Skeletal Muscle Gene Expression Profile in Response to Caloric Restriction and Aging: A Role for SirT1
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Dietary Regime
2.3. Transcriptome Sequencing and Data Analysis
2.4. GO and KEGG Analyses
2.5. Custom Functional Analysis
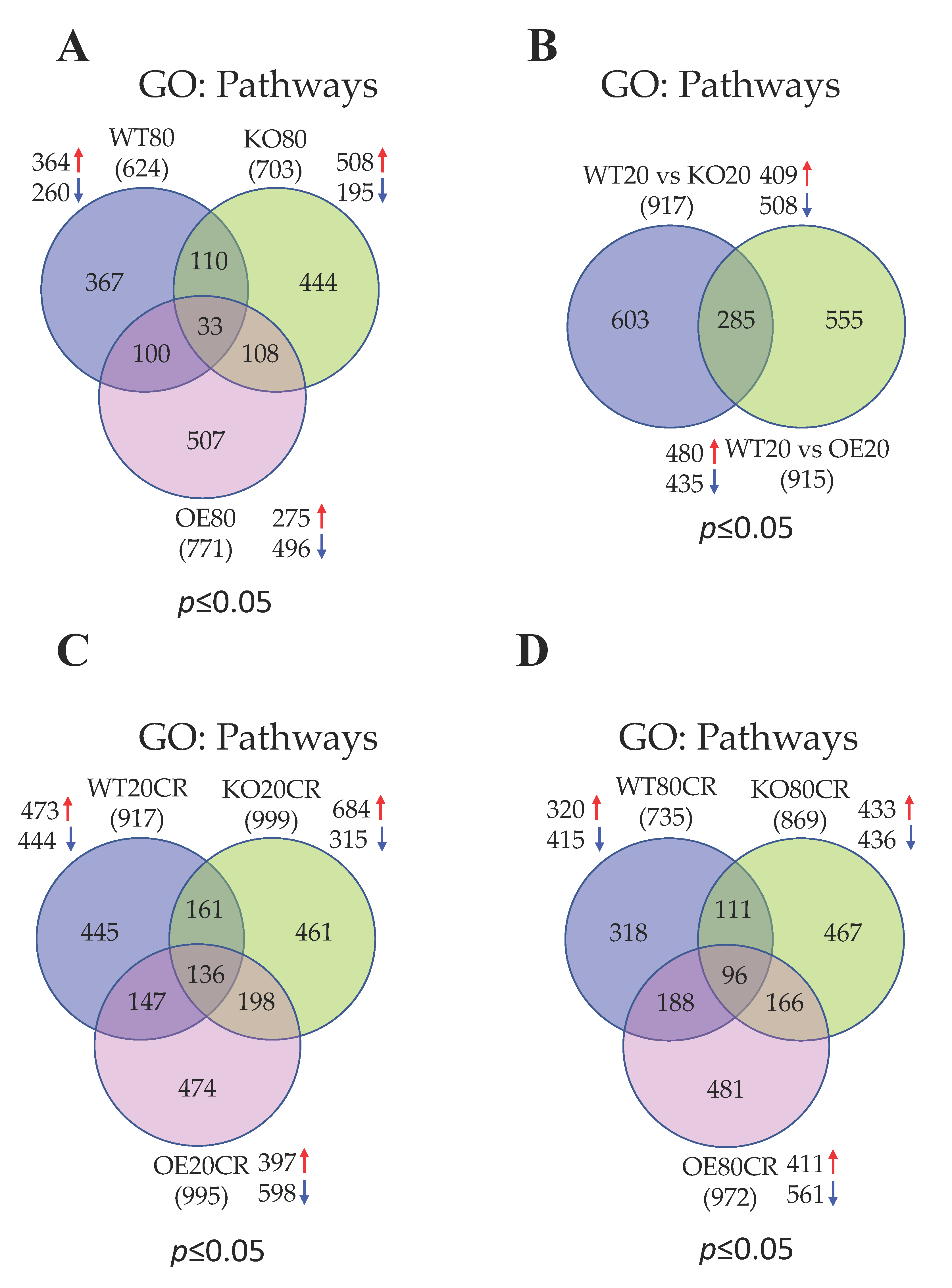
3. Results
3.1. Summary of Skeletal Muscle Gene Expression Profile in 20 and 80 wk Old WT, SirT1-KO, and SirT1-OE Mice
3.2. Aging Augments Muscle Contraction and Cellular Transcription and Translation Pathways and Represses Ubiquitin and Energy Homeostasis Related Pathways in the Skeletal Muscle of WT Mice
3.3. Aging Promotes Cellular Transcriptions and Represses Mitochondrial Functions in the Skeletal Muscle of SirT1 KO Mice
3.4. GTPase Signaling, DNA Binding Activity, and Ion Channel Functions Are Activated Whereas Apoptotic Signaling, and Cellular Metabolism Are Suppressed in SirT1 OE Mice in Aging
3.5. SirT1 Loss-of-Function Increases Mitochondrial Functions, Metabolism and Ubiquitin Proteasomal Activity, and Decreases Muscle Function and Insulin Signaling in the Skeletal Muscle of Young Mice
3.6. SirT1 Gain-of-Function Upregulates Signal Transduction and Downregulates Lipid Metabolism in the Skeletal Muscle of Young Mice
3.7. Most of the Top Pathways Altered by CR Are Associated with Translational and Muscle Contraction Process, Which Are Suppressed in the Skeletal Muscle of Young WT Mice
3.8. CR Promotes Muscle Contraction and Inhibits Mitochondrial Function Related Pathways in the Skeletal Muscle of Young SirT1 KO Mice
3.9. CR Promotes Gene Expression Events and Inhibits ECM and Cytoskeleton in the Skeletal Muscle of Young SirT1 OE Mice
3.10. CR Positively Regulates Transcription and Translation and Negatively Regulates ECM and Cytoskeleton in the Skeletal Muscle of Aged WT Mice
3.11. CR Promotes Mitochondrial β-Oxidation, Fatty Acid Metabolism, and Muscle Contraction and Blocks Cellular Gene Expression in the Skeletal Muscle of Aged SirT1 KO Mice
3.12. SirT1 OE Promotes Ubiquitination Signaling and Downregulated Muscle Structure and Contraction Related Proteins Following CR in the Skeletal Muscle of Aged Mice
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kennedy, B.K.; Austriaco, N.R., Jr.; Zhang, J.; Guarente, L. Mutation in the silencing gene SIR4 can delay aging in S. cerevisiae. Cell 1995, 80, 485–496. [Google Scholar] [CrossRef]
- Kaeberlein, M.; McVey, M.; Guarente, L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 1999, 13, 2570–2580. [Google Scholar] [CrossRef]
- Guarente, L.; Picard, F. Calorie restriction—The SIR2 connection. Cell 2005, 120, 473–482. [Google Scholar] [CrossRef] [PubMed]
- Morris, B.J. Seven sirtuins for seven deadly diseases of aging. Free Radic. Biol. Med. 2013, 56, 133–171. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, R.M.; Fraga, M.F. Aging and cancer: Are sirtuins the link? Future Oncol. 2010, 6, 905–915. [Google Scholar] [CrossRef] [PubMed]
- Schug, T.T.; Li, X. Sirtuin 1 in lipid metabolism and obesity. Ann. Med. 2011, 43, 198–211. [Google Scholar] [CrossRef]
- Park, S.J.; Ahmad, F.; Philp, A.; Baar, K.; Williams, T.; Luo, H.; Ke, H.; Rehmann, H.; Taussig, R.; Brown, A.L.; et al. Resveratrol ameliorates aging-related metabolic phenotypes by inhibiting cAMP phosphodiesterases. Cell 2012, 148, 421–433. [Google Scholar] [CrossRef] [PubMed]
- Donmez, G.; Outeiro, T.F. SIRT1 and SIRT2: Emerging targets in neurodegeneration. EMBO Mol. Med. 2013, 5, 344–352. [Google Scholar] [CrossRef]
- Mohamed, J.S.; Hajira, A.; Pardo, P.S.; Boriek, A.M. MicroRNA-149 inhibits PARP-2 and promotes mitochondrial biogenesis via SIRT-1/PGC-1alpha network in skeletal muscle. Diabetes 2014, 63, 1546–1559. [Google Scholar] [CrossRef] [PubMed]
- Mohamed, J.S.; Wilson, J.C.; Myers, M.J.; Sisson, K.J.; Alway, S.E. Dysregulation of SIRT-1 in aging mice increases skeletal muscle fatigue by a PARP-1-dependent mechanism. Aging 2014, 6, 820–834. [Google Scholar] [CrossRef]
- Alway, S.E.; McCrory, J.L.; Kearcher, K.; Vickers, A.; Frear, B.; Gilleland, D.L.; Bonner, D.E.; Thomas, J.M.; Donley, D.A.; Lively, M.W.; et al. Resveratrol Enhances Exercise-Induced Cellular and Functional Adaptations of Skeletal Muscle in Older Men and Women. J. Gerontol. A Biol. Sci. Med. Sci. 2017, 72, 1595–1606. [Google Scholar] [CrossRef] [PubMed]
- Haramizu, S.; Asano, S.; Butler, D.C.; Stanton, D.A.; Hajira, A.; Mohamed, J.S.; Alway, S.E. Dietary resveratrol confers apoptotic resistance to oxidative stress in myoblasts. J. Nutr. Biochem. 2017, 50, 103–115. [Google Scholar] [CrossRef] [PubMed]
