Genome-Wide Analysis of Sex Disparities in the Genetic Architecture of Lung and Colorectal Cancers
Abstract
1. Introduction
2. Materials and Methods
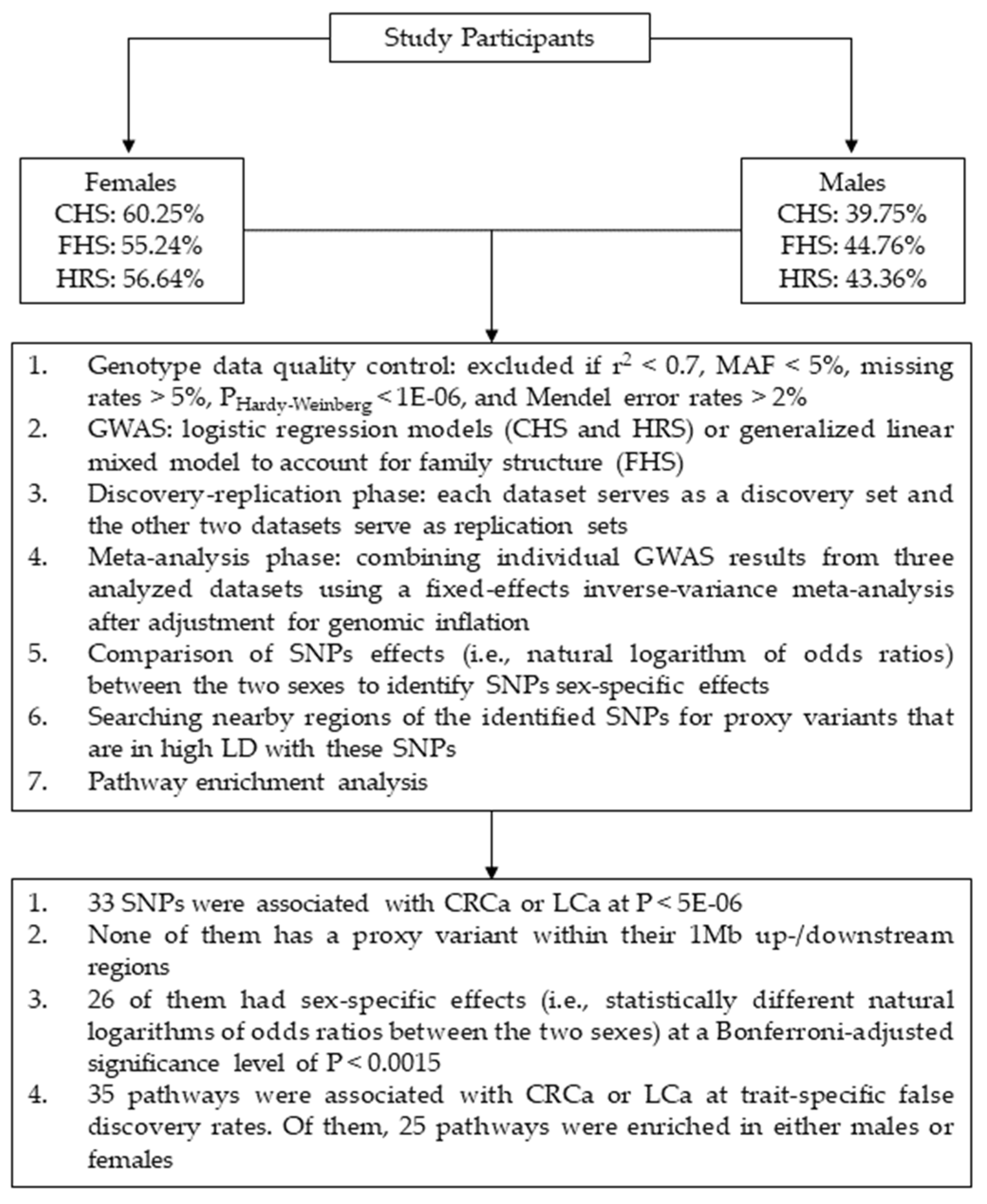
2.1. Study Participants
2.2. Genotype Data and Quality Control (QC)
2.3. GWAS
2.3.1. Genetic Models
2.3.2. Discovery and Replication Analyses
2.3.3. Novel Associations
2.3.4. Sex-specific Associations
2.4. Pathway Enrichment Analysis
3. Results
3.1. Fixed-Effects Covariates
3.2. GWAS
3.3. Pathway Enrichment Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cook, M.B.; Dawsey, S.M.; Freedman, N.D.; Inskip, P.D.; Wichner, S.M.; Quraishi, S.M.; Devesa, S.S.; McGlynn, K.A. Sex Disparities in Cancer Incidence by Period and Age. Cancer Epidemiol. Biomark. Prev. 2009, 18, 1174–1182. [Google Scholar] [CrossRef]
- Cook, M.B.; McGlynn, K.A.; Devesa, S.S.; Freedman, N.D.; Anderson, W.F. Sex Disparities in Cancer Mortality and Survival. Cancer Epidemiol. Biomark. Prev. 2011, 20, 1629–1637. [Google Scholar] [CrossRef] [PubMed]
- Edgren, G.; Liang, L.; Adami, H.-O.; Chang, E.T. Enigmatic Sex Disparities in Cancer Incidence. Eur. J. Epidemiol. 2012, 27, 187–196. [Google Scholar] [CrossRef] [PubMed]
- Li, C.H.; Haider, S.; Shiah, Y.-J.; Thai, K.; Boutros, P.C. Sex Differences in Cancer Driver Genes and Biomarkers. Cancer Res. 2018, 78, 5527–5537. [Google Scholar] [CrossRef] [PubMed]
- Rubin, J.B.; Lagas, J.S.; Broestl, L.; Sponagel, J.; Rockwell, N.; Rhee, G.; Rosen, S.F.; Chen, S.; Klein, R.S.; Imoukhuede, P.; et al. Sex Differences in Cancer Mechanisms. Biol. Sex Differ. 2020, 11, 17. [Google Scholar] [CrossRef]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA A Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef]
- Ferguson, M.K.; Skosey, C.; Hoffman, P.C.; Golomb, H.M. Sex-Associated Differences in Presentation and Survival in Patients with Lung Cancer. J. Clin. Oncol 1990, 8, 1402–1407. [Google Scholar] [CrossRef]
- Singh, S.; Parulekar, W.; Murray, N.; Feld, R.; Evans, W.K.; Tu, D.; Shepherd, F.A. Influence of Sex on Toxicity and Treatment Outcome in Small-Cell Lung Cancer. J. Clin. Oncol. 2005, 23, 850–856. [Google Scholar] [CrossRef]
- Yang, Y.; Wang, G.; He, J.; Ren, S.; Wu, F.; Zhang, J.; Wang, F. Gender Differences in Colorectal Cancer Survival: A Meta-Analysis. Int. J. Cancer 2017, 141, 1942–1949. [Google Scholar] [CrossRef]
- Pal, S.K.; Hurria, A. Impact of Age, Sex, and Comorbidity on Cancer Therapy and Disease Progression. J. Clin. Oncol. 2010, 28, 4086–4093. [Google Scholar] [CrossRef] [PubMed]
- Kiyohara, C.; Ohno, Y. Sex Differences in Lung Cancer Susceptibility: A Review. Gend. Med. 2010, 7, 381–401. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.-E.; Paik, H.Y.; Yoon, H.; Lee, J.E.; Kim, N.; Sung, M.-K. Sex- and Gender-Specific Disparities in Colorectal Cancer Risk. World J. Gastroenterol. 2015, 21, 5167–5175. [Google Scholar] [CrossRef]
- Benedix, F.; Kube, R.; Meyer, F.; Schmidt, U.; Gastinger, I.; Lippert, H. Colon/Rectum Carcinomas (Primary Tumor) Study Group Comparison of 17,641 Patients with Right- and Left-Sided Colon Cancer: Differences in Epidemiology, Perioperative Course, Histology, and Survival. Dis. Colon Rectum 2010, 53, 57–64. [Google Scholar] [CrossRef]
- Hansen, I.O.; Jess, P. Possible Better Long-Term Survival in Left versus Right-Sided Colon Cancer—A Systematic Review. Dan Med. J. 2012, 59, A4444. [Google Scholar]
- Cheng, T.-Y.D.; Cramb, S.M.; Baade, P.D.; Youlden, D.R.; Nwogu, C.; Reid, M.E. The International Epidemiology of Lung Cancer: Latest Trends, Disparities, and Tumor Characteristics. J. Thorac. Oncol. 2016, 11, 1653–1671. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, I.B.; Raine, T.; Wiles, K.; Goodhart, A.; Hall, C.; O’Neill, H. Oxford Handbook of Clinical Medicine, 10th ed.; Sinharay, R., Furmedge, D., Eds.; Oxford University Press: Oxford UK, 2019; ISBN 978-0-19-883490-8. [Google Scholar]
