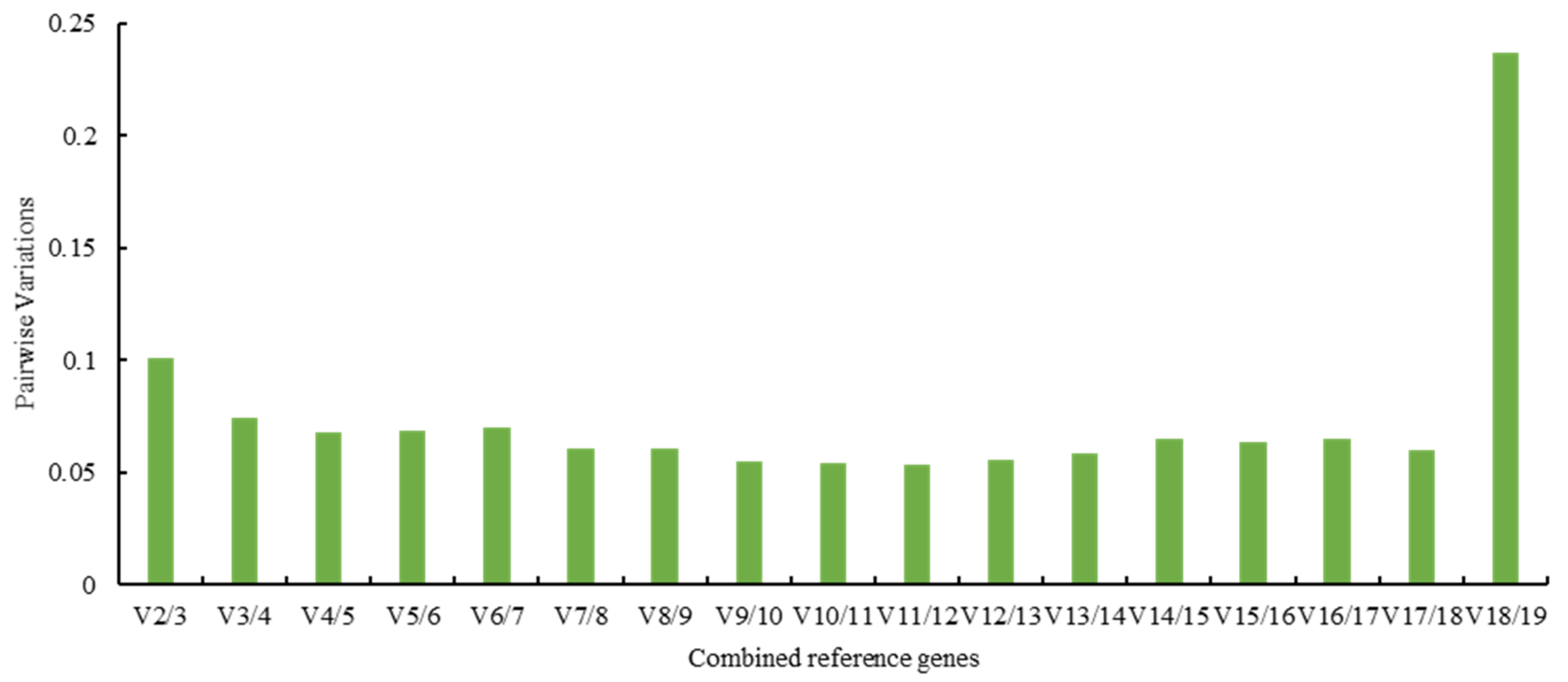
3.3. Analysis of Reference Gene Stability Using Three Bioinformatic Programs
3.4. Validation of the Identified Reference Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Wong, M.L.; Medrano, J.F. Real-time PCR for mRNA quantitation. Biotechniques 2005, 39, 75–85. [Google Scholar] [CrossRef]
- Nolan, T.; Hands, R.E.; Bustin, S.A. Quantification of mRNA using real-time RT-PCR. Nat. Protoc. 2006, 1, 1559–1582. [Google Scholar] [CrossRef]
- Artico, S.; Nardeli, S.M.; Brilhante, O.; Grossi-de-Sa, M.; Alves-Ferreira, M. Identification and evaluation of new reference genes in Gossypium hirsutum for accurate normalization of real-time quantitative RT-PCR data. BMC Plant Biol. 2010, 10, 49. [Google Scholar] [CrossRef]
- Wen, L.; Tan, B.; Guo, W.W. Estimating transgene copy number in precocious trifoliate orange by TaqMan real-time PCR. Plant Cell Tissue Organ 2012, 109, 363–371. [Google Scholar] [CrossRef]
- Su, X.Y.; Shi, Y.B.; Yang, X.M.; Wang, G.B.; Cao, F.L. Selection and validation of reference genes for quantitative real-time PCR analysis in Ginkgo biloba. Plant Physiol. J. 2019, 55, 875–882. [Google Scholar]
- Yoo, W.G.; Kim, T.I.; Li, S.Y.; Kwon, O.S.; Cho, P.Y.; Kim, T.-S.; Kim, K.J.; Hong, S.-J. Reference genes for quantitative analysis on Clonorchis sinensis gene expression by real-time PCR. Parasitol. Res. 2009, 104, 321–328. [Google Scholar] [CrossRef]
- Huang, T.; Long, J.M.; Liu, S.W.; Yang, Z.W.; Zhu, Q.J.; Zhao, X.L.; Peng, C.C. Selection and validation of reference genes for mRNA expression by quantitative real-time PCR analysis in Neolamarckia cadamba. Sci. Rep. 2018, 8, 9311. [Google Scholar] [CrossRef]
- Nicot, N.; Hausman, J.F.; Hoffmann, L.; Evers, D. Housekeeping gene selection for real-time RT-PCR normalization in potato during biotic and abiotic stress. J. Exp. Bot. 2005, 56, 2907–2914. [Google Scholar] [CrossRef] [PubMed]
- Patankar, H.V.; MAssaha, D.V.; Al-Yahyai, R.; Sunkar, R.; Yaish, M.W. Identification of reference genes for quantitative real-time PCR in Date Palm (Phoenix dactylifera L.) subjected to drought and salinity. PLoS ONE 2006, 11, e0166216. [Google Scholar] [CrossRef]
- Qu, R.J.; Miao, Y.J.; Cui, Y.J.; Cao, Y.; Zhou, Y.W.; Tang, X.Q.; Yang, J.; Wang, F.Y. Selection of reference genes for the quantitative real-time PCR normalization of gene expression in Isatis indigotica fortune. BMC Mol. Biol. 2019, 20, 9. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Han, J.; Hu, Y.; Yang, J. Selection of reference genes for quantitative real-time PCR during flower development in Tree Peony (Paeonia suffruticosa Andr.). Front. Plant Sci. 2016, 7, 516. [Google Scholar] [CrossRef] [PubMed]