- Myers, M.J.; Shepherd, D.L.; Durr, A.J.; Stanton, D.S.; Mohamed, J.S.; Hollander, J.M.; Alway, S.E. The role of SIRT1 in skeletal muscle function and repair of older mice. J. Cachexia Sarcopenia Muscle 2019, 10, 929–949. [Google Scholar] [CrossRef] [PubMed]
- Vaziri, H.; Dessain, S.K.; Ng Eaton, E.; Imai, S.I.; Frye, R.A.; Pandita, T.K.; Guarente, L.; Weinberg, R.A. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell 2001, 107, 149–159. [Google Scholar] [CrossRef]
- Luo, J.; Nikolaev, A.Y.; Imai, S.; Chen, D.; Su, F.; Shiloh, A.; Guarente, L.; Gu, W. Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell 2001, 107, 137–148. [Google Scholar] [CrossRef]
- Rodgers, J.T.; Lerin, C.; Haas, W.; Gygi, S.P.; Spiegelman, B.M.; Puigserver, P. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature 2005, 434, 113–118. [Google Scholar] [CrossRef] [PubMed]
- Dean, W. The Retardation of Aging and Diseases of Aging by Dietary Restriction—Weindruch,R, Walford,RL. J. Am. Geriatr. Soc. 1990, 38, 736. [Google Scholar]
- Lin, S.J.; Defossez, P.A.; Guarente, L. Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science 2000, 289, 2126–2128. [Google Scholar] [CrossRef]
- Masoro, E.J. Caloric restriction and aging: An update. Exp. Gerontol. 2000, 35, 299–305. [Google Scholar] [CrossRef]
- Roth, G.S.; Ingram, D.K.; Lane, M.A. Caloric restriction in primates and relevance to humans. Healthy Aging Funct. Longev. 2001, 928, 305–315. [Google Scholar] [CrossRef]
- Lin, S.J.; Kaeberlein, M.; Andalis, A.A.; Sturtz, L.A.; Defossez, P.A.; Culotta, V.C.; Fink, G.R.; Guarente, L. Calorie restriction extends Saccharomyces cerevisiae lifespan by increasing respiration. Nature 2002, 418, 344–348. [Google Scholar] [CrossRef] [PubMed]
- Cohen, H.Y.; Miller, C.; Bitterman, K.J.; Wall, N.R.; Hekking, B.; Kessler, B.; Howitz, K.T.; Gorospe, M.; de Cabo, R.; Sinclair, D.A. Calorie restriction promotes mammalian cell survival by inducing the SIRT1 deacetylase. Science 2004, 305, 390–392. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.J.; Ford, E.; Haigis, M.; Liszt, G.; Guarente, L. Calorie restriction extends yeast life span by lowering the level of NADH. Genes Dev. 2004, 18, 12–16. [Google Scholar] [CrossRef] [PubMed]
- Opie, L.H. Caloric restriction models reverse metabolic syndrome. J. Am. Coll. Cardiol. 2009, 53, 899–900. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lakowski, B.; Hekimi, S. The genetics of caloric restriction in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 1998, 95, 13091–13096. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Thomas, E.L.; Kapahi, P. HIF-1 modulates dietary restriction-mediated lifespan extension via IRE-1 in Caenorhabditis elegans. PLoS Genet 2009, 5, e1000486. [Google Scholar] [CrossRef]
- Rogina, B.; Helfand, S.L. Sir2 mediates longevity in the fly through a pathway related to calorie restriction. Proc. Natl. Acad. Sci. USA 2004, 101, 15998–16003. [Google Scholar] [CrossRef]
- Wood, J.G.; Rogina, B.; Lavu, S.; Howitz, K.; Helfand, S.L.; Tatar, M.; Sinclair, D. Sirtuin activators mimic caloric restriction and delay ageing in metazoans. Nature 2004, 430, 686–689. [Google Scholar] [CrossRef]
- Herranz, D.; Munoz-Martin, M.; Canamero, M.; Mulero, F.; Martinez-Pastor, B.; Fernandez-Capetillo, O.; Serrano, M. Sirt1 improves healthy ageing and protects from metabolic syndrome-associated cancer. Nat. Commun. 2010, 1, 3. [Google Scholar] [CrossRef]
- Holloszy, J.O.; Fontana, L. Caloric restriction in humans. Exp. Gerontol. 2007, 42, 709–712. [Google Scholar] [CrossRef]
- Marzetti, E.; Lawler, J.M.; Hiona, A.; Manini, T.; Seo, A.Y.; Leeuwenburgh, C. Modulation of age-induced apoptotic signaling and cellular remodeling by exercise and calorie restriction in skeletal muscle. Free Radic. Biol. Med. 2008, 44, 160–168. [Google Scholar] [CrossRef] [PubMed]
- Collins, C.A.; Zammit, P.S.; Ruiz, A.P.; Morgan, J.E.; Partridge, T.A. A population of myogenic stem cells that survives skeletal muscle aging. Stem Cells 2007, 25, 885–894. [Google Scholar] [CrossRef] [PubMed]
- Neal, A.; Boldrin, L.; Morgan, J.E. The satellite cell in male and female, developing and adult mouse muscle: Distinct stem cells for growth and regeneration. PLoS ONE 2012, 7, e37950. [Google Scholar] [CrossRef] [PubMed]
- Alway, S.E.; Myers, M.J.; Mohamed, J.S. Regulation of satellite cell function in sarcopenia. Front. Aging Neurosci. 2014, 6, 246. [Google Scholar] [CrossRef] [PubMed]
- Cerletti, M.; Jang, Y.C.; Finley, L.W.; Haigis, M.C.; Wagers, A.J. Short-term calorie restriction enhances skeletal muscle stem cell function. Cell Stem Cell 2012, 10, 515–519. [Google Scholar] [CrossRef] [PubMed]
