The Cause of Hereditary Hearing Loss in GJB2 Heterozygotes—A Comprehensive Study of the GJB2/DFNB1 Region
Abstract
1. Introduction
2. Materials and Methods
3. Results
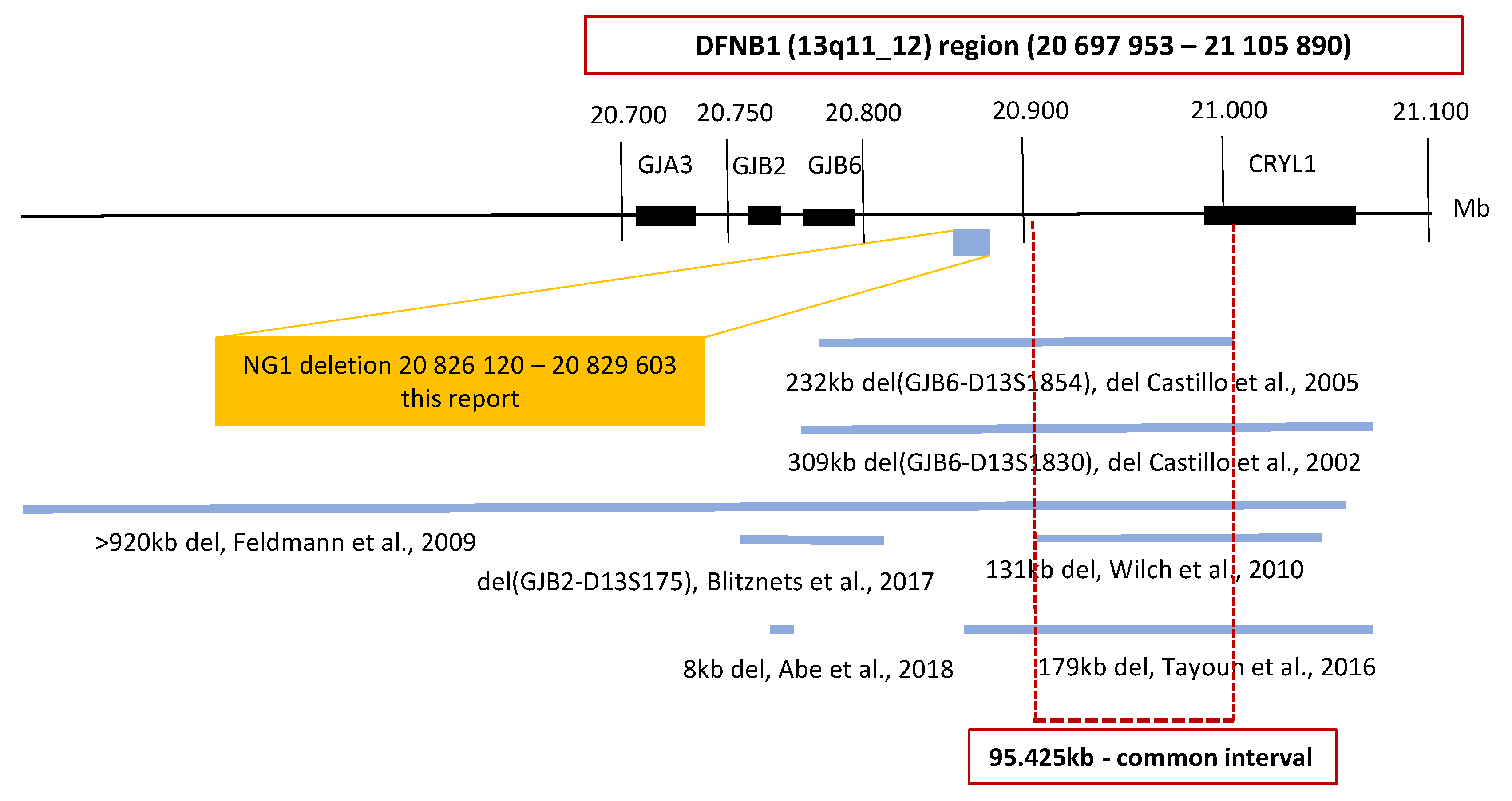
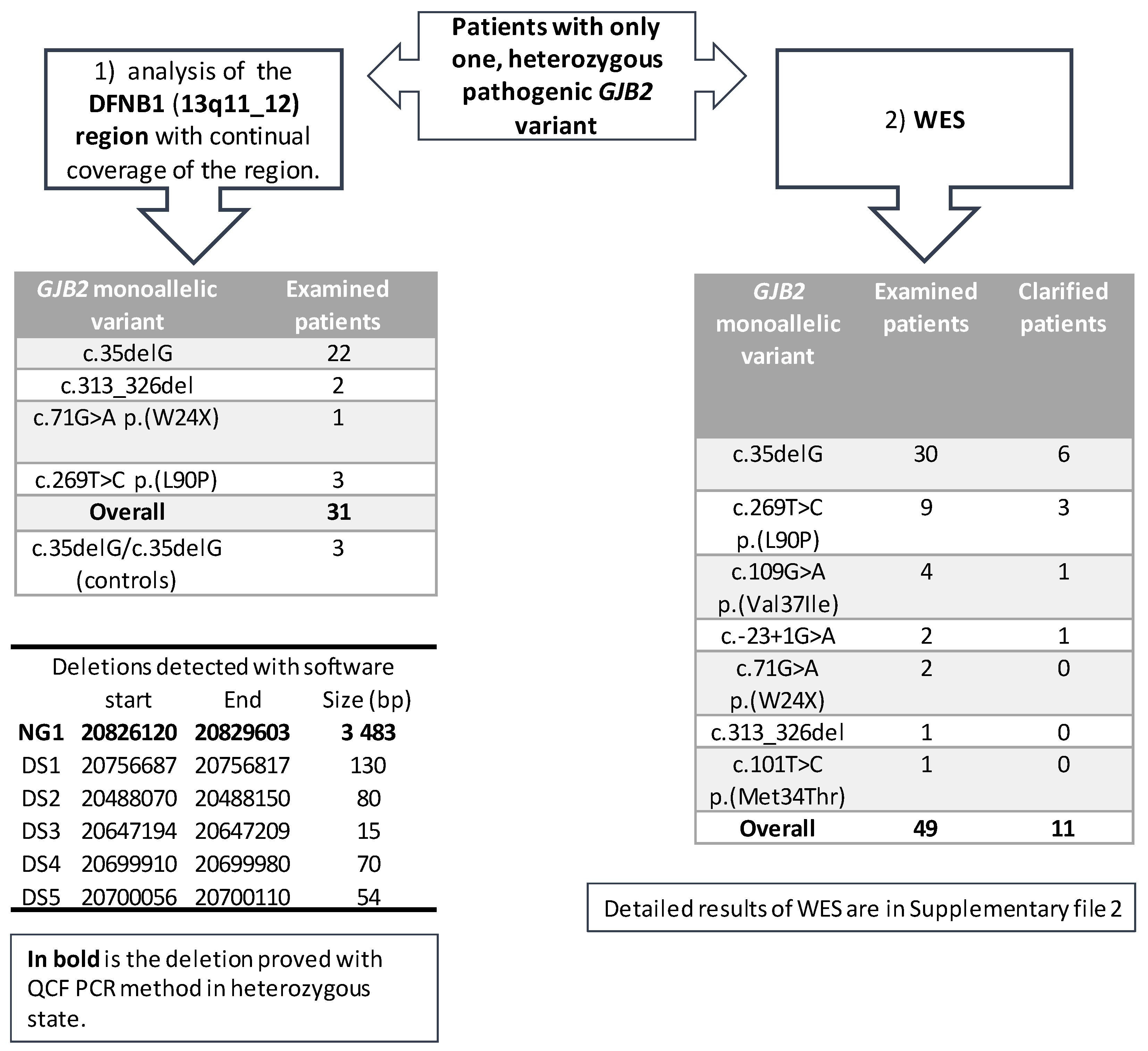
3.1. Continuous Examination of the DFNB1 (13q11-q12)
3.2. Haplotype Analysis of the DFNB1 (13q11-q12)
3.3. WES
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Morton, C.C.; Nance, W.E. Newborn hearing screening—A silent revolution. N. Engl. J. Med. 2006, 354, 2151–2164. [Google Scholar] [CrossRef] [PubMed]
- Shearer, A.E.; Hildebrand, M.S.; Smith, R.J.H. Hereditary Hearing Loss and Deafness Overview. In GeneReviews®; Adam, M.P., Ardinger, H.H., Pagon, R.A., Wallace, S.E., Bean, L.J.H., Stephens, K., Amemiya, A., Eds.; University of Washington: Seattle, WA, USA, 1993. [Google Scholar]
- Seeman, P.; Malikova, M.; Raskova, D.; Bendova, O.; Groh, D.; Kubalkova, M.; Sakmaryova, I.; Seemanova, E.; Kabelka, Z. Spectrum and frequencies of mutations in the GJB2 (Cx26) gene among 156 Czech patients with pre-lingual deafness. Clin. Genet. 2004, 66, 152–157. [Google Scholar] [CrossRef]
- Chan, D.K.; Chang, K.W. GJB2-associated hearing loss: Systematic review of worldwide prevalence, genotype, and auditory phenotype. Laryngoscope 2014, 124, E34–E53. [Google Scholar] [CrossRef] [PubMed]
- Tsukada, K.; Nishio, S.Y.; Hattori, M.; Usami, S. Ethnic-specific spectrum of GJB2 and SLC26A4 mutations: Their origin and a literature review. Ann. Otol. Rhinol. Laryngol. 2015, 124 (Suppl. 1), 61S–76S. [Google Scholar] [CrossRef] [PubMed]
- Seeman, P.; Sakmaryova, I. High prevalence of the IVS 1 + 1 G to A/GJB2 mutation among Czech hearing impaired patients with monoallelic mutation in the coding region of GJB2. Clin. Genet. 2006, 69, 410–413. [Google Scholar] [CrossRef]
- Karczewski, K.J.; Francioli, L.C.; Tiao, G.; Cummings, B.B.; Alfoldi, J.; Wang, Q.; Collins, R.L.; Laricchia, K.M.; Ganna, A.; Birnbaum, D.P.; et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 2020, 581, 434–443. [Google Scholar] [CrossRef] [PubMed]
- Del Castillo, I.; Moreno-Pelayo, M.A.; Del Castillo, F.J.; Brownstein, Z.; Marlin, S.; Adina, Q.; Cockburn, D.J.; Pandya, A.; Siemering, K.R.; Chamberlin, G.P.; et al. Prevalence and evolutionary origins of the del(GJB6-D13S1830) mutation in the DFNB1 locus in hearing-impaired subjects: A multicenter study. Am. J. Hum. Genet. 2003, 73, 1452–1458. [Google Scholar] [CrossRef] [PubMed]