- Farach-Carson, M.C.; Lin, S.-H.; Nalty, T.; Satcher, R.L. Sex Differences and Bone Metastases of Breast, Lung, and Prostate Cancers: Do Bone Homing Cancers Favor Feminized Bone Marrow? Front. Oncol. 2017, 7, 163. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.; Warrington, N.M.; Rubin, J.B. Why Does Jack, and Not Jill, Break His Crown? Sex Disparity in Brain Tumors. Biol. Sex Differ. 2012, 3, 3. [Google Scholar] [CrossRef]
- Koo, J.H.; Jalaludin, B.; Wong, S.K.C.; Kneebone, A.; Connor, S.J.; Leong, R.W.L. Improved Survival in Young Women with Colorectal Cancer. Off. J. Am. Coll. Gastroenterol. 2008, 103, 1488–1495. [Google Scholar] [CrossRef]
- Sloan, J.A.; Goldberg, R.M.; Sargent, D.J.; Vargas-Chanes, D.; Nair, S.; Cha, S.S.; Novotny, P.J.; Poon, M.A.; O’Connell, M.J.; Loprinzi, C.L. Women Experience Greater Toxicity with Fluorouracil-Based Chemotherapy for Colorectal Cancer. J. Clin. Oncol. 2002, 20, 1491–1498. [Google Scholar] [CrossRef]
- Ober, C.; Loisel, D.A.; Gilad, Y. Sex-Specific Genetic Architecture of Human Disease. Nat. Rev. Genet. 2008, 9, 911. [Google Scholar] [CrossRef]
- Dorak, M.T.; Karpuzoglu, E. Gender Differences in Cancer Susceptibility: An Inadequately Addressed Issue. Front. Genet. 2012, 3, 268. [Google Scholar] [CrossRef] [PubMed]
- Galvan, A.; Ioannidis, J.P.A.; Dragani, T.A. Beyond Genome-Wide Association Studies: Genetic Heterogeneity and Individual Predisposition to Cancer. Trends Genet. 2010, 26, 132–141. [Google Scholar] [CrossRef] [PubMed]
- Vogelstein, B.; Papadopoulos, N.; Velculescu, V.E.; Zhou, S.; Diaz, L.A.; Kinzler, K.W. Cancer Genome Landscapes. Science 2013, 339, 1546–1558. [Google Scholar] [CrossRef] [PubMed]
- Ulivi, P.; Scarpi, E.; Chiadini, E.; Marisi, G.; Valgiusti, M.; Capelli, L.; Casadei Gardini, A.; Monti, M.; Ruscelli, S.; Frassineti, G.L.; et al. Right- vs. Left-Sided Metastatic Colorectal Cancer: Differences in Tumor Biology and Bevacizumab Efficacy. Int. J. Mol. Sci. 2017, 18, 1240. [Google Scholar] [CrossRef] [PubMed]
- Gaj, P.; Maryan, N.; Hennig, E.E.; Ledwon, J.K.; Paziewska, A.; Majewska, A.; Karczmarski, J.; Nesteruk, M.; Wolski, J.; Antoniewicz, A.A.; et al. Pooled Sample-Based GWAS: A Cost-Effective Alternative for Identifying Colorectal and Prostate Cancer Risk Variants in the Polish Population. PLoS ONE 2012, 7, e35307. [Google Scholar] [CrossRef]
- Timofeeva, M.N.; Hung, R.J.; Rafnar, T.; Christiani, D.C.; Field, J.K.; Bickeböller, H.; Risch, A.; McKay, J.D.; Wang, Y.; Dai, J.; et al. Influence of Common Genetic Variation on Lung Cancer Risk: Meta-Analysis of 14 900 Cases and 29 485 Controls. Hum. Mol. Genet. 2012, 21, 4980–4995. [Google Scholar] [CrossRef] [PubMed]
- Shepherd, F.A.; Rodrigues Pereira, J.; Ciuleanu, T.; Tan, E.H.; Hirsh, V.; Thongprasert, S.; Campos, D.; Maoleekoonpiroj, S.; Smylie, M.; Martins, R.; et al. Erlotinib in Previously Treated Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2005, 353, 123–132. [Google Scholar] [CrossRef] [PubMed]
- Holm, B.; Mellemgaard, A.; Skov, T.; Skov, B.G. Different Impact of Excision Repair Cross-Complementation Group 1 on Survival in Male and Female Patients with Inoperable Non-Small-Cell Lung Cancer Treated with Carboplatin and Gemcitabine. J. Clin. Oncol. 2009, 27, 4254–4259. [Google Scholar] [CrossRef]
- Warren, R.S.; Atreya, C.E.; Niedzwiecki, D.; Weinberg, V.K.; Donner, D.B.; Mayer, R.J.; Goldberg, R.M.; Compton, C.C.; Zuraek, M.B.; Ye, C.; et al. Association of TP53 Mutational Status and Gender with Survival after Adjuvant Treatment for Stage III Colon Cancer: Results of CALGB 89803. Clin. Cancer Res. 2013, 19, 5777–5787. [Google Scholar] [CrossRef]
- Ning, Y.; Gerger, A.; Zhang, W.; Hanna, D.L.; Yang, D.; Winder, T.; Wakatsuki, T.; Labonte, M.J.; Stintzing, S.; Volz, N.; et al. Plastin Polymorphisms Predict Gender- and Stage-Specific Colon Cancer Recurrence after Adjuvant Chemotherapy. Mol. Cancer Ther. 2014, 13, 528–539. [Google Scholar] [CrossRef]