- Sudhakar Reddy, P.; Srinivas Reddy, D.; Sivasakthi, K.; Bhatnagar-Mathur, P.; Vadez, V.; Sharma, K.K. Evaluation of Sorghum [Sorghum bicolor (L.)] reference genes in various tissues and under abiotic stress conditions for quantitative real-time PCR data normalization. Front. Plant Sci. 2016, 7, 529. [Google Scholar] [CrossRef]
- Yu, Y.T.; Zhang, G.; Chen, Y.K.; Bai, Q.Q.; Gao, C.S.; Zeng, L.B.; Li, Z.M.; Cheng, Y.; Chen, J.; Sun, X.P.; et al. Selection of reference genes for qPCR analyses of gene expression in ramie leaves and roots across eleven abiotic/biotic treatments. Sci. Rep. 2019, 9, 20004. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Z.; Sun, X.B.; Liu, X.Q.; Li, C.; He, L.S.; Chen, S.P.; Su, J.L. Selection of reliable reference genes for gene expression studies on Rhododendron molle G. Don. Front. Plant Sci. 2016, 7, 1547. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.B.; Zhang, R.; Zhou, Z.C. Pollen dispersal, mating patterns and pollen contamination in an insect-pollinated seed orchard of Schima superba Gardn. Et champ. New For. 2017, 48, 431–444. [Google Scholar] [CrossRef]
- Yang, H.B.; Zhang, R.; Song, P.; Zhou, Z.C. The floral biology, breeding system and pollination efficiency of Schima superba Gardn. Et Champ. Forests 2017, 8, 404. [Google Scholar] [CrossRef]
- Zhou, Z.C.; Zhang, R.; Fan, H.H.; Chu, X.L. Schima Superba; Chinese Science Press: Beijing, China, 2020; pp. 1–9. [Google Scholar]
- Stevens, P.F.; Dressler, S.; Weitzman, A.L. Theaceae. In Flowering Plants Dicotyledons; Springer: Berlin, Germany, 2004; pp. 463–471. [Google Scholar]
- Sun, M.L.; Wang, Y.S.; Yang, D.Q.; Wei, C.L.; Gao, L.P.; Xia, T.; Shan, Y.; Luo, Y. Reference genes for real-time fluorescence quantitative PCR in Camellia sinensis. Chin. Bull. Bot. 2010, 45, 579–587. [Google Scholar]
- Zhou, C.F.; Lin, P.; Yao, X.H.; Wang, K.L.; Chang, J.; Han, X.J. Selection of reference genes for quantitative real-time PCR in six oil-tea camellia based on RNA-seq. Mol. Biol. 2013, 47, 836–851. [Google Scholar] [CrossRef]
- Liu, X.F.; Yu, B.; Huang, L.L.; Sun, Y.B. Screening and Validation of Reference Genes of Camellia azalea by Quantitative Real-time PCR. Guangdong Agric. Sci. 2020, 47, 203–211. [Google Scholar]