- Etienne, J.; Liu, C.; Skinner, C.M.; Conboy, M.J.; Conboy, I.M. Skeletal muscle as an experimental model of choice to study tissue aging and rejuvenation. Skelet. Muscle 2020, 10, 4. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [PubMed]
- Smyth, G.K. Limma: Linear models for microarray data. In Bioinformatics and Computational Biology Solutions Using R and Bioconductor; Gentleman, R., Carey, V., Dudoit, S., Irizarry, R., Hubereds, W., Eds.; Springer: New York, NY, USA, 2005; pp. 397–420. [Google Scholar] [CrossRef]
- Law, C.W.; Chen, Y.; Shi, W.; Smyth, G.K. Voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014, 15, R29. [Google Scholar] [CrossRef]
- Liu, R.; Holik, A.Z.; Su, S.; Jansz, N.; Chen, K.; Leong, H.S.; Blewitt, M.E.; Asselin-Labat, M.L.; Smyth, G.K.; Ritchie, M.E. Why weight? Modelling sample and observational level variability improves power in RNA-seq analyses. Nucleic Acids Res. 2015, 43, e97. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, M.E.; Diyagama, D.; Neilson, J.; van Laar, R.; Dobrovic, A.; Holloway, A.; Smyth, G.K. Empirical array quality weights in the analysis of microarray data. BMC Bioinform. 2006, 7, 261. [Google Scholar] [CrossRef] [PubMed]
- Kovaka, S.; Zimin, A.V.; Pertea, G.M.; Razaghi, R.; Salzberg, S.L.; Pertea, M. Transcriptome assembly from long-read RNA-seq alignments with StringTie2. Genome Biol. 2019, 20, 278. [Google Scholar] [CrossRef] [PubMed]
- Reimand, J.; Kull, M.; Peterson, H.; Hansen, J.; Vilo, J. g:Profiler—A web-based toolset for functional profiling of gene lists from large-scale experiments. Nucleic Acids Res. 2007, 35, W193–W200. [Google Scholar] [CrossRef] [PubMed]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S.; Hattori, M.; Aoki-Kinoshita, K.F.; Itoh, M.; Kawashima, S.; Katayama, T.; Araki, M.; Hirakawa, M. From genomics to chemical genomics: New developments in KEGG. Nucleic Acids Res. 2006, 34, D354–D357. [Google Scholar] [CrossRef]
- Joshi-Tope, G.; Gillespie, M.; Vastrik, I.; D’Eustachio, P.; Schmidt, E.; de Bono, B.; Jassal, B.; Gopinath, G.R.; Wu, G.R.; Matthews, L.; et al. Reactome: A knowledgebase of biological pathways. Nucleic Acids Res. 2005, 33, D428–D432. [Google Scholar] [CrossRef]
- Matys, V.; Kel-Margoulis, O.V.; Fricke, E.; Liebich, I.; Land, S.; Barre-Dirrie, A.; Reuter, I.; Chekmenev, D.; Krull, M.; Hornischer, K.; et al. TRANSFAC and its module TRANSCompel: Transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006, 34, D108–D110. [Google Scholar] [CrossRef]
- Kwon, S.; Seok, S.; Yau, P.; Li, X.; Kemper, B.; Kemper, J.K. Obesity and aging diminish sirtuin 1 (SIRT1)-mediated deacetylation of SIRT3, leading to hyperacetylation and decreased activity and stability of SIRT3. J. Biol. Chem. 2017, 292, 17312–17323. [Google Scholar] [CrossRef]
- Pardo, P.S.; Boriek, A.M. The physiological roles of Sirt1 in skeletal muscle. Aging 2011, 3, 430–437. [Google Scholar] [CrossRef]
- Lyu, A.K.; Zhu, S.Y.; Chen, J.L.; Zhao, Y.X.; Pu, D.; Luo, C.; Lyu, Q.; Fan, Z.; Sun, Y.; Wu, J.; et al. Inhibition of TLR9 attenuates skeletal muscle fibrosis in aged sarcopenic mice via the p53/SIRT1 pathway. Exp. Gerontol. 2019, 122, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Braidy, N.; Guillemin, G.J.; Mansour, H.; Chan-Ling, T.; Poljak, A.; Grant, R. Age related changes in NAD+ metabolism oxidative stress and Sirt1 activity in wistar rats. PLoS ONE 2011, 6, e19194. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.K.; Klopp, R.G.; Weindruch, R.; Prolla, T.A. Gene expression profile of aging and its retardation by caloric restriction. Science 1999, 285, 1390–1393. [Google Scholar] [CrossRef] [PubMed]
- Goyns, M.H.; Charlton, M.A.; Dunford, J.E.; Lavery, W.L.; Merry, B.J.; Salehi, M.; Simoes, D.C. Differential display analysis of gene expression indicates that age-related changes are restricted to a small cohort of genes. Mech. Ageing Dev. 1998, 101, 73–90. [Google Scholar] [CrossRef]
- Gong, H.; Pang, J.; Han, Y.; Dai, Y.; Dai, D.; Cai, J.; Zhang, T.M. Age-dependent tissue expression patterns of Sirt1 in senescence-accelerated mice. Mol. Med. Rep. 2014, 10, 3296–3302. [Google Scholar] [CrossRef] [PubMed]
- Schenk, S.; McCurdy, C.E.; Philp, A.; Chen, M.Z.; Holliday, M.J.; Bandyopadhyay, G.K.; Osborn, O.; Baar, K.; Olefsky, J.M. Sirt1 enhances skeletal muscle insulin sensitivity in mice during caloric restriction. J. Clin. Investig. 2011, 121, 4281–4288. [Google Scholar] [CrossRef] [PubMed]
- Fujiwara, D.; Iwahara, N.; Sebori, R.; Hosoda, R.; Shimohama, S.; Kuno, A.; Horio, Y. SIRT1 deficiency interferes with membrane resealing after cell membrane injury. PLoS ONE 2019, 14, e0218329. [Google Scholar] [CrossRef]