- Schwab, M.; Zanger, U.M.; Marx, C.; Schaeffeler, E.; Klein, K.; Dippon, J.; Kerb, R.; Blievernicht, J.; Fischer, J.; Hofmann, U.; et al. Role of Genetic and Nongenetic Factors for Fluorouracil Treatment-Related Severe Toxicity: A Prospective Clinical Trial by the German 5-FU Toxicity Study Group. J. Clin. Oncol. 2008, 26, 2131–2138. [Google Scholar] [CrossRef] [PubMed]
- Clayton, J.A.; Collins, F.S. Policy: NIH to Balance Sex in Cell and Animal Studies. Nat. News 2014, 509, 282–283. [Google Scholar] [CrossRef] [PubMed]
- Leslie, R.; O’Donnell, C.J.; Johnson, A.D. GRASP: Analysis of Genotype-Phenotype Results from 1390 Genome-Wide Association Studies and Corresponding Open Access Database. Bioinformatics 2014, 30, i185–i194. [Google Scholar] [CrossRef] [PubMed]
- MacArthur, J.; Bowler, E.; Cerezo, M.; Gil, L.; Hall, P.; Hastings, E.; Junkins, H.; McMahon, A.; Milano, A.; Morales, J.; et al. The New NHGRI-EBI Catalog of Published Genome-Wide Association Studies (GWAS Catalog). Nucleic Acids Res. 2017, 45, D896–D901. [Google Scholar] [CrossRef] [PubMed]
- Fried, L.P.; Borhani, N.O.; Enright, P.; Furberg, C.D.; Gardin, J.M.; Kronmal, R.A.; Kuller, L.H.; Manolio, T.A.; Mittelmark, M.B.; Newman, A. The Cardiovascular Health Study: Design and Rationale. Ann. Epidemiol. 1991, 1, 263–276. [Google Scholar] [CrossRef]
- Dawber, T.R.; Meadors, G.F.; Moore, F.E. Epidemiological Approaches to Heart Disease: The Framingham Study. Am. J. Public Health Nations Health 1951, 41, 279–286. [Google Scholar] [CrossRef]
- Feinleib, M.; Kannel, W.B.; Garrison, R.J.; McNamara, P.M.; Castelli, W.P. The Framingham Offspring Study: Design and Preliminary Data. Prev. Med. 1975, 4, 518–525. [Google Scholar] [CrossRef]
- Sonnega, A.; Faul, J.D.; Ofstedal, M.B.; Langa, K.M.; Phillips, J.W.; Weir, D.R. Cohort Profile: The Health and Retirement Study (HRS). Int. J. Epidemiol. 2014, 43, 576–585. [Google Scholar] [CrossRef] [PubMed]
- Nazarian, A.; Yashin, A.I.; Kulminski, A.M. Genome-Wide Analysis of Genetic Predisposition to Alzheimer’s Disease and Related Sex Disparities. Alzheimer’s Res. Ther. 2019, 11, 5. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Conomos, M.P.; Miller, M.B.; Thornton, T.A. Robust Inference of Population Structure for Ancestry Prediction and Correction of Stratification in the Presence of Relatedness. Genet. Epidemiol. 2015, 39, 276–293. [Google Scholar] [CrossRef] [PubMed]
- McArdle, P.F.; O’Connell, J.R.; Pollin, T.I.; Baumgarten, M.; Shuldiner, A.R.; Peyser, P.A.; Mitchell, B.D. Accounting for Relatedness in Family Based Genetic Association Studies. Hum. Hered. 2007, 64, 234–242. [Google Scholar] [CrossRef] [PubMed]
- Bates, D.; Mächler, M.; Bolker, B.; Walker, S. Fitting Linear Mixed-Effects Models Using Lme4. J. Stat. Softw. 2015, 67, 1–48. [Google Scholar] [CrossRef]
- Nazarian, A.; Arbeev, K.G.; Kulminski, A.M. The Impact of Disregarding Family Structure on Genome-Wide Association Analysis of Complex Diseases in Cohorts with Simple Pedigrees. J. Appl. Genet. 2020, 61, 75–86. [Google Scholar] [CrossRef]
- Mägi, R.; Morris, A.P. GWAMA: Software for Genome-Wide Association Meta-Analysis. BMC Bioinform. 2010, 11, 288. [Google Scholar] [CrossRef]
- Machiela, M.J.; Chanock, S.J. LDlink: A Web-Based Application for Exploring Population-Specific Haplotype Structure and Linking Correlated Alleles of Possible Functional Variants. Bioinformatics 2015, 31, 3555–3557. [Google Scholar] [CrossRef]
- Allison, P.D. Comparing Logit and Probit Coefficients across Groups. Soc. Methods Res. 1999, 28, 186–208. [Google Scholar] [CrossRef]
- Yoon, S.; Nguyen, H.C.T.; Yoo, Y.J.; Kim, J.; Baik, B.; Kim, S.; Kim, J.; Kim, S.; Nam, D. Efficient Pathway Enrichment and Network Analysis of GWAS Summary Data Using GSA-SNP2. Nucleic Acids Res. 2018, 46, e60. [Google Scholar] [CrossRef]
- Yang, J.; Lee, S.H.; Goddard, M.E.; Visscher, P.M. GCTA: A Tool for Genome-Wide Complex Trait Analysis. Am. J. Hum. Genet. 2011, 88, 76–82. [Google Scholar] [CrossRef]