- Yang, H.B. Characteristics of the reproductive biology and the mating system in seed orchard of Schima superba. Chin. Acad. For. 2017. [Google Scholar]
- Zhang, R.; Yang, H.B.; Zhou, Z.C.; Shen, B.; Xiao, J.J.; Wang, B.S. A high-density genetic map of Schima superba based on its chromosomal characteristics. BMC Plant Biol. 2019, 19, 41. [Google Scholar] [CrossRef]
- Takata, N.; Awano, T.; Nakata, M.T.; Sano, Y.; Sakamoto, S.; Mitsuda, N.; Taniguchi, T. Populus NST/SND orthologs are key regulators of secondary cell wall formation in wood fibers, phloem fibers and xylem ray parenchyma cells. Tree Physiol. 2019, 39, 514–525. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Andersen, C.L.; Jensen, J.L.; Orntoft, T.F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper-Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef]
- Xu, L.F.; Xu, H.; Cao, Y.W.; Yang, P.P.; Feng, Y.Y.; Tang, Y.C.; Yuan, S.X.; Ming, J. Validation of reference genes for quantitative real-time PCR during bicolor tepal development in Asiatic Hybrid Lilies (Lilium spp.). Front. Plant Sci. 2017, 8, 669. [Google Scholar] [CrossRef] [PubMed]
- Han, X.J.; Lu, M.Z.; Chen, Y.C.; Zhan, Z.Y.; Cui, Q.Q.; Wang, Y.D. Selection of reliable reference genes for gene expression studies using real-Time PCR in Tung Tree during seed development. PLoS ONE 2012, 7, e43084. [Google Scholar] [CrossRef] [PubMed]
- Li, M.M. Phylogenetic and Biogeographic Studies of Theaceae with a Special Reference to Stewartia; Zhejiang University: Hangzhou, China, 2011. [Google Scholar]
- Delporte, M.; Legrand, G.; Hilbert, J.-L.; Gagneul, D. Selection and validation of reference genes for quantitative real-time PCR analysis of gene expression in Cichorium intybus. Front. Plant Sci. 2015, 6, 651. [Google Scholar] [CrossRef]
- Mangeot-Peter, L.; Legay, S.; Hausman, J.F.; Esposito, S.; Guerriero, G. Identification of reference genes for RT-qPCR data normalization in Cannabis sativa stem tissues. Int. J. Mol. Sci. 2016, 17, 1556. [Google Scholar] [CrossRef]
- Zhao, J.M.; Zhou, M.; Meng, Y. Identification and validation of reference genes for RT-qPCR analysis in Switchgrass under heavy metal stresses. Genes 2020, 11, 502. [Google Scholar] [CrossRef] [PubMed]
- Souer, E.; Van Houwelingen, A.; Kloos, D.; Mol, J.; Koes, R. The no apical meristem gene of petunia is required for pattern formation in embryos and flowers and is expressed at meristem and primordia boundaries. Cell 1996, 85, 159–170. [Google Scholar] [CrossRef]