- Del Castillo, F.J.; Rodriguez-Ballesteros, M.; Alvarez, A.; Hutchin, T.; Leonardi, E.; de Oliveira, C.A.; Azaiez, H.; Brownstein, Z.; Avenarius, M.R.; Marlin, S.; et al. A novel deletion involving the connexin-30 gene, del(GJB6-d13s1854), found in trans with mutations in the GJB2 gene (connexin-26) in subjects with DFNB1 non-syndromic hearing impairment. J. Med. Genet. 2005, 42, 588–594. [Google Scholar] [CrossRef] [PubMed]
- Seeman, P.; Bendova, O.; Raskova, D.; Malikova, M.; Groh, D.; Kabelka, Z. Double heterozygosity with mutations involving both the GJB2 and GJB6 genes is a possible, but very rare, cause of congenital deafness in the Czech population. Ann. Hum. Genet. 2005, 69, 9–14. [Google Scholar] [CrossRef]
- Feldmann, D.; Le Marechal, C.; Jonard, L.; Thierry, P.; Czajka, C.; Couderc, R.; Ferec, C.; Denoyelle, F.; Marlin, S.; Fellmann, F. A new large deletion in the DFNB1 locus causes nonsyndromic hearing loss. Eur. J. Med. Genet. 2009, 52, 195–200. [Google Scholar] [CrossRef]
- Del Castillo, I.; Villamar, M.; Moreno-Pelayo, M.A.; del Castillo, F.J.; Alvarez, A.; Telleria, D.; Menendez, I.; Moreno, F. A deletion involving the connexin 30 gene in nonsyndromic hearing impairment. N. Engl. J. Med. 2002, 346, 243–249. [Google Scholar] [CrossRef] [PubMed]
- Wilch, E.; Azaiez, H.; Fisher, R.A.; Elfenbein, J.; Murgia, A.; Birkenhager, R.; Bolz, H.; Da Silva-Costa, S.M.; Del Castillo, I.; Haaf, T.; et al. A novel DFNB1 deletion allele supports the existence of a distant cis-regulatory region that controls GJB2 and GJB6 expression. Clin. Genet. 2010, 78, 267–274. [Google Scholar] [CrossRef]
- Tayoun, A.N.; Mason-Suares, H.; Frisella, A.L.; Bowser, M.; Duffy, E.; Mahanta, L.; Funke, B.; Rehm, H.L.; Amr, S.S. Targeted Droplet-Digital PCR as a Tool for Novel Deletion Discovery at the DFNB1 Locus. Hum. Mutat. 2016, 37, 119–126. [Google Scholar] [CrossRef]
- Rodriguez-Paris, J.; Schrijver, I. The digenic hypothesis unraveled: The GJB6 del(GJB6-D13S1830) mutation causes allele-specific loss of GJB2 expression in cis. Biochem. Biophys. Res. Commun. 2009, 389, 354–359. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Paris, J.; Tamayo, M.L.; Gelvez, N.; Schrijver, I. Allele-specific impairment of GJB2 expression by GJB6 deletion del(GJB6-D13S1854). PLoS ONE 2011, 6, e21665. [Google Scholar] [CrossRef]