- Bakshi, A.; Zhu, Z.; Vinkhuyzen, A.A.E.; Hill, W.D.; McRae, A.F.; Visscher, P.M.; Yang, J. Fast Set-Based Association Analysis Using Summary Data from GWAS Identifies Novel Gene Loci for Human Complex Traits. Sci. Rep. 2016, 6, 32894. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene Set Enrichment Analysis: A Knowledge-Based Approach for Interpreting Genome-Wide Expression Profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Nishimura, D. BioCarta. Biotech Softw. Internet Rep. 2001, 2, 117–120. [Google Scholar] [CrossRef]
- Schaefer, C.F.; Anthony, K.; Krupa, S.; Buchoff, J.; Day, M.; Hannay, T.; Buetow, K.H. PID: The Pathway Interaction Database. Nucleic Acids Res. 2009, 37, D674–D679. [Google Scholar] [CrossRef]
- Fabregat, A.; Jupe, S.; Matthews, L.; Sidiropoulos, K.; Gillespie, M.; Garapati, P.; Haw, R.; Jassal, B.; Korninger, F.; May, B.; et al. The Reactome Pathway Knowledgebase. Nucleic Acids Res. 2018, 46, D649–D655. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B (Methodol. ) 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Wellcome Trust Case Control Consortium Genome-Wide Association Study of 14,000 Cases of Seven Common Diseases and 3000 Shared Controls. Nature 2007, 447, 661–678. [CrossRef]
- Luo, W.; Schork, N.J.; Marschke, K.B.; Ng, S.-C.; Hermann, T.W.; Zhang, J.; Sanders, J.M.; Tooker, P.; Malo, N.; Zapala, M.A.; et al. Identification of Polymorphisms Associated with Hypertriglyceridemia and Prolonged Survival Induced by Bexarotene in Treating Non-Small Cell Lung Cancer. Anticancer Res. 2011, 31, 2303–2311. [Google Scholar]
- Uhlen, M.; Zhang, C.; Lee, S.; Sjöstedt, E.; Fagerberg, L.; Bidkhori, G.; Benfeitas, R.; Arif, M.; Liu, Z.; Edfors, F.; et al. A Pathology Atlas of the Human Cancer Transcriptome. Science 2017, 357, 2507. [Google Scholar] [CrossRef]
- Galichon, P.; Mesnard, L.; Hertig, A.; Stengel, B.; Rondeau, E. Unrecognized Sequence Homologies May Confound Genome-Wide Association Studies. Nucleic Acids Res. 2012, 40, 4774–4782. [Google Scholar] [CrossRef]
- Boraska, V.; Jerončić, A.; Colonna, V.; Southam, L.; Nyholt, D.R.; Rayner, N.W.; Perry, J.R.B.; Toniolo, D.; Albrecht, E.; Ang, W.; et al. Genome-Wide Meta-Analysis of Common Variant Differences between Men and Women. Hum. Mol. Genet. 2012, 21, 4805–4815. [Google Scholar] [CrossRef]
- Colussi, D.; Brandi, G.; Bazzoli, F.; Ricciardiello, L. Molecular Pathways Involved in Colorectal Cancer: Implications for Disease Behavior and Prevention. Int. J. Mol. Sci. 2013, 14, 16365–16385. [Google Scholar] [CrossRef]
- Markman, J.L.; Shiao, S.L. Impact of the Immune System and Immunotherapy in Colorectal Cancer. J. Gastrointest. Oncol. 2015, 6, 208–223. [Google Scholar] [CrossRef]
- Eymin, B.; Gazzeri, S. Role of Cell Cycle Regulators in Lung Carcinogenesis. Cell Adh. Migr. 2010, 4, 114–123. [Google Scholar] [CrossRef]
- Jafri, M.A.; Ansari, S.A.; Alqahtani, M.H.; Shay, J.W. Roles of Telomeres and Telomerase in Cancer, and Advances in Telomerase-Targeted Therapies. Genome Med. 2016, 8, 69. [Google Scholar] [CrossRef]
- Zhang, W.; Mao, J.-H.; Zhu, W.; Jain, A.K.; Liu, K.; Brown, J.B.; Karpen, G.H. Centromere and Kinetochore Gene Misexpression Predicts Cancer Patient Survival and Response to Radiotherapy and Chemotherapy. Nat. Commun. 2016, 7, 12619. [Google Scholar] [CrossRef]
| Chr | Gene | SNP | Pos | A1 | PHRS | PFHS | PCHS | Effects | Freq | OR (se) | PMETA | PQ | i2 | N | 1Mb? | Proxy? | Region? | Gene? | Prognostic? |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CRCa-F | |||||||||||||||||||
| 8q22.3 | SNX31 | rs1078186 | 100613251 | C | 4.19 × 10−4 | 9.48 × 10−4 | 2.21 × 10−1 | --- | 0.574 | 0.615 (0.056) | 1.60 × 10−6 | 4.49 × 10−1 | 0 | 8590 | N | N | S | N | O |
| 8q24.23 | KHDRBS3 | rs118174020 | 135843271 | C | NA | 3.92 × 10−4 | 8.73 × 10−4 | ?-- | 0.949 | 0.357 (0.062) | 1.41 × 10−6 | 8.49 × 10−1 | 0 | 4752 | N | N | N | O | O |
| 10q26.3 | GLRX3 | rs3118492 | 130435775 | A | 2.74 × 10−3 | 8.05 × 10−3 | 1.69 × 10−2 | +++ | 0.229 | 1.672 (0.168) | 4.15 × 10−6 | 9.71 × 10−1 | 0 | 8544 | N | N | N | N | O |