- Mao, C.J.; He, J.M.; Liu, L.N.; Deng, Q.M.; Yao, X.Y.; Liu, C.M.; Qiao, Y.L.; Li, P.; Ming, F. OsNAC2 integrates auxin and cytokinin pathways to modulate rice root development. Plant Biotechnol. J. 2020, 18, 429–442. [Google Scholar] [CrossRef] [PubMed]
- Zhong, R.Q.; Demura, T.K.; Ye, Z.H. SND1, a NAC domain transcription factor, is a key regulator of secondary wall synthesis in fibers of Arabidopsis. Plant Cell 2006, 18, 3158–3170. [Google Scholar] [CrossRef]


| Gene | Amplification Efficiency (%) | Correlation Coefficient (R2) | Ct Value |
|---|---|---|---|
| ColGAPDH [22] | 106.14 | 0.9991 | 21.562 ± 0.865 |
| SsuACT | 99.17 | 0.9981 | 23.877 ± 1.059 |
| SsuCal7 | 106.74 | 0.9998 | 21.984 ± 0.619 |
| SsuCas | 104.35 | 0.9977 | 23.388 ± 1.467 |
| SsueIF5 | 103.76 | 0.9996 | 25.185 ± 0.807 |
| SsuGAPDH | 97.39 | 0.9996 | 20.608 ± 0.903 |
| SsuGTP | 100.02 | 0.9929 | 23.709 ± 4.902 |
| SsuHis | 109.7 | 0.9991 | 21.455 ± 1.150 |
| SsuMDH | 102.6 | 0.9994 | 20.113 ± 0.546 |
| SsuMet2 | 97.27 | 0.9995 | 22.875 ± 0.808 |
| SsuRIB | 94.92 | 0.9931 | 22.073 ± 1.003 |
| SsuTUA1 | 93.47 | 0.9991 | 25.556 ± 1.004 |
| SsuTUA2 | 104.27 | 0.9986 | 18.032 ± 0.984 |
| SsuTUB | 103.17 | 0.9984 | 22.314 ± 0.848 |
| SsuUBC1 | 105.46 | 0.9907 | 25.320 ± 0.817 |
| SsuUBC17 | 107.89 | 0.9993 | 21.644 ± 1.555 |
| SsuUBC2 | 104.03 | 0.9985 | 21.556 ± 1.681 |
| SsuUBCJ2 | 109.03 | 0.9975 | 24.512 ± 1.180 |
| SsuUDP | 108.71 | 0.9989 | 24.877 ± 1.782 |
| Gene | Leaf | Bud | Fruit | Phloem | Root | Xylem | Average | Min | Max | Range |
|---|---|---|---|---|---|---|---|---|---|---|
| SsuACT | 21.125 ± 2.752 | 20.818 ± 0.668 | 22.681 ± 1.065 | 21.606 ± 0.604 | 22.504 ± 1.687 | 20.638 ± 0.700 | 21.562 | 20.638 | 22.681 | 2.043 |
| SsuTUA1 | 23.116 ± 3.300 | 22.855 ± 1.102 | 24.844 ± 1.528 | 23.801 ± 0.875 | 25.474 ± 0.979 | 23.173 ± 0.766 | 23.877 | 22.855 | 25.474 | 2.62 |
| SsuTUA2 | 21.717 ± 1.408 | 21.637 ± 1.138 | 22.489 ± 1.343 | 21.927 ± 0.284 | 22.918 ± 1.060 | 21.214 ± 0.388 | 21.984 | 21.214 | 22.918 | 1.704 |
| SsubTUB | 23.802 ± 2.168 | 22.222 ± 0.634 | 24.350 ± 0.855 | 22.667 ± 0.281 | 25.599 ± 1.185 | 21.687 ± 0.812 | 23.388 | 21.687 | 25.599 | 3.911 |
| ColGAPDH | 24.569 ± 1.992 | 24.188 ± 1.022 | 26.065 ± 1.048 | 26.214 ± 0.373 | 25.079 ± 0.822 | 24.997 ± 0.272 | 25.185 | 24.188 | 26.214 | 2.026 |
| SsuCal7 | 20.469 ± 2.372 | 19.505 ± 0.960 | 21.703 ± 0.636 | 20.858 ± 0.228 | 21.444 ± 1.328 | 19.672 ± 1.033 | 20.608 | 19.505 | 21.703 | 2.198 |
| SsuCas | 22.702 ± 3.140 | 20.807 ± 0.850 | 33.379 ± 1.396 | 21.377 ± 0.187 | 23.713 ± 2.040 | 20.280 ± 0.637 | 23.709 | 20.28 | 33.379 | 13.099 |