- Ross, J.A.; Levy, Y.; Svensson, K.; Philp, A.; Schenk, S.; Ochala, J. SIRT1 regulates nuclear number and domain size in skeletal muscle fibers. J. Cell. Physiol. 2018, 233, 7157–7163. [Google Scholar] [CrossRef] [PubMed]
- Price, N.L.; Gomes, A.P.; Ling, A.J.; Duarte, F.V.; Martin-Montalvo, A.; North, B.J.; Agarwal, B.; Ye, L.; Ramadori, G.; Teodoro, J.S.; et al. SIRT1 is required for AMPK activation and the beneficial effects of resveratrol on mitochondrial function. Cell Metab. 2012, 15, 675–690. [Google Scholar] [CrossRef]
- Gurd, B.J. Deacetylation of PGC-1alpha by SIRT1: Importance for skeletal muscle function and exercise-induced mitochondrial biogenesis. Appl. Physiol. Nutr. Metab. Physiol. Appl. Nutr. Metab. 2011, 36, 589–597. [Google Scholar] [CrossRef]
- Philp, A.; Chen, A.; Lan, D.; Meyer, G.A.; Murphy, A.N.; Knapp, A.E.; Olfert, I.M.; McCurdy, C.E.; Marcotte, G.R.; Hogan, M.C.; et al. Sirtuin 1 (SIRT1) deacetylase activity is not required for mitochondrial biogenesis or peroxisome proliferator-activated receptor-γ coactivator-1alpha (PGC-1alpha) deacetylation following endurance exercise. J. Biol. Chem. 2011, 286, 30561–30570. [Google Scholar] [CrossRef]
- Wang, F.; Chan, C.H.; Chen, K.; Guan, X.; Lin, H.K.; Tong, Q. Deacetylation of FOXO3 by SIRT1 or SIRT2 leads to Skp2-mediated FOXO3 ubiquitination and degradation. Oncogene 2012, 31, 1546–1557. [Google Scholar] [CrossRef]
- Chabi, B.; Adhihetty, P.J.; O’Leary, M.F.; Menzies, K.J.; Hood, D.A. Relationship between Sirt1 expression and mitochondrial proteins during conditions of chronic muscle use and disuse. J. Appl. Physiol. 2009, 107, 1730–1735. [Google Scholar] [CrossRef] [PubMed]
- Gurd, B.J.; Yoshida, Y.; Lally, J.; Holloway, G.P.; Bonen, A. The deacetylase enzyme SIRT1 is not associated with oxidative capacity in rat heart and skeletal muscle and its overexpression reduces mitochondrial biogenesis. J. Physiol. 2009, 587, 1817–1828. [Google Scholar] [CrossRef]
- White, A.T.; McCurdy, C.E.; Philp, A.; Hamilton, D.L.; Johnson, C.D.; Schenk, S. Skeletal muscle-specific overexpression of SIRT1 does not enhance whole-body energy expenditure or insulin sensitivity in young mice. Diabetologia 2013, 56, 1629–1637. [Google Scholar] [CrossRef] [PubMed]
- Vila, L.; Roca, C.; Elias, I.; Casellas, A.; Lage, R.; Franckhauser, S.; Bosch, F. AAV-mediated Sirt1 overexpression in skeletal muscle activates oxidative capacity but does not prevent insulin resistance. Mol. Ther. Methods Clin. Dev. 2016, 5, 16072. [Google Scholar] [CrossRef] [PubMed]
- Svensson, K.; Tahvilian, S.; Martins, V.F.; Dent, J.R.; Lemanek, A.; Barooni, N.; Greyslak, K.; McCurdy, C.E.; Schenk, S. Combined overexpression of SIRT1 and knockout of GCN5 in adult skeletal muscle does not affect glucose homeostasis or exercise performance in mice. Am. J. Physiol. Endocrinol. Metab. 2020, 318, E145–E151. [Google Scholar] [CrossRef]
- White, A.T.; Philp, A.; Fridolfsson, H.N.; Schilling, J.M.; Murphy, A.N.; Hamilton, D.L.; McCurdy, C.E.; Patel, H.H.; Schenk, S. High-fat diet-induced impairment of skeletal muscle insulin sensitivity is not prevented by SIRT1 overexpression. Am. J. Physiol. Endocrinol. Metab. 2014, 307, E764–E772. [Google Scholar] [CrossRef]
- Banks, A.S.; Kon, N.; Knight, C.; Matsumoto, M.; Gutierrez-Juarez, R.; Rossetti, L.; Gu, W.; Accili, D. SirT1 gain of function increases energy efficiency and prevents diabetes in mice. Cell Metab. 2008, 8, 333–341. [Google Scholar] [CrossRef]
- Wu, D.; Qiu, Y.; Gao, X.; Yuan, X.B.; Zhai, Q. Overexpression of SIRT1 in mouse forebrain impairs lipid/glucose metabolism and motor function. PLoS ONE 2011, 6, e21759. [Google Scholar] [CrossRef]
- Alway, S.E.; Bennett, B.T.; Wilson, J.C.; Sperringer, J.; Mohamed, J.S.; Edens, N.K.; Pereira, S.L. Green tea extract attenuates muscle loss and improves muscle function during disuse, but fails to improve muscle recovery following unloading in aged rats. J. Appl. Physiol. 2015, 118, 319–330. [Google Scholar] [CrossRef]
- Alway, S.E.; Mohamed, J.S.; Myers, M.J. Mitochondria Initiate and Regulate Sarcopenia. Exerc. Sport Sci. Rev. 2017, 45, 58–69. [Google Scholar] [CrossRef] [PubMed]
- Donniacuo, M.; Urbanek, K.; Nebbioso, A.; Sodano, L.; Gallo, L.; Altucci, L.; Rinaldi, B. Cardioprotective effect of a moderate and prolonged exercise training involves sirtuin pathway. Life Sci. 2019, 222, 140–147. [Google Scholar] [CrossRef] [PubMed]
- Furukawa, N.; Ongusaha, P.; Jahng, W.J.; Araki, K.; Choi, C.S.; Kim, H.J.; Lee, Y.H.; Kaibuchi, K.; Kahn, B.B.; Masuzaki, H.; et al. Role of Rho-kinase in regulation of insulin action and glucose homeostasis. Cell Metab. 2005, 2, 119–129. [Google Scholar] [CrossRef] [PubMed]