| 10q26.3 | GLRX3 | rs372813 | 130442933 | A | 2.89 × 10−3 | 6.08 × 10−3 | 1.68 × 10−2 | +++ | 0.233 | 1.661 (0.163) | 3.25 × 10−6 | 9.75 × 10−1 | 0 | 8592 | N | N | N | N | O |
| 10q26.3 | GLRX3 | rs359063 | 130452326 | T | 4.64 × 10−3 | 3.89 × 10−3 | 1.39 × 10−2 | +++ | 0.230 | 1.669 (0.164) | 2.86 × 10−6 | 9.14 × 10−1 | 0 | 8545 | N | N | N | N | O |
| 10q26.3 | GLRX3 | rs307385 | 130452402 | T | 3.61 × 10−3 | 3.50 × 10−3 | 1.39 × 10−2 | +++ | 0.230 | 1.684 (0.167) | 2.16 × 10−6 | 9.23 × 10−1 | 0 | 8543 | N | N | N | N | O |
| LCa-F | |||||||||||||||||||
| 2p21 | LOC102723824 | rs7593032 * | 42122050 | T | 4.63 × 10−6 | 3.32 × 10−1 | 3.68 × 10−2 | --- | 0.713 | 0.631 (0.057) | 3.66 × 10−6 | 9.14 × 10−2 | 0.582 | 8577 | N | N | S | N | N |
| 5q13.2 | LINC02056 | rs42775 | 72584687 | T | 2.43 × 10−4 | 1.97 × 10−2 | 8.54 × 10−2 | +++ | 0.325 | 1.561 (0.138) | 4.98 × 10−6 | 6.74 × 10−1 | 0 | 8516 | S | N | S | N | N |
| 10q21.1 | PRKG1 | rs11000463 * | 52014359 | T | 4.60 × 10−6 | 3.77 × 10−2 | 7.65 × 10−1 | --+ | 0.903 | 0.558 (0.067) | 1.75 × 10−5 | 3.31 × 10−2 | 0.707 | 8591 | N | N | N | O | N |
| 10q21.1 | PRKG1 | rs11000467 * | 52015312 | G | 4.67 × 10−6 | 2.87 × 10−2 | 7.65 × 10−1 | --+ | 0.905 | 0.548 (0.066) | 1.37 × 10−5 | 3.41 × 10−2 | 0.704 | 8570 | N | N | N | O | N |
| 10q26.13 | ZRANB1 | rs76972397 | 124961380 | T | 1.70 × 10−4 | NA | 1.43 × 10−3 | -?- | 0.930 | 0.432 (0.063) | 9.62 × 10−7 | 6.72 × 10−1 | 0 | 5770 | N | N | G | O | N |
| 18q11.2 | HRH4 | rs482962 | 24532843 | A | 2.41 × 10−2 | 2.04 × 10−3 | 4.52 × 10−3 | +++ | 0.262 | 1.607 (0.151) | 4.90 × 10−6 | 4.75 × 10−1 | 0 | 8585 | N | N | S | N | N |
| CRCa-M | |||||||||||||||||||
| 3p24.2 | THRB | rs57751578 | 24486090 | T | 5.16 × 10−4 | 5.80 × 10−2 | 4.39 × 10−3 | --- | 0.911 | 0.483 (0.064) | 1.89 × 10−6 | 6.98 × 10−1 | 0 | 6491 | N | N | N | O | O |
| 5q31.1 | FSTL4 | rs4631227 | 133152748 | A | 1.81 × 10−4 | 5.77 × 10−2 | 1.49 × 10−2 | +++ | 0.293 | 1.663 (0.163) | 3.20 × 10−6 | 4.76 × 10−1 | 0 | 6481 | N | N | G | O | N |
| 6q14.3 | HTR1E | rs72907251 | 86515650 | C | 7.49 × 10−5 | 1.55 × 10−3 | 4.64 × 10−1 | --- | 0.937 | 0.446 (0.063) | 1.06 × 10−6 | 3.73 × 10−1 | 0 | 6496 | N | N | N | O | N |
| 13q12.11 | MPHOSPH8 | rs9579517 * | 19603453 | C | 2.43 × 10−6 | 8.91 × 10−1 | 2.62 × 10−2 | --- | 0.900 | 0.519 (0.066) | 8.88 × 10−6 | 3.31 × 10−2 | 0.707 | 6490 | N | N | N | N | N |
| 13q12.11 | MPHOSPH8 | rs56357430 * | 19632421 | C | 2.43 × 10−6 | 9.20 × 10−1 | 3.69 × 10−2 | -+- | 0.900 | 0.528 (0.068) | 1.77 × 10−5 | 2.25 × 10−2 | 0.736 | 6498 | N | N | N | N | N |
| LCa-M | |||||||||||||||||||
| 5p15.33 | LINC01377 | rs12657742 | 3123661 | A | 5.98 × 10−3 | 2.66 × 10−3 | 2.27 × 10−2 | --- | 0.864 | 0.555 (0.062) | 3.67 × 10−6 | 9.63 × 10−1 | 0 | 6490 | N | N | G | O | N |
| 5q23.2 | LINC02039 | rs77914729 | 126003571 | C | 6.07 × 10−3 | 1.33 × 10−3 | 2.67 × 10−2 | --- | 0.929 | 0.475 (0.065) | 2.67 × 10−6 | 8.09 × 10−1 | 0 | 6495 | N | N | S | N | N |
| 5q23.2 | LINC02039 | rs75874914 | 126003878 | C | 6.25 × 10−3 | 1.64 × 10−3 | 2.66 × 10−2 | --- | 0.929 | 0.478 (0.065) | 3.24 × 10−6 | 8.29 × 10−1 | 0 | 6496 | N | N | S | N | N |
| 5q23.2 | LINC02039 | rs11954381 | 126006435 | G | 5.32 × 10−3 | 1.73 × 10−3 | 2.66 × 10−2 | --- | 0.929 | 0.477 (0.065) | 2.81 × 10−6 | 8.63 × 10−1 | 0 | 6491 | N | N | S | N | N |
| 6q23.3 | MAP7 | rs9399183 | 136496350 | A | 1.03 × 10−5 | 7.56 × 10−2 | 1.33 × 10−1 | --- | 0.833 | 0.585 (0.060) | 3.33 × 10−6 | 2.19 × 10−1 | 0.341 | 6494 | N | N | N | N | O |
| 6q23.3 | MAP7 | rs3799451 | 136508794 | A | 4.91 × 10−6 | 5.78 × 10−2 | 1.61 × 10−1 | --- | 0.852 | 0.565 (0.060) | 1.62 × 10−6 | 2.01 × 10−1 | 0.377 | 6495 | N | N | N | N | O |
| 6q23.3 | MAP7 | rs3799453 | 136518050 | G | 4.70 × 10−6 | 5.78 × 10−2 | 2.42 × 10−1 | --- | 0.853 | 0.570 (0.061) | 2.46 × 10−6 | 1.72 × 10−1 | 0.432 | 6495 | N | N | N | N | O |
| 6q23.3 | MAP7 | rs3799454 | 136518078 | T | 6.90 × 10−6 | 6.85 × 10−2 | 2.16 × 10−1 | --- | 0.835 | 0.586 (0.060) | 3.58 × 10−6 | 1.76 × 10−1 | 0.425 | 6498 | N | N | N | N | O |