| SsueIF5 | 21.820 ± 2.500 | 19.810 ± 1.122 | 21.700 ± 0.709 | 21.423 ± 0.266 | 23.243 ± 1.744 | 20.736 ± 0.619 | 21.455 | 19.81 | 23.243 | 3.433 |
| SsuGAPDH | 19.325 ± 2.828 | 19.818 ± 1.308 | 20.596 ± 0.941 | 20.723 ± 0.325 | 20.409 ± 0.326 | 19.810 ± 0.278 | 20.113 | 19.325 | 20.723 | 1.398 |
| SsuGTP | 22.843 ± 2.547 | 21.975 ± 1.044 | 22.920 ± 1.006 | 22.966 ± 0.437 | 24.304 ± 1.194 | 22.239 ± 0.692 | 22.875 | 21.975 | 24.304 | 2.328 |
| SsuHis | 21.538 ± 2.536 | 20.623 ± 0.710 | 22.414 ± 0.705 | 22.775 ± 0.268 | 23.434 ± 2.293 | 21.654 ± 0.458 | 22.073 | 20.623 | 23.434 | 2.811 |
| SsuMDH | 24.656 ± 3.161 | 24.456 ± 0.432 | 26.864 ± 1.025 | 25.655 ± 0.211 | 26.601 ± 0.786 | 25.104 ± 0.760 | 25.556 | 24.456 | 26.864 | 2.408 |
| SsuMet2 | 17.014 ± 2.966 | 16.752 ± 0.789 | 17.975 ± 1.264 | 19.275 ± 0.189 | 18.652 ± 1.393 | 18.522 ± 0.908 | 18.032 | 16.752 | 19.275 | 2.523 |
| SsuRIB | 21.848 ± 2.851 | 21.358 ± 0.871 | 23.163 ± 1.038 | 22.122 ± 0.454 | 23.539 ± 2.217 | 21.856 ± 0.544 | 22.314 | 21.358 | 23.539 | 2.181 |
| SsuUBC1 | 24.886 ± 3.431 | 24.180 ± 0.886 | 25.768 ± 0.564 | 26.410 ± 0.186 | 25.820 ± 0.990 | 24.857 ± 0.396 | 25.32 | 24.18 | 26.41 | 2.229 |
| SsuUBC17 | 22.491 ± 3.106 | 20.030 ± 1.158 | 21.798 ± 1.360 | 21.316 ± 0.288 | 24.137 ± 2.867 | 20.090 ± 0.560 | 21.644 | 20.03 | 24.137 | 4.107 |
| SsuUBC2 | 22.343 ± 2.593 | 20.277 ± 0.247 | 21.973 ± 1.268 | 20.856 ± 0.569 | 24.277 ± 2.711 | 19.607 ± 0.508 | 21.556 | 19.607 | 24.277 | 4.67 |
| SsuUBCJ2 | 24.718 ± 2.937 | 23.014 ± 0.667 | 24.883 ± 0.758 | 24.601 ± 0.248 | 26.375 ± 2.294 | 23.482 ± 0.350 | 24.512 | 23.014 | 26.375 | 3.361 |
| SsuUDP | 25.303 ± 3.170 | 23.309 ± 1.093 | 25.399 ± 0.965 | 24.038 ± 0.669 | 27.973 ± 2.695 | 23.242 ± 0.684 | 24.877 | 23.242 | 27.973 | 4.73 |
| Gene | Geometric Mean | Average Mean | Minimum | Maximum | SD | CV | r | p-Value |
|---|---|---|---|---|---|---|---|---|
| SsuACT | 21.55 | 21.56 | 20.64 | 22.68 | 0.7 | 3.25 | 0.987 | 0.001 |
| SsuUBCJ2 | 24.49 | 24.51 | 23.01 | 26.38 | 0.84 | 3.44 | 0.987 | 0.001 |
| SsuCal7 | 20.59 | 20.61 | 19.51 | 21.7 | 0.73 | 3.53 | 0.977 | 0.001 |
| SsuHis | 22.05 | 22.07 | 20.62 | 23.43 | 0.8 | 3.63 | 0.93 | 0.007 |
| SsuRIB | 22.3 | 22.31 | 21.36 | 23.54 | 0.69 | 3.1 | 0.928 | 0.008 |
| SsuUDP | 24.83 | 24.88 | 23.24 | 27.97 | 1.35 | 5.42 | 0.926 | 0.008 |
| SsuTUA1 | 23.86 | 23.88 | 22.85 | 25.47 | 0.85 | 3.58 | 0.92 | 0.009 |
| SsuUBC17 | 21.6 | 21.64 | 20.03 | 24.14 | 1.17 | 5.38 | 0.919 | 0.01 |