- Wilch, E.; Zhu, M.; Burkhart, K.B.; Regier, M.; Elfenbein, J.L.; Fisher, R.A.; Friderici, K.H. Expression of GJB2 and GJB6 is reduced in a novel DFNB1 allele. Am. J. Hum. Genet. 2006, 79, 174–179. [Google Scholar] [CrossRef] [PubMed]
- Common, J.E.; Bitner-Glindzicz, M.; O’Toole, E.A.; Barnes, M.R.; Jenkins, L.; Forge, A.; Kelsell, D.P. Specific loss of connexin 26 expression in ductal sweat gland epithelium associated with the deletion mutation del(GJB6-D13S1830). Clin. Exp. Dermatol. 2005, 30, 688–693. [Google Scholar] [CrossRef] [PubMed]
- Oza, A.M.; DiStefano, M.T.; Hemphill, S.E.; Cushman, B.J.; Grant, A.R.; Siegert, R.K.; Shen, J.; Chapin, A.; Boczek, N.J.; Schimmenti, L.A.; et al. Expert specification of the ACMG/AMP variant interpretation guidelines for genetic hearing loss. Hum. Mutat. 2018, 39, 1593–1613. [Google Scholar] [CrossRef]
- Mohiyuddin, M.; Mu, J.C.; Li, J.; Bani Asadi, N.; Gerstein, M.B.; Abyzov, A.; Wong, W.H.; Lam, H.Y. MetaSV: An accurate and integrative structural-variant caller for next generation sequencing. Bioinformatics 2015, 31, 2741–2744. [Google Scholar] [CrossRef]
- Fan, X.; Abbott, T.E.; Larson, D.; Chen, K. BreakDancer: Identification of Genomic Structural Variation from Paired-End Read Mapping. Curr. Protoc. Bioinform. 2014, 45, 15–16. [Google Scholar] [CrossRef]
- Abyzov, A.; Urban, A.E.; Snyder, M.; Gerstein, M. CNVnator: An approach to discover, genotype, and characterize typical and atypical CNVs from family and population genome sequencing. Genome Res. 2011, 21, 974–984. [Google Scholar] [CrossRef]
- Abyzov, A.; Li, S.; Kim, D.R.; Mohiyuddin, M.; Stutz, A.M.; Parrish, N.F.; Mu, X.J.; Clark, W.; Chen, K.; Hurles, M.; et al. Analysis of deletion breakpoints from 1,092 humans reveals details of mutation mechanisms. Nat. Commun. 2015, 6, 7256. [Google Scholar] [CrossRef] [PubMed]
- Ye, K.; Schulz, M.H.; Long, Q.; Apweiler, R.; Ning, Z. Pindel: A pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics 2009, 25, 2865–2871. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Schulz-Trieglaff, O.; Shaw, R.; Barnes, B.; Schlesinger, F.; Kallberg, M.; Cox, A.J.; Kruglyak, S.; Saunders, C.T. Manta: Rapid detection of structural variants and indels for germline and cancer sequencing applications. Bioinformatics 2016, 32, 1220–1222. [Google Scholar] [CrossRef] [PubMed]