| 6q23.3 | MAP7 | rs3799462 | 136524611 | G | 1.10 × 10−5 | 5.78 × 10−2 | 2.42 × 10−1 | --- | 0.853 | 0.577 (0.062) | 4.49 × 10−6 | 2.12 × 10−1 | 0.356 | 6498 | N | N | N | N | O |
| 8p23.2 | CSMD1 | rs13261356 | 4605119 | C | 1.04 × 10−3 | 8.05 × 10−4 | NA | --? | 0.900 | 0.482 (0.065) | 3.23 × 10−6 | 7.69 × 10−1 | 0 | 5209 | G | N | G | SO | N |
| 10p12.31 | MIR4675 | rs11012129 | 20545511 | T | 2.15 × 10−3 | 1.68 × 10−5 | NA | --? | 0.945 | 0.375 (0.060) | 3.21 × 10−7 | 2.14 × 10−1 | 0.352 | 5207 | N | N | S | O | N |
| 18q21.31 | ATP8B1 | rs2437037 | 57795758 | A | 2.39 × 10−4 | NA | 4.50 × 10−3 | -?- | 0.909 | 0.449 (0.066) | 3.95 × 10−6 | 8.71 × 10−1 | 0 | 4216 | N | N | N | N | O |
| 21q21.3 | GRIK1 | rs363433 | 29588811 | C | 8.08 × 10−4 | 1.03 × 10−2 | 1.20 × 10−2 | --- | 0.863 | 0.539 (0.061) | 1.14 × 10−6 | 9.30 × 10−1 | 0 | 6490 | N | N | N | O | O |
| 21q21.3 | GRIK1 | rs363432 | 29591270 | G | 3.23 × 10−3 | 1.03 × 10−2 | 1.12 × 10−2 | --- | 0.863 | 0.554 (0.063) | 4.10 × 10−6 | 9.03 × 10−1 | 0 | 6495 | N | N | N | O | O |
| 21q21.3 | GRIK1 | rs2832405 | 29591425 | T | 5.58 × 10−4 | 1.24 × 10−2 | 1.98 × 10−2 | --- | 0.854 | 0.548 (0.061) | 1.50 × 10−6 | 9.16 × 10−1 | 0 | 6497 | N | N | N | O | O |
| 21q21.3 | GRIK1 | rs363472 | 29595793 | G | 1.77 × 10−3 | 1.29 × 10−2 | 1.78 × 10−2 | --- | 0.856 | 0.562 (0.062) | 4.23 × 10−6 | 9.26 × 10−1 | 0 | 6490 | N | N | N | O | O |
| Chr | Gene | SNP | Pos | A1 | Males | Females | Comparison | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Freq | OR | se | P-Value | Freq | OR | se | P-Value | Chi-Square | P-Value | |||||
| CRCa-F | ||||||||||||||
| 8q22.3 | SNX31 | rs1078186 | 100613251 | C | 0.568 | 0.989 | 0.097 | 9.19 × 10−1 | 0.574 | 0.615 | 0.056 | 1.60 × 10−6 | 18.033 | 2.17 × 10−5 |
| 8q24.23 | KHDRBS3 | rs118174020 + | 135843271 | C | NA | NA | NA | NA | 0.949 | 0.357 | 0.062 | 1.41 × 10−6 | NA | NA |
| 10q26.3 | GLRX3 | rs3118492 + | 130435775 | A | 0.235 | 1.032 | 0.119 | 8.07 × 10−1 | 0.229 | 1.672 | 0.168 | 4.15 × 10−6 | 5.507 | 1.89 × 10−2 |
| 10q26.3 | GLRX3 | rs372813 + | 130442933 | A | 0.238 | 1.037 | 0.118 | 7.79 × 10−1 | 0.233 | 1.661 | 0.163 | 3.25 × 10−6 | 5.497 | 1.90 × 10−2 |
| 10q26.3 | GLRX3 | rs359063 + | 130452326 | T | 0.235 | 1.061 | 0.121 | 6.43 × 10−1 | 0.230 | 1.669 | 0.164 | 2.86 × 10−6 | 4.935 | 2.63 × 10−2 |
| 10q26.3 | GLRX3 | rs307385 + | 130452402 | T | 0.234 | 1.066 | 0.122 | 6.21 × 10−1 | 0.230 | 1.684 | 0.167 | 2.16 × 10−6 | 4.923 | 2.65 × 10−2 |
| LCa-F | ||||||||||||||
| 2p21 | LOC102723824 | rs7593032 | 42122050 | T | 0.711 | 1.206 | 0.122 | 9.73 × 10−2 | 0.713 | 0.631 | 0.057 | 3.66 × 10−6 | 23.096 | 1.54 × 10−6 |
| 5q13.2 | LOC102477328 | rs42775 + | 72584687 | T | 0.326 | 0.958 | 0.093 | 6.92 × 10−1 | 0.325 | 1.561 | 0.138 | 4.98 × 10−6 | 8.542 | 3.47 × 10−3 |
| 10q21.1 | PRKG1 | rs11000463 | 52014359 | T | 0.903 | 1.266 | 0.199 | 2.09 × 10−1 | 0.903 | 0.558 | 0.067 | 1.75 × 10−5 | 15.245 | 9.44 × 10−5 |
| 10q21.1 | PRKG1 | rs11000467 | 52015312 | G | 0.904 | 1.252 | 0.201 | 2.43 × 10−1 | 0.905 | 0.548 | 0.066 | 1.37 × 10−5 | 15.283 | 9.25 × 10−5 |
| 10q26.13 | ZRANB1 | rs76972397 | 124961380 | T | 0.935 | 0.945 | 0.193 | 8.27 × 10−1 | 0.930 | 0.432 | 0.063 | 9.62 × 10−7 | 14.928 | 1.12 × 10−4 |
| 18q11.2 | HRH4 | rs482962+ | 24532843 | A | 0.252 | 1.132 | 0.116 | 2.76 × 10−1 | 0.262 | 1.607 | 0.151 | 4.90 × 10−6 | 3.390 | 6.56 × 10−2 |
| CRCa-M | ||||||||||||||
| 3p24.2 | THRB | rs57751578 | 24486090 | T | 0.911 | 0.483 | 0.064 | 1.89 × 10−6 | 0.909 | 1.131 | 0.175 | 5.03 × 10−1 | 20.847 | 4.97 × 10−6 |
| 5q31.1 | FSTL4 | rs4631227 | 133152748 | A | 0.293 | 1.663 | 0.163 | 3.20 × 10−6 | 0.293 | 0.892 | 0.090 | 3.09 × 10−1 | 11.149 | 8.41 × 10−4 |
| 6q14.3 | HTR1E | rs72907251 | 86515650 | C | 0.937 | 0.446 | 0.063 | 1.06 × 10−6 | 0.935 | 0.774 | 0.122 | 1.74 × 10−1 | 16.133 | 5.91 × 10−5 |
| 13q12.11 | MPHOSPH8 | rs9579517 | 19603453 | C | 0.900 | 0.519 | 0.066 | 8.88 × 10−6 | 0.901 | 0.919 | 0.13 | 6.11 × 10−1 | 15.390 | 8.74 × 10−5 |