- Sylow, L.; Jensen, T.E.; Kleinert, M.; Hojlund, K.; Kiens, B.; Wojtaszewski, J.; Prats, C.; Schjerling, P.; Richter, E.A. Rac1 signaling is required for insulin-stimulated glucose uptake and is dysregulated in insulin-resistant murine and human skeletal muscle. Diabetes 2013, 62, 1865–1875. [Google Scholar] [CrossRef] [PubMed]
- Ueda, S.; Kitazawa, S.; Ishida, K.; Nishikawa, Y.; Matsui, M.; Matsumoto, H.; Aoki, T.; Nozaki, S.; Takeda, T.; Tamori, Y.; et al. Crucial role of the small GTPase Rac1 in insulin-stimulated translocation of glucose transporter 4 to the mouse skeletal muscle sarcolemma. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2010, 24, 2254–2261. [Google Scholar] [CrossRef]


| Sample Information | ||||||||||||
| Sample ID | WT20AL | WT20CR | WT80AL | WT80CR | KO20AL | KO20CR | KO80AL | KO80CR | OE20AL | OE20CR | OE80AL | OE80CR |
| Sample size | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 |
| Database | ||||||||||||
| Genome assembly | https://useast.ensembl.org/Mus_musculus/Info/Index: GRCm38.p6 (GCA_000001635.8) | |||||||||||
| Gene annotation | ftp://ftp.ensembl.org/pub/release-102/gtf/mus_musculus//Mus_musculus.GRCm38.102.gtf.gz | |||||||||||
| lncRNA | ftp://ftp.sanger.ac.uk/pub/gencode/Gencode_mouse/release_M13/gencode.vM13.long_noncoding_RNAs.gtf.gz (v13) | |||||||||||
| Gene Orthology (GO) | http://www.geneontology.org/ (accessed on 1 January 2021) | |||||||||||
| KEGG | http://www.genome.jp/kegg/pathway.html (accessed on 18 January 2021) | |||||||||||
| Bioinformatics software | ||||||||||||
| Quality control | FastQC (vs. 0.11.9) | |||||||||||
| Adaptors remove | Trimmomatic (vs. 0.39) | |||||||||||
| Mapping | HISAT2 (vs. 2.1.0) | |||||||||||
| Transcripts assembly | StringTie (vs. 2.1.1) | |||||||||||
| Differential expression analysis | Limma (vs. 3.42) | |||||||||||
| GO and KEGG enrichment analysis | g:Profiler version e102_eg49_p15_7a9b4d6 | |||||||||||
| Coding potential | CPC (Coding Potential Calculator) (vs. 0.9) | |||||||||||
| Coding potential | CNCI (Coding-Non-Coding Index) (vs. 2.0) | |||||||||||
| Mice | Age | Diet | Genes | Up | Down | Upregulated Pathways | Downregulated Pathways |
|---|---|---|---|---|---|---|---|
| WT | O | AL | 74 | 39 | 35 | Muscle contraction; Cellular transcription and translation | Ubiquitin; Energy homeostasis |
| KO | O | AL | 1146 | 145 | 1001 | Cellular Transcriptions | Mitochondrial functions |
| OE | O | AL | 284 | 61 | 223 | GTPase signaling; DNA binding activity; Ion channel functions | Apoptotic signaling; Metabolism |
| KO | Y | AL | 566 | 80 | 486 | Mitochondrial functions; Metabolism; Ubiquitin activity | Muscle function; Insulin signaling |
| OE | Y | AL | 48 | 14 | 34 | Signal transduction; Transcription corepressor activity | Lipid metabolism |
| WT | Y | CR | 802 | 125 | 677 | Phosphatidylinositol binding | Translational; Muscle contraction |
| KO | Y | CR | 1296 | 229 | 1067 | Muscle contraction | Mitochondrial function |
| OE | Y | CR | 994 | 83 | 911 | Gene expression | ECM; Cytoskeletal proteins |
| WT | O | CR | 339 | 92 | 247 | Transcription and translation | ECM; Cytoskeletal proteins |
| KO | O | CR | 465 | 133 | 332 | Mitochondrial β-oxidation; fatty acid metabolism; muscle contraction | Cellular gene expression |
| OE | O | CR | 1465 | 174 | 1291 | Ubiquitination signaling | Muscle structure; Contraction |
| No. | Genes | logFC | Adj.P.Val | No. | Genes | logFC | Adj.P.Val |
|---|---|---|---|---|---|---|---|
| WT80 | 2 | Leng8 | −2.25 | 0.05 | |||
| 1 | Gm6472 | 3.10 | 0.00 | 3 | Sppl2a | −2.21 | 0.05 |
| 2 | Rps13-ps1 | 2.85 | 0.00 | 4 | Arid1b | −2.21 | 0.05 |
| 3 | Gm15920 | 2.81 | 0.00 | 5 | Agpat1 | −2.16 | 0.05 |
| 4 | Ngp | 2.50 | 0.01 | 6 | Dnajc18 | −2.14 | 0.04 |
| 5 | Ltf | 2.41 | 0.00 | 7 | Rock2 | −2.10 | 0.05 |
| 6 | S100a8 | 2.41 | 0.03 | 8 | Fto | −2.09 | 0.04 |
| 7 | S100a9 | 2.38 | 0.03 | 9 | Atp6v0a2 | −2.09 | 0.04 |
| 8 | Igkc | 2.33 | 0.00 | 10 | St13 | −2.08 | 0.05 |
| 9 | Gm9855 | 2.20 | 0.00 | 11 | Plin4 | −2.08 | 0.05 |
| 10 | Chil3 | 2.04 | 0.05 | 12 | Crk | −2.08 | 0.04 |
| KO80 | 13 | Cbx5 | -2.07 | 0.03 | |||
| 1 | Gm5537 | 2.87 | 0.00 | 14 | Baz2a | −2.05 | 0.04 |
| 2 | Hmgb1-ps8 | 2.71 | 0.00 | 15 | Phc3 | −2.03 | 0.04 |
| 3 | Lcn2 | 2.53 | 0.00 | 16 | Fbxo38 | −2.02 | 0.03 |
| 4 | Gm9794 | 2.40 | 0.02 | 17 | Wfikkn2 | −2.02 | 0.05 |
| 5 | Pakap | 2.15 | 0.03 | 18 | St3gal3 | −2.01 | 0.04 |
| 6 | Mt2 | 2.12 | 0.02 | 19 | Slc9a8 | −2.01 | 0.04 |
| 7 | Igkc | 2.06 | 0.02 | 20 | AU022252 | −2.01 | 0.05 |
| OE80 | 21 | Erap1 | -2.00 | 0.05 | |||
| 1 | Gm28438 | 2.83 | 0.04 | 22 | Ckap5 | −2.00 | 0.04 |
| 2 | Mup10 | 2.69 | 0.00 | 23 | Mlh3 | −2.00 | 0.04 |
| 3 | Myo15 | 2.53 | 0.00 | 24 | Ccdc97 | −2.00 | 0.03 |
| KO80 | |||||||
| 1 | Zfp148 | −2.27 | 0.05 | ||||