- Layer, R.M.; Chiang, C.; Quinlan, A.R.; Hall, I.M. LUMPY: A probabilistic framework for structural variant discovery. Genome Biol. 2014, 15, R84. [Google Scholar] [CrossRef] [PubMed]
- Kronenberg, Z.N.; Osborne, E.J.; Cone, K.R.; Kennedy, B.J.; Domyan, E.T.; Shapiro, M.D.; Elde, N.C.; Yandell, M. Wham: Identifying Structural Variants of Biological Consequence. PLoS Comput. Biol. 2015, 11, e1004572. [Google Scholar] [CrossRef] [PubMed]
- Talevich, E.; Shain, A.H.; Botton, T.; Bastian, B.C. CNVkit: Genome-Wide Copy Number Detection and Visualization from Targeted DNA Sequencing. PLoS Comput. Biol. 2016, 12, e1004873. [Google Scholar] [CrossRef]
- MacDonald, J.R.; Ziman, R.; Yuen, R.K.; Feuk, L.; Scherer, S.W. The Database of Genomic Variants: A curated collection of structural variation in the human genome. Nucleic Acids Res. 2014, 42, D986–D992. [Google Scholar] [CrossRef] [PubMed]
- Kerkhof, J.; Schenkel, L.C.; Reilly, J.; McRobbie, S.; Aref-Eshghi, E.; Stuart, A.; Rupar, C.A.; Adams, P.; Hegele, R.A.; Lin, H.; et al. Clinical Validation of Copy Number Variant Detection from Targeted Next-Generation Sequencing Panels. J. Mol. Diagn. 2017, 19, 905–920. [Google Scholar] [CrossRef]
- Safka Brozkova, D.; Poisson Markova, S.; Meszarosova, A.U.; Jencik, J.; Cejnova, V.; Cada, Z.; Lastuvkova, J.; Raskova, D.; Seeman, P. Spectrum and frequencies of non GJB2 gene mutations in Czech patients with early non-syndromic hearing loss detected by gene panel NGS and whole-exome sequencing. Clin. Genet. 2020, 98, 548–554. [Google Scholar] [CrossRef]
- Likar, T.; Hasanhodzic, M.; Teran, N.; Maver, A.; Peterlin, B.; Writzl, K. Diagnostic outcomes of exome sequencing in patients with syndromic or non-syndromic hearing loss. PLoS ONE 2018, 13, e0188578. [Google Scholar] [CrossRef]
- Sloan-Heggen, C.M.; Bierer, A.O.; Shearer, A.E.; Kolbe, D.L.; Nishimura, C.J.; Frees, K.L.; Ephraim, S.S.; Shibata, S.B.; Booth, K.T.; Campbell, C.A.; et al. Comprehensive genetic testing in the clinical evaluation of 1119 patients with hearing loss. Hum. Genet. 2016, 135, 441–450. [Google Scholar] [CrossRef] [PubMed]