| 13q12.11 | MPHOSPH8 | rs56357430 | 19632421 | C | 0.900 | 0.528 | 0.068 | 1.77 × 10−5 | 0.901 | 0.915 | 0.128 | 5.89 × 10−1 | 14.294 | 1.56 × 10−4 |
| LCa-M | ||||||||||||||
| 5p15.33 | LOC102467074 | rs12657742 | 3123661 | A | 0.864 | 0.555 | 0.062 | 3.67 × 10−6 | 0.868 | 0.975 | 0.118 | 8.56 × 10−1 | 17.809 | 2.44 × 10−5 |
| 5q23.2 | LOC102546228 | rs77914729 | 126003571 | C | 0.929 | 0.475 | 0.065 | 2.67 × 10−6 | 0.932 | 1.047 | 0.173 | 8.16 × 10−1 | 18.396 | 1.79 × 10−5 |
| 5q23.2 | LOC102546228 | rs75874914 | 126003878 | C | 0.929 | 0.478 | 0.065 | 3.24 × 10−6 | 0.932 | 1.095 | 0.182 | 6.51 × 10−1 | 18.307 | 1.88 × 10−5 |
| 5q23.2 | LOC102546228 | rs11954381 | 126006435 | G | 0.929 | 0.477 | 0.065 | 2.81 × 10−6 | 0.932 | 1.133 | 0.191 | 5.41 × 10−1 | 18.408 | 1.78 × 10−5 |
| 6q23.3 | MAP7 | rs9399183 | 136496350 | A | 0.833 | 0.585 | 0.060 | 3.33 × 10−6 | 0.838 | 0.863 | 0.097 | 2.43 × 10−1 | 11.669 | 6.36 × 10−4 |
| 6q23.3 | MAP7 | rs3799451 | 136508794 | A | 0.852 | 0.565 | 0.060 | 1.62 × 10−6 | 0.854 | 0.833 | 0.095 | 1.57 × 10−1 | 11.877 | 5.68 × 10−4 |
| 6q23.3 | MAP7 | rs3799453 | 136518050 | G | 0.853 | 0.570 | 0.061 | 2.46 × 10−6 | 0.854 | 0.832 | 0.095 | 1.54 × 10−1 | 11.287 | 7.81 × 10−4 |
| 6q23.3 | MAP7 | rs3799454 | 136518078 | T | 0.835 | 0.586 | 0.060 | 3.58 × 10−6 | 0.840 | 0.852 | 0.095 | 2.03 × 10−1 | 11.010 | 9.06 × 10−4 |
| 6q23.3 | MAP7 | rs3799462 | 136524611 | G | 0.853 | 0.577 | 0.062 | 4.49 × 10−6 | 0.855 | 0.829 | 0.095 | 1.47 × 10−1 | 10.291 | 1.34 × 10−3 |
| 8p23.2 | CSMD1 | rs13261356 | 4605119 | C | 0.900 | 0.482 | 0.065 | 3.23 × 10−6 | 0.903 | 0.852 | 0.128 | 3.68 × 10−1 | 15.839 | 6.90 × 10−5 |
| 10p12.31 | MIR4675 | rs11012129 | 20545511 | T | 0.945 | 0.375 | 0.060 | 3.21 × 10−7 | 0.945 | 1.465 | 0.288 | 1.23 × 10−1 | 21.542 | 3.46 × 10−6 |
| 18q21.31 | ATP8B1 | rs2437037 | 57795758 | A | 0.909 | 0.449 | 0.066 | 3.95 × 10−6 | 0.910 | 1.075 | 0.185 | 7.30 × 10−1 | 19.804 | 8.58 × 10−6 |
| 21q21.3 | GRIK1 | rs363433 | 29588811 | C | 0.863 | 0.539 | 0.061 | 1.14 × 10−6 | 0.861 | 0.882 | 0.102 | 3.40 × 10−1 | 17.206 | 3.35 × 10−5 |
| 21q21.3 | GRIK1 | rs363432 | 29591270 | G | 0.863 | 0.554 | 0.063 | 4.10 × 10−6 | 0.861 | 0.879 | 0.102 | 3.26 × 10−1 | 14.882 | 1.14 × 10−4 |
| 21q21.3 | GRIK1 | rs2832405 | 29591425 | T | 0.854 | 0.548 | 0.061 | 1.50 × 10−6 | 0.853 | 0.924 | 0.106 | 5.45 × 10−1 | 18.197 | 1.99 × 10−5 |
| 21q21.3 | GRIK1 | rs363472 | 29595793 | G | 0.856 | 0.562 | 0.062 | 4.23 × 10−6 | 0.855 | 0.895 | 0.102 | 3.89 × 10−1 | 15.071 | 1.04 × 10−4 |
| Pathway | Pathway Source | GSEA ID | Size | Males | Females | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Count | z-Score | p-Value | q-Value | Count | z-Score | p-Value | q-Value | ||||
| CRCa | |||||||||||
| Regulation of IFNA signaling | REACTOME | M982 | 24 | 24 | 8.33 | 0 | 0 | 24 | 3.64 | 1.36 × 10−4 | 1.72 × 10−1 |
| Antigen processing and presentation | KEGG | M16004 | 89 | 84 | 7.69 | 6.83 × 10−15 | 4.32 × 10−12 | NS | NS | NS | NS |
| Natural killer cell mediated cytotoxicity | KEGG | M5669 | 137 | 135 | 6.71 | 9.61 × 10−12 | 4.05 × 10−9 | NS | NS | NS | NS |
| Regulation of autophagy | KEGG | M6382 | 35 | 35 | 5.67 | 7.09 × 10−9 | 2.24 × 10−6 | NS | NS | NS | NS |
| TRAF6 mediated IRF7 activation | REACTOME | M936 | 30 | 30 | 5.63 | 9.24 × 10−9 | 2.34 × 10−6 | NS | NS | NS | NS |
| Autoimmune thyroid disease | KEGG | M13103 | 53 | 50 | 5.62 | 9.64 × 10−9 | 2.34 × 10−6 | NS | NS | NS | NS |
| CD8/TCR downstream signaling | PID | M272 | 65 | 65 | 4.97 | 3.27 × 10−7 | 5.91 × 10−5 | NS | NS | NS | NS |
| Interferon alpha/beta signaling | REACTOME | M973 | 64 | 62 | 4.78 | 8.82 × 10−7 | 1.40 × 10−4 | NS | NS | NS | NS |
| Toll-like receptor signaling pathway | KEGG | M3261 | 102 | 100 | 4.54 | 2.76 × 10−6 | 3.88 × 10−4 | 100 | 3.51 | 2.26 × 10−4 | 1.72 × 10−1 |
| RIG-I-like receptor signaling pathway | KEGG | M15913 | 71 | 69 | 4.29 | 8.86 × 10−6 | 1.12 × 10−3 | NS | NS | NS | NS |