| No. | Genes | log2FC | Adj.P.Val | No. | Genes | log2FC | Adj.P.Val |
|---|---|---|---|---|---|---|---|
| SirT1 KO | 26 | Cd81 | −2.15 | 0.04 | |||
| 1 | Gm15920 | 3.09 | 0.00 | 27 | Atp5j | −2.13 | 0.03 |
| 2 | Rps13-ps1 | 2.93 | 0.00 | 28 | Prpf38b | −2.11 | 0.05 |
| 3 | Gm15459 | 2.88 | 0.00 | 29 | Sec62 | −2.10 | 0.03 |
| 4 | Gm2539 | 2.69 | 0.00 | 30 | Atp5h | −2.10 | 0.05 |
| 5 | Gm19791 | 2.68 | 0.00 | 31 | Mrpl18 | −2.09 | 0.03 |
| 6 | Rps12l1 | 2.53 | 0.02 | 32 | Dnajc21 | −2.09 | 0.05 |
| 7 | Gm7993 | 2.44 | 0.00 | 33 | Eif2b4 | −2.08 | 0.05 |
| 8 | Mrpl48-ps | 2.32 | 0.00 | 34 | Cbfb | −2.08 | 0.04 |
| 9 | Gm9855 | 2.31 | 0.00 | 35 | Ndufs3 | −2.07 | 0.04 |
| 10 | Gm14586 | 2.28 | 0.01 | 36 | Ewsr1 | −2.06 | 0.03 |
| 11 | Rtkn2 | 2.03 | 0.00 | 37 | Serf2 | −2.06 | 0.02 |
| 12 | Ighm | 2.00 | 0.00 | 38 | Six1 | −2.05 | 0.03 |
| 13 | Vdac3 | −2.40 | 0.05 | 39 | Tceb1 | −2.05 | 0.02 |
| 14 | Cisd2 | −2.35 | 0.05 | 40 | Aplp2 | −2.04 | 0.02 |
| 15 | Ndufs6 | −2.28 | 0.04 | 41 | Gpx4 | −2.03 | 0.02 |
| 16 | Rps25 | −2.24 | 0.05 | 42 | Osgepl1 | −2.03 | 0.04 |
| 17 | Hnrnpa2b1 | −2.21 | 0.04 | 43 | Fxr1 | −2.03 | 0.02 |
| 18 | Cox7c | −2.21 | 0.03 | 44 | Rps14 | −2.02 | 0.05 |
| 19 | Rhoa | −2.21 | 0.04 | 45 | Ppp2r4 | −2.01 | 0.04 |
| 20 | Maf | −2.18 | 0.05 | 46 | Thumpd1 | −2.01 | 0.03 |
| 21 | Ndufb7 | −2.18 | 0.03 | 47 | Zmat2 | −2.01 | 0.04 |
| 22 | Pkia | −2.18 | 0.04 | 48 | Uba3 | −2.01 | 0.04 |
| 23 | Psmd12 | −2.16 | 0.04 | 49 | Thumpd1 | −2.15 | 0.04 |
| 24 | Mtch2 | −2.15 | 0.03 | 50 | Zmat2 | −2.13 | 0.03 |
| 25 | Apobec2 | −2.15 | 0.05 | 51 | Uba3 | −2.11 | 0.05 |
| No. | Genes | log2FC | Adj. P. Val | No. | Genes | log2FC | Adj. P. Val |
|---|---|---|---|---|---|---|---|
| WT20 | KO80 | ||||||
| 1 | Ddx3y | 3.25 | 0.00 | 1 | Gm6472 | 0.01 | 2.69 |
| 2 | Eif2s3y | 3.01 | 0.00 | 2 | Padi1 | 0.01 | 2.14 |
| 3 | Rpl3-ps1 | 2.81 | 0.00 | OE80 | |||
| 4 | Slc15a5 | 2.45 | 0.00 | 1 | Eif2s3y | 0.00 | 3.01 |
| 5 | Uty | 2.30 | 0.00 | 2 | Ddx3y | 0.00 | 2.99 |
| 6 | Vmn1r65 | 2.15 | 0.00 | 3 | Gm12191 | 0.01 | 2.47 |
| KO20 | 4 | Slc15a5 | 0.00 | 2.42 | |||
| 1 | Gm5537 | 2.93 | 0.00 | 5 | Uty | 0.00 | 2.40 |
| 2 | Igha | 2.44 | 0.00 | 6 | Kcnf1 | 0.00 | 2.29 |
| 3 | Ddit4 | 2.14 | 0.00 | 7 | Sult1e1 | 0.00 | 2.17 |
| 4 | Gm26992 | 2.08 | 0.00 | 8 | RP24-258B3.4 | 0.00 | 2.03 |
| 5 | Gm9794 | 2.05 | 0.04 | 9 | C7 | 0.00 | 2.00 |
| 6 | Lcn2 | 2.03 | 0.00 | 10 | 1700001O22Rik | 0.00 | 2.00 |
| OE20 | |||||||
| 1 | Doc2b | 0.00 | 2.22 | ||||
| 2 | Gm21541 | 0.02 | 2.13 | ||||
| No. | Genes | log2FC | Adj. P. Val | No. | Genes | log2FC | Adj. P. Val |
|---|---|---|---|---|---|---|---|
| WT20 | 41 | Dnajc27 | 0.04 | −2.00 | |||
| 1 | Fth1 | 0.05 | −2.43 | 42 | Trappc8 | 0.05 | −2.00 |
| 2 | Ank | 0.05 | −2.40 | KO20 | |||
| 3 | Unc45b | 0.05 | −2.38 | 1 | Tnrc6b | 0.04 | −2.27 |
| 4 | Ehbp1l1 | 0.05 | −2.35 | 2 | Usp24 | 0.04 | −2.26 |
| 5 | Rnf11 | 0.05 | −2.31 | 3 | Prpf4 | 0.05 | −2.19 |
| 6 | Rxra | 0.05 | −2.29 | 4 | Got2 | 0.04 | −2.18 |
| 7 | Kif1b | 0.04 | −2.29 | 5 | Thrap3 | 0.04 | −2.16 |
| 8 | Srl | 0.03 | −2.28 | 6 | Ralgapb | 0.05 | −2.15 |
| 9 | Ankrd40 | 0.04 | −2.25 | 7 | Ppp2ca | 0.05 | −2.15 |
| 10 | Klhl24 | 0.05 | −2.23 | 8 | Bptf | 0.05 | −2.15 |
| 11 | Sbno1 | 0.05 | −2.23 | 9 | Camsap1 | 0.05 | −2.14 |
| 12 | Setd7 | 0.05 | −2.22 | 10 | Osbp | 0.04 | −2.14 |
| 13 | Ap2a1 | 0.05 | −2.21 | 11 | Epb41 | 0.03 | −2.14 |
| 14 | Cds2 | 0.05 | −2.20 | 12 | Zer1 | 0.05 | −2.13 |
| 15 | Tmem64 | 0.03 | −2.18 | 13 | Eif4g3 | 0.04 | −2.10 |
| 16 | Wwp1 | 0.03 | −2.17 | 14 | Asb11 | 0.04 | −2.07 |
| 17 | Otud4 | 0.05 | −2.17 | 15 | Cep104 | 0.04 | −2.06 |
| 18 | Ppp2r2a | 0.05 | −2.16 | 16 | Echs1 | 0.03 | −2.06 |
| 19 | Wdtc1 | 0.05 | −2.16 | 17 | Dnajc13 | 0.04 | −2.06 |
| 20 | Ube2n | 0.05 | −2.15 | 18 | Ptpn1 | 0.05 | −2.05 |
| 21 | Cyhr1 | 0.04 | −2.15 | 19 | Pum1 | 0.04 | −2.05 |
| 22 | Nipbl | 0.05 | −2.13 | 20 | Ap3d1 | 0.05 | −2.05 |
| 23 | Pdpr | 0.02 | −2.11 | 21 | Snx17 | 0.03 | −2.04 |
| 24 | Pdzrn3 | 0.05 | −2.10 | 22 | Eif4g1 | 0.03 | −2.04 |
| 25 | Ulk1 | 0.04 | −2.09 | 23 | Lrch3 | 0.05 | −2.04 |
| 26 | Usp13 | 0.02 | −2.08 | 24 | Ubap2l | 0.05 | −2.04 |
| 27 | Isca1 | 0.02 | −2.06 | 25 | Aldh6a1 | 0.02 | −2.01 |
| 28 | Pja2 | 0.05 | −2.05 | 26 | Tceb3 | 0.04 | −2.01 |
| 29 | Vwa8 | 0.03 | −2.05 | OE20 | |||
| 30 | Tnrc6a | 0.03 | −2.04 | 1 | Tcp1 | 0.05 | −2.36 |
| 31 | Mtus1 | 0.04 | −2.04 | 2 | Arhgef6 | 0.05 | −2.35 |
| 32 | Ergic2 | 0.05 | −2.03 | 3 | Mbnl2 | 0.04 | −2.33 |
| 33 | Tubb4b | 0.03 | −2.02 | 4 | Rplp2 | 0.04 | −2.29 |
| 34 | Tbc1d16 | 0.05 | −2.02 | 5 | Jtb | 0.05 | −2.28 |
| 35 | Timp3 | 0.05 | −2.01 | 6 | Synrg | 0.04 | −2.27 |