| SsuTUA2 | 21.98 | 21.98 | 21.21 | 22.92 | 0.48 | 2.18 | 0.905 | 0.013 |
| SsuUBC2 | 21.5 | 21.56 | 19.61 | 24.28 | 1.31 | 6.07 | 0.887 | 0.018 |
| SsuTUB | 23.35 | 23.39 | 21.69 | 25.6 | 1.2 | 5.11 | 0.858 | 0.029 |
| SsuCas | 23.35 | 23.71 | 20.28 | 33.38 | 3.22 | 13.6 | 0.83 | 0.041 |
| SsuMDH | 25.54 | 25.56 | 24.46 | 26.86 | 0.82 | 3.2 | 0.824 | 0.044 |
| SsueIF5 | 21.43 | 21.46 | 19.81 | 23.24 | 0.8 | 3.72 | 0.761 | 0.079 |
| SsuGTP | 22.86 | 22.87 | 21.98 | 24.3 | 0.52 | 2.28 | 0.742 | 0.092 |
| SsuUBC1 | 25.31 | 25.32 | 24.18 | 26.41 | 0.68 | 2.68 | 0.741 | 0.092 |
| SsuGAPDH | 20.11 | 20.11 | 19.32 | 20.72 | 0.46 | 2.3 | 0.634 | 0.176 |
| ColGAPDH | 25.17 | 25.19 | 24.19 | 26.21 | 0.64 | 2.53 | 0.602 | 0.206 |
| SsuMet2 | 18.01 | 18.03 | 16.75 | 19.27 | 0.78 | 4.35 | 0.513 | 0.296 |
| Gene | geNorm | NormFinder | BestKeeper | Geometric Mean | Combined Ranking |
|---|---|---|---|---|---|
| SsuACT | 3 | 1 | 1 | 1.44 | 1 |
| SsuRIB | 1 | 2 | 5 | 2.15 | 2 |
| SsuTUA1 | 1 | 4 | 6 | 2.88 | 3 |
| SsuCal7 | 4 | 3 | 3 | 3.3 | 4 |
| SsuMDH | 2 | 5 | 8 | 4.31 | 5 |
| SsuUBCJ2 | 8 | 6 | 2 | 4.58 | 6 |
| SsuTUA2 | 5 | 7 | 7 | 6.26 | 7 |
| SsuHis | 7 | 9 | 4 | 6.32 | 8 |
| SsuGTP | 6 | 8 | 10 | 7.83 | 9 |
| SsueIF5 | 9 | 10 | 9 | 9.32 | 10 |
| SsuUBC1 | 10 | 12 | 11 | 10.97 | 11 |
| SsuGAPDH | 11 | 13 | 12 | 11.97 | 12 |
| ColGAPDH | 12 | 14 | 13 | 12.97 | 13 |
| SsuTUB | 14 | 11 | 16 | 13.51 | 14 |
| SsuMet2 | 13 | 18 | 14 | 14.85 | 15 |
| SsuUBC17 | 15 | 15 | 15 | 15 | 16 |
| SsuUBC2 | 16 | 16 | 17 | 16.33 | 17 |
| SsuUDP | 17 | 17 | 18 | 17.33 | 18 |
| SsuCas | 18 | 19 | 19 | 18.66 | 19 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, Z.; Zhang, R.; Zhou, Z. Identification and Validation of Reference Genes for Gene Expression Analysis in Schima superba. Genes 2021, 12, 732. https://doi.org/10.3390/genes12050732
Yang Z, Zhang R, Zhou Z. Identification and Validation of Reference Genes for Gene Expression Analysis in Schima superba. Genes. 2021; 12(5):732. https://doi.org/10.3390/genes12050732
Chicago/Turabian StyleYang, Zhongyi, Rui Zhang, and Zhichun Zhou. 2021. "Identification and Validation of Reference Genes for Gene Expression Analysis in Schima superba" Genes 12, no. 5: 732. https://doi.org/10.3390/genes12050732
APA StyleYang, Z., Zhang, R., & Zhou, Z. (2021). Identification and Validation of Reference Genes for Gene Expression Analysis in Schima superba. Genes, 12(5), 732. https://doi.org/10.3390/genes12050732