| Translocation of ZAP-70 to immunological synapse | REACTOME | M722 | 14 | 13 | 4.15 | 1.66 × 10−5 | 1.91 × 10−3 | NS | NS | NS | NS |
| JAK-STAT signaling pathway | KEGG | M17411 | 155 | 146 | 4.07 | 2.32 × 10−5 | 2.44 × 10−3 | NS | NS | NS | NS |
| Immunoregulatory interactions between a lymphoid and a non-lymphoid cell | REACTOME | M8240 | 70 | 64 | 3.91 | 4.68 × 10−5 | 4.56 × 10−3 | NS | NS | NS | NS |
| Generation of second messenger molecules | REACTOME | M16523 | 27 | 26 | 3.84 | 6.05 × 10−5 | 5.46 × 10−3 | NS | NS | NS | NS |
| Cytosolic DNA-sensing pathway | KEGG | M11844 | 56 | 55 | 3.69 | 1.10 × 10−4 | 9.28 × 10−3 | NS | NS | NS | NS |
| Phosphorylation of CD3 and TCR zeta chains | REACTOME | M12494 | 16 | 15 | 3.68 | 1.18 × 10−4 | 9.33 × 10−3 | NS | NS | NS | NS |
| Interferon signaling | REACTOME | M983 | 159 | 152 | 3.67 | 1.21 × 10−4 | 9.33 × 10−3 | NS | NS | NS | NS |
| P2Y receptors | REACTOME | M10960 | 12 | 12 | 3.59 | 1.66 × 10−4 | 1.17 × 10−2 | NS | NS | NS | NS |
| PD-1 signaling | REACTOME | M18810 | 18 | 17 | 3.41 | 3.19 × 10−4 | 2.13 × 10−2 | NS | NS | NS | NS |
| LCa | |||||||||||
| RNA polymerase I promoter opening | REACTOME | M884 | 62 | 59 | 5.52 | 1.67 × 10−8 | 2.11 × 10−5 | 59 | 5.78 | 3.83 × 10−9 | 4.84 × 10−6 |
| Meiotic recombination | REACTOME | M1011 | 86 | 82 | 4.83 | 6.77 × 10−7 | 4.28 × 10−4 | 82 | 4.19 | 1.38 × 10−5 | 2.49 × 10−3 |
| Amyloid fiber formation | REACTOME | M1076 | 83 | 79 | 4.68 | 1.42 × 10−6 | 4.97 × 10−4 | 79 | 4.63 | 1.80 × 10−6 | 5.69 × 10−4 |
| RNA polymerase I transcription | REACTOME | M728 | 89 | 83 | 4.72 | 1.18 × 10−6 | 4.97 × 10−4 | 83 | 4.25 | 1.08 × 10−5 | 2.28 × 10−3 |
| Interleukin-7 signaling | REACTOME | M542 | 11 | 11 | 4.46 | 4.04 × 10−6 | 1.02 × 10−3 | NS | NS | NS | NS |
| Meiosis | REACTOME | M529 | 116 | 110 | 4.26 | 1.02 × 10−5 | 2.16 × 10−3 | 110 | 3.47 | 2.61 × 10−4 | 3.31 × 10−2 |
| Packaging of telomere ends | REACTOME | M17695 | 48 | 48 | 4.19 | 1.42 × 10−5 | 2.57 × 10−3 | 48 | 5.47 | 2.20 × 10−8 | 1.39 × 10−5 |
| RNA polymerase I, RNA polymerase III, and mitochondrial transcription | REACTOME | M858 | 122 | 116 | 3.99 | 3.36 × 10−5 | 5.31 × 10−3 | NS | NS | NS | NS |
| Meiotic synapsis | REACTOME | M1061 | 73 | 71 | 3.88 | 5.20 × 10−5 | 7.32 × 10−3 | 71 | 4.31 | 8.14 × 10−6 | 2.06 × 10−3 |
| A6B1 and A6B4 integrin signaling | PID | M239 | 46 | 46 | 3.83 | 6.29 × 10−5 | 7.95 × 10−3 | NS | NS | NS | NS |
| Interleukin-7 signaling | BIOCARTA | M1296 | 17 | 17 | 3.62 | 1.49 × 10−4 | 1.71 × 10−2 | NS | NS | NS | NS |
| Interleukin-2 family signaling | REACTOME | M1012 | 41 | 39 | 3.58 | 1.70 × 10−4 | 1.79 × 10−2 | NS | NS | NS | NS |
| Deposition of new CENPA-containing nucleosomes at the centromere | REACTOME | M871 | 64 | 61 | 3.44 | 2.88 × 10−4 | 2.81 × 10−2 | 61 | 3.79 | 7.53 × 10−5 | 1.12 × 10−2 |
| Telomere maintenance | REACTOME | M4052 | 75 | NS | NS | NS | NS | 75 | 5.04 | 2.37 × 10−7 | 1.00 × 10−4 |
| Systemic lupus erythematosus | KEGG | M4741 | 140 | NS | NS | NS | NS | 133 | 3.81 | 7.09 × 10−5 | 1.12 × 10−2 |
| Chromosome maintenance | REACTOME | M868 | 122 | NS | NS | NS | NS | 117 | 3.4 | 3.42 × 10−4 | 3.94 × 10−2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nazarian, A.; Kulminski, A.M. Genome-Wide Analysis of Sex Disparities in the Genetic Architecture of Lung and Colorectal Cancers. Genes 2021, 12, 686. https://doi.org/10.3390/genes12050686
Nazarian A, Kulminski AM. Genome-Wide Analysis of Sex Disparities in the Genetic Architecture of Lung and Colorectal Cancers. Genes. 2021; 12(5):686. https://doi.org/10.3390/genes12050686
Chicago/Turabian StyleNazarian, Alireza, and Alexander M. Kulminski. 2021. "Genome-Wide Analysis of Sex Disparities in the Genetic Architecture of Lung and Colorectal Cancers" Genes 12, no. 5: 686. https://doi.org/10.3390/genes12050686
APA StyleNazarian, A., & Kulminski, A. M. (2021). Genome-Wide Analysis of Sex Disparities in the Genetic Architecture of Lung and Colorectal Cancers. Genes, 12(5), 686. https://doi.org/10.3390/genes12050686