| 37 | Fbxo11 | 0.05 | −2.01 | 7 | Apoo | 0.05 | −2.26 |
| 36 | Dhx32 | 0.03 | −2.01 | 8 | Ctsl | 0.05 | −2.25 |
| 37 | Ryk | 0.04 | −2.00 | 9 | Rps15 | 0.04 | −2.25 |
| 38 | Sf3b2 | 0.04 | −2.24 | 10 | Pdk2 | 0.03 | −2.09 |
| 39 | Dlg1 | 0.04 | −2.00 | 11 | Dnaja2 | 0.04 | −2.23 |
| 12 | Nrd1 | 0.04 | −2.22 | 46 | Eprs | 0.04 | −2.08 |
| 13 | Ppp6c | 0.05 | −2.22 | 47 | Phf2 | 0.04 | −2.08 |
| 14 | Rps18 | 0.04 | −2.22 | 48 | Fbxw2 | 0.03 | −2.08 |
| 15 | Tex2 | 0.05 | −2.20 | 49 | Dnajc7 | 0.05 | −2.08 |
| 16 | Luc7l2 | 0.05 | −2.20 | 50 | Ndel1 | 0.03 | −2.08 |
| 17 | Mtdh | 0.04 | −2.19 | 51 | Pnn | 0.04 | −2.07 |
| 18 | Rpl35 | 0.04 | −2.17 | 52 | Csde1 | 0.03 | −2.07 |
| 19 | Atp2b1 | 0.03 | −2.17 | 53 | Ddx1 | 0.03 | −2.07 |
| 20 | Rab18 | 0.05 | −2.17 | 54 | Atp8a1 | 0.04 | −2.07 |
| 21 | Acacb | 0.04 | −2.17 | 55 | Nap1l1 | 0.03 | −2.06 |
| 22 | Zbtb18 | 0.05 | −2.16 | 56 | Rnf150 | 0.04 | −2.06 |
| 23 | Ppp1r2 | 0.03 | −2.16 | 57 | Rpl23 | 0.03 | −2.06 |
| 24 | Usp15 | 0.03 | −2.16 | 58 | Ppp3cb | 0.05 | −2.06 |
| 25 | Serinc1 | 0.03 | −2.16 | 59 | Msrb1 | 0.04 | −2.06 |
| 26 | Agpat3 | 0.04 | −2.16 | 60 | Rps6 | 0.02 | −2.06 |
| 27 | Zfand3 | 0.04 | −2.15 | 61 | Mylk2 | 0.04 | −2.05 |
| 28 | Zc3h14 | 0.05 | −2.15 | 62 | Sgca | 0.05 | −2.05 |
| 29 | Rpl15 | 0.03 | −2.15 | 63 | Rps11 | 0.05 | −2.05 |
| 30 | Atf7 | 0.04 | −2.14 | 64 | Rap1gds1 | 0.03 | −2.04 |
| 31 | Atp6v1g1 | 0.03 | −2.14 | 65 | Scaf1 | 0.05 | −2.04 |
| 32 | Dcaf6 | 0.05 | −2.13 | 66 | Fem1a | 0.04 | −2.04 |
| 33 | Homer1 | 0.04 | −2.12 | 67 | Pfdn5 | 0.02 | −2.03 |
| 34 | Smad4 | 0.05 | −2.12 | 68 | Bmi1 | 0.02 | −2.03 |
| 35 | Fgfr1 | 0.04 | −2.12 | 69 | Ccar1 | 0.04 | −2.03 |
| 36 | Set | 0.03 | −2.11 | 70 | Abhd16a | 0.04 | −2.03 |
| 37 | Nsmaf | 0.05 | −2.11 | 71 | Golga3 | 0.03 | −2.03 |
| 38 | Impad1 | 0.05 | −2.11 | 72 | Arf1 | 0.05 | −2.02 |
| 39 | Sugt1 | 0.04 | −2.11 | 73 | Fyco1 | 0.02 | −2.02 |
| 40 | Scn4a | 0.05 | −2.10 | 74 | Prkcq | 0.02 | −2.02 |
| 41 | Zfand5 | 0.03 | −2.10 | 75 | Wfdc1 | 0.03 | −2.02 |
| 42 | Map1lc3b | 0.04 | −2.10 | 76 | Ubxn4 | 0.03 | −2.00 |
| 43 | Vps13d | 0.05 | −2.10 | 77 | Drap1 | 0.05 | −2.00 |
| 45 | Adipor1 | 0.04 | −2.09 | ||||
| No. | Genes | logFC | Adj.P.Val | No. | Genes | logFC | Adj.P.Val |
|---|---|---|---|---|---|---|---|
| KO80 | 26 | Ltn1 | 0.05 | −2.40 | |||
| 1 | Gm6472 | 2.69 | 0.01 | 27 | Ipo7 | 0.04 | −2.39 |
| 2 | Padi1 | 2.14 | 0.01 | 28 | Btbd1 | 0.02 | −2.39 |
| OE80 | 29 | Snx5 | 0.03 | −2.39 | |||
| 1 | Eif2s3y | 3.01 | 0.00 | 30 | Atf7ip | 0.05 | −2.38 |
| 2 | Ddx3y | 2.99 | 0.00 | 31 | Srp72 | 0.04 | −2.37 |
| 3 | Gm12191 | 2.47 | 0.01 | 32 | Araf | 0.05 | −2.36 |
| 4 | Slc15a5 | 2.42 | 0.00 | 33 | Rnf10 | 0.03 | −2.35 |
| 5 | Uty | 2.40 | 0.00 | 34 | Tmed2 | 0.05 | −2.35 |
| 6 | Kcnf1 | 2.29 | 0.00 | 35 | Jph1 | 0.03 | −2.35 |
| 7 | Sult1e1 | 2.17 | 0.00 | 36 | Lmtk2 | 0.05 | −2.35 |
| 8 | RP24-258B3.4 | 2.03 | 0.00 | 37 | Obscn | 0.04 | −2.35 |
| 9 | C7 | 2.00 | 0.00 | 38 | Ncor1 | 0.03 | −2.34 |
| 10 | 1700001O22Rik | 2.00 | 0.00 | 39 | Dhx15 | 0.04 | −2.34 |
| OE80 | 40 | Mtmr3 | 0.03 | −2.33 | |||
| 1 | Tsnax | 0.05 | −2.64 | 41 | Map2k7 | 0.03 | −2.32 |
| 2 | Fto | 0.05 | −2.62 | 42 | Eif4g1 | 0.03 | −2.32 |
| 3 | Matr3 | 0.05 | −2.60 | 43 | Eif2b5 | 0.05 | −2.32 |
| 4 | Ywhag | 0.05 | −2.59 | 44 | Actr1a | 0.05 | −2.31 |
| 5 | Hdlbp | 0.04 | −2.58 | 45 | Adipor2 | 0.03 | −2.30 |
| 6 | Setd3 | 0.05 | −2.56 | 46 | Tmbim6 | 0.02 | −2.30 |
| 7 | Map3k7 | 0.05 | −2.53 | 47 | Insr | 0.03 | −2.29 |
| 8 | Tmem38a | 0.05 | −2.51 | 48 | Ufl1 | 0.04 | −2.29 |
| 9 | Myh4 | 0.03 | −2.51 | 49 | Dctn2 | 0.02 | −2.29 |
| 10 | Polr1d | 0.05 | −2.51 | 50 | Rad23b | 0.03 | −2.28 |
| 11 | Rsl1d1 | 0.05 | −2.50 | 51 | Cnot10 | 0.05 | −2.28 |
| 12 | Trpc4ap | 0.05 | −2.50 | 52 | Cnot1 | 0.04 | −2.28 |
| 13 | Clint1 | 0.05 | −2.50 | 53 | Isca1 | 0.04 | −2.28 |
| 14 | Spag7 | 0.04 | −2.50 | 54 | Sumo1 | 0.02 | −2.28 |
| 15 | Maf1 | 0.05 | −2.49 | 55 | Arglu1 | 0.05 | −2.28 |
| 16 | Nudt3 | 0.04 | −2.47 | 56 | Dusp27 | 0.02 | −2.27 |
| 17 | Ddx6 | 0.04 | −2.46 | 57 | Eif2a | 0.03 | −2.26 |
| 18 | Dlg1 | 0.02 | −2.46 | 58 | Sox6 | 0.05 | −2.26 |
| 19 | Rfx7 | 0.04 | −2.45 | 59 | Cnbp | 0.05 | −2.26 |
| 20 | Chd4 | 0.02 | −2.43 | 60 | Tom1 | 0.03 | −2.26 |
| 21 | Rfk | 0.02 | −2.42 | 61 | Atg4a | 0.04 | −2.26 |
| 22 | Wdtc1 | 0.05 | −2.41 | 62 | Usp4 | 0.03 | −2.25 |
| 23 | Ar | 0.05 | −2.41 | 63 | Ei24 | 0.02 | −2.25 |
| 24 | Stau1 | 0.03 | −2.41 | 64 | Ccdc43 | 0.03 | −2.25 |
| 25 | Akt2 | 0.03 | −2.41 | 62 | Eif3l | 0.05 | −2.25 |
| 66 | Pitpnb | 0.03 | −2.24 | 111 | Klf9 | 0.01 | −2.11 |
| 67 | Paip2b | 0.03 | −2.24 | 112 | Eif3j2 | 0.03 | −2.11 |