- Gandia, M.; Del Castillo, F.J.; Rodriguez-Alvarez, F.J.; Garrido, G.; Villamar, M.; Calderon, M.; Moreno-Pelayo, M.A.; Moreno, F.; del Castillo, I. A novel splice-site mutation in the GJB2 gene causing mild postlingual hearing impairment. PLoS ONE 2013, 8, e73566. [Google Scholar] [CrossRef] [PubMed]
- Stanghellini, I.; Genovese, E.; Palma, S.; Ravani, A.; Falcinelli, C.; Guarnaccia, M.C.; Percesepe, A. New and rare GJB2 alleles in patients with nonsyndromic sensorineural hearing impairment: A genotype/auditory phenotype correlation. Genet. Test Mol. Biomark. 2014, 18, 839–844. [Google Scholar] [CrossRef] [PubMed]
- Walsh, T.; Shahin, H.; Elkan-Miller, T.; Lee, M.K.; Thornton, A.M.; Roeb, W.; Abu Rayyan, A.; Loulus, S.; Avraham, K.B.; King, M.C.; et al. Whole exome sequencing and homozygosity mapping identify mutation in the cell polarity protein GPSM2 as the cause of nonsyndromic hearing loss DFNB82. Am. J. Hum. Genet. 2010, 87, 90–94. [Google Scholar] [CrossRef]
- Bademci, G.; Abad, C.; Incesulu, A.; Rad, A.; Alper, O.; Kolb, S.M.; Cengiz, F.B.; Diaz-Horta, O.; Silan, F.; Mihci, E.; et al. MPZL2 is a novel gene associated with autosomal recessive nonsyndromic moderate hearing loss. Hum. Genet. 2018, 137, 479–486. [Google Scholar] [CrossRef]
- Wesdorp, M.; Murillo-Cuesta, S.; Peters, T.; Celaya, A.M.; Oonk, A.; Schraders, M.; Oostrik, J.; Gomez-Rosas, E.; Beynon, A.J.; Hartel, B.P.; et al. MPZL2, Encoding the Epithelial Junctional Protein Myelin Protein Zero-like 2, Is Essential for Hearing in Man and Mouse. Am. J. Hum. Genet. 2018, 103, 74–88. [Google Scholar] [CrossRef]
- Tekin, M.; Chioza, B.A.; Matsumoto, Y.; Diaz-Horta, O.; Cross, H.E.; Duman, D.; Kokotas, H.; Moore-Barton, H.L.; Sakoori, K.; Ota, M.; et al. SLITRK6 mutations cause myopia and deafness in humans and mice. J. Clin. Investig. 2013, 123, 2094–2102. [Google Scholar] [CrossRef]
- Morlet, T.; Rabinowitz, M.R.; Looney, L.R.; Riegner, T.; Greenwood, L.A.; Sherman, E.A.; Achilly, N.; Zhu, A.; Yoo, E.; O’Reilly, R.C.; et al. A homozygous SLITRK6 nonsense mutation is associated with progressive auditory neuropathy in humans. Laryngoscope 2014, 124, E95–E103. [Google Scholar] [CrossRef] [PubMed]