| 68 | Gapvd1 | 0.04 | −2.24 | 113 | Cds2 | 0.02 | −2.11 |
| 69 | Synrg | 0.01 | −2.23 | 114 | Trip12 | 0.01 | −2.11 |
| 70 | Rab3gap2 | 0.05 | −2.23 | 115 | Eif1 | 0.05 | −2.11 |
| 71 | Usp7 | 0.03 | −2.22 | 116 | Cap2 | 0.04 | −2.11 |
| 72 | Eif4a1 | 0.04 | −2.21 | 117 | Chmp4b | 0.02 | −2.11 |
| 73 | Mfap1b | 0.02 | −2.21 | 118 | Srsf10 | 0.04 | −2.10 |
| 74 | Pcyt1a | 0.01 | −2.20 | 119 | Atp6v1c1 | 0.02 | −2.10 |
| 75 | Adipor1 | 0.03 | −2.20 | 120 | Atmin | 0.03 | −2.10 |
| 76 | Tcap | 0.02 | −2.20 | 121 | Phkb | 0.02 | −2.10 |
| 77 | Pacsin3 | 0.05 | −2.19 | 122 | Arnt | 0.04 | −2.09 |
| 78 | Kat7 | 0.03 | −2.18 | 123 | Mbtps1 | 0.02 | −2.09 |
| 79 | Ddx5 | 0.04 | −2.18 | 124 | Dnajb5 | 0.01 | −2.09 |
| 80 | Chmp1a | 0.03 | −2.18 | 125 | Kpna3 | 0.01 | −2.09 |
| 81 | Nphp1 | 0.03 | −2.18 | 126 | Ttc9c | 0.05 | −2.09 |
| 82 | Txnl1 | 0.04 | −2.17 | 127 | Sympk | 0.02 | −2.09 |
| 83 | Pdxdc1 | 0.03 | −2.17 | 128 | Txndc16 | 0.05 | −2.09 |
| 84 | Axin1 | 0.05 | −2.17 | 129 | Nup98 | 0.05 | −2.09 |
| 85 | Usp15 | 0.03 | −2.17 | 130 | Dhx29 | 0.01 | −2.09 |
| 86 | Wac | 0.05 | −2.16 | 131 | Ccni | 0.01 | −2.09 |
| 87 | Marf1 | 0.05 | −2.16 | 132 | Ttc3 | 0.02 | −2.09 |
| 88 | Tmem214 | 0.05 | −2.16 | 133 | Psmd11 | 0.03 | −2.08 |
| 89 | Bcap29 | 0.03 | −2.16 | 134 | Limd1 | 0.01 | −2.08 |
| 90 | E430025E21Rik | 0.04 | −2.16 | 135 | Plaa | 0.03 | −2.08 |
| 91 | Det1 | 0.05 | −2.16 | 136 | Mybpc2 | 0.02 | −2.08 |
| 92 | Oxa1l | 0.03 | −2.15 | 137 | Dedd | 0.05 | −2.08 |
| 93 | Mrps30 | 0.05 | −2.15 | 138 | Zbtb18 | 0.03 | −2.08 |
| 94 | Eif2b2 | 0.05 | −2.15 | 139 | Ubl7 | 0.05 | −2.07 |
| 95 | Prpf4b | 0.03 | −2.15 | 140 | Hsp90ab1 | 0.03 | −2.07 |
| 96 | Fbrs | 0.05 | −2.14 | 141 | Sra1 | 0.04 | −2.07 |
| 97 | Son | 0.01 | −2.14 | 142 | Rab3gap1 | 0.04 | −2.07 |
| 98 | Psmb1 | 0.02 | −2.14 | 143 | Sgta | 0.02 | −2.07 |
| 99 | 2310036O22Rik | 0.04 | −2.14 | 144 | Smad4 | 0.05 | −2.07 |
| 100 | Mtor | 0.03 | −2.14 | 145 | Ift172 | 0.02 | −2.06 |
| 101 | Tmem30a | 0.02 | −2.14 | 146 | Csde1 | 0.04 | −2.06 |
| 102 | 0610009O20Rik | 0.03 | −2.13 | 147 | Fam63a | 0.03 | −2.06 |
| 103 | Polr2a | 0.01 | −2.13 | 148 | Arhgap35 | 0.04 | −2.06 |
| 104 | Psmc3 | 0.02 | −2.13 | 149 | Phrf1 | 0.05 | −2.06 |
| 105 | Zc3h4 | 0.05 | −2.13 | 150 | Kif1bp | 0.02 | −2.06 |
| 106 | Mgat4b | 0.05 | −2.13 | 151 | Abcf1 | 0.02 | −2.06 |
| 107 | Cab39 | 0.05 | −2.13 | 152 | Tm9sf2 | 0.03 | −2.06 |
| 108 | Ppfia1 | 0.04 | −2.12 | 153 | Cep120 | 0.04 | −2.05 |
| 109 | Camta2 | 0.01 | −2.12 | 154 | Clptm1 | 0.03 | −2.05 |
| 110 | Syap1 | 0.03 | −2.12 | 155 | Psmd3 | 0.02 | −2.05 |
| 156 | Thrap3 | 0.01 | −2.05 | 174 | 1110037F02Rik | 0.02 | −2.02 |
| 157 | Cnot4 | 0.04 | −2.05 | 175 | Ubqln2 | 0.05 | −2.02 |
| 158 | Sh3pxd2a | 0.02 | −2.04 | 176 | Phf14 | 0.02 | −2.02 |
| 159 | Stx12 | 0.01 | −2.04 | 177 | Akap9 | 0.02 | −2.02 |
| 160 | Rac1 | 0.01 | −2.04 | 178 | Per2 | 0.04 | −2.02 |
| 161 | Npepl1 | 0.03 | −2.04 | 179 | Rbm5 | 0.03 | −2.02 |
| 162 | Tsc1 | 0.04 | −2.04 | 180 | Ewsr1 | 0.01 | −2.01 |
| 163 | Dnajc2 | 0.04 | −2.04 | 181 | Gcn1l1 | 0.01 | −2.01 |
| 164 | Gpx4 | 0.03 | −2.04 | 182 | Fryl | 0.02 | −2.01 |
| 165 | Crot | 0.05 | −2.03 | 183 | Eea1 | 0.04 | −2.01 |
| 166 | Msrb1 | 0.05 | −2.03 | 184 | Las1l | 0.02 | −2.01 |
| 167 | Ikzf5 | 0.02 | −2.03 | 185 | Dnajb11 | 0.01 | −2.01 |
| 168 | Ulk1 | 0.03 | −2.03 | 186 | Tpd52l1 | 0.04 | −2.00 |
| 169 | Glg1 | 0.01 | −2.03 | 187 | Psmc2 | 0.03 | −2.00 |
| 170 | Crbn | 0.01 | −2.03 | 188 | Gna11 | 0.02 | −2.00 |
| 171 | Ubr1 | 0.04 | −2.03 | 189 | Osgep | 0.05 | −2.00 |
| 172 | Ubxn6 | 0.02 | −2.02 | 190 | Tmem43 | 0.02 | −2.00 |
| 173 | Sdf4 | 0.02 | −2.02 | ||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Myers, M.J.; Shaik, F.; Shaik, F.; Alway, S.E.; Mohamed, J.S. Skeletal Muscle Gene Expression Profile in Response to Caloric Restriction and Aging: A Role for SirT1. Genes 2021, 12, 691. https://doi.org/10.3390/genes12050691
Myers MJ, Shaik F, Shaik F, Alway SE, Mohamed JS. Skeletal Muscle Gene Expression Profile in Response to Caloric Restriction and Aging: A Role for SirT1. Genes. 2021; 12(5):691. https://doi.org/10.3390/genes12050691
Chicago/Turabian StyleMyers, Matthew J., Fathima Shaik, Fahema Shaik, Stephen E. Alway, and Junaith S. Mohamed. 2021. "Skeletal Muscle Gene Expression Profile in Response to Caloric Restriction and Aging: A Role for SirT1" Genes 12, no. 5: 691. https://doi.org/10.3390/genes12050691
APA StyleMyers, M. J., Shaik, F., Shaik, F., Alway, S. E., & Mohamed, J. S. (2021). Skeletal Muscle Gene Expression Profile in Response to Caloric Restriction and Aging: A Role for SirT1. Genes, 12(5), 691. https://doi.org/10.3390/genes12050691