- Du, W.; Han, M.K.; Wang, D.Y.; Han, B.; Zong, L.; Lan, L.; Yang, J.; Shen, Q.; Xie, L.Y.; Yu, L.; et al. A POU3F4 Mutation Causes Nonsyndromic Hearing Loss in a Chinese X-linked Recessive Family. Chin. Med. J. 2017, 130, 88–92. [Google Scholar] [CrossRef]
- Scheidecker, S.; Bar, S.; Stoetzel, C.; Geoffroy, V.; Lannes, B.; Rinaldi, B.; Fischer, F.; Becker, H.D.; Pelletier, V.; Pagan, C.; et al. Mutations in KARS cause a severe neurological and neurosensory disease with optic neuropathy. Hum. Mutat. 2019, 40, 1826–1840. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Lin, Y.; Xu, J.; Che, T.; Li, L.; Yang, T.; Wu, H. Molecular epidemiology of Chinese Han deaf patients with bi-allelic and mono-allelic GJB2 mutations. Orphanet. J. Rare Dis. 2020, 15, 29. [Google Scholar] [CrossRef] [PubMed]
- Moisan, S.; Le Nabec, A.; Quillevere, A.; Le Marechal, C.; Ferec, C. Characterization of GJB2 cis-regulatory elements in the DFNB1 locus. Hum. Genet. 2019, 138, 1275–1286. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Safka Brozkova, D.; Uhrova Meszarosova, A.; Lassuthova, P.; Varga, L.; Staněk, D.; Borecká, S.; Laštůvková, J.; Čejnová, V.; Rašková, D.; Lhota, F.; et al. The Cause of Hereditary Hearing Loss in GJB2 Heterozygotes—A Comprehensive Study of the GJB2/DFNB1 Region. Genes 2021, 12, 684. https://doi.org/10.3390/genes12050684
Safka Brozkova D, Uhrova Meszarosova A, Lassuthova P, Varga L, Staněk D, Borecká S, Laštůvková J, Čejnová V, Rašková D, Lhota F, et al. The Cause of Hereditary Hearing Loss in GJB2 Heterozygotes—A Comprehensive Study of the GJB2/DFNB1 Region. Genes. 2021; 12(5):684. https://doi.org/10.3390/genes12050684
Chicago/Turabian StyleSafka Brozkova, Dana, Anna Uhrova Meszarosova, Petra Lassuthova, Lukáš Varga, David Staněk, Silvia Borecká, Jana Laštůvková, Vlasta Čejnová, Dagmar Rašková, Filip Lhota, and et al. 2021. "The Cause of Hereditary Hearing Loss in GJB2 Heterozygotes—A Comprehensive Study of the GJB2/DFNB1 Region" Genes 12, no. 5: 684. https://doi.org/10.3390/genes12050684
APA StyleSafka Brozkova, D., Uhrova Meszarosova, A., Lassuthova, P., Varga, L., Staněk, D., Borecká, S., Laštůvková, J., Čejnová, V., Rašková, D., Lhota, F., Gašperíková, D., & Seeman, P. (2021). The Cause of Hereditary Hearing Loss in GJB2 Heterozygotes—A Comprehensive Study of the GJB2/DFNB1 Region. Genes, 12(5), 684. https://doi.org/10.3390/genes12050684





