Novel GRHL2 Gene Variant Associated with Hearing Loss: A Case Report and Review of the Literature
Abstract
1. Introduction
2. Materials and Methods
3. Results
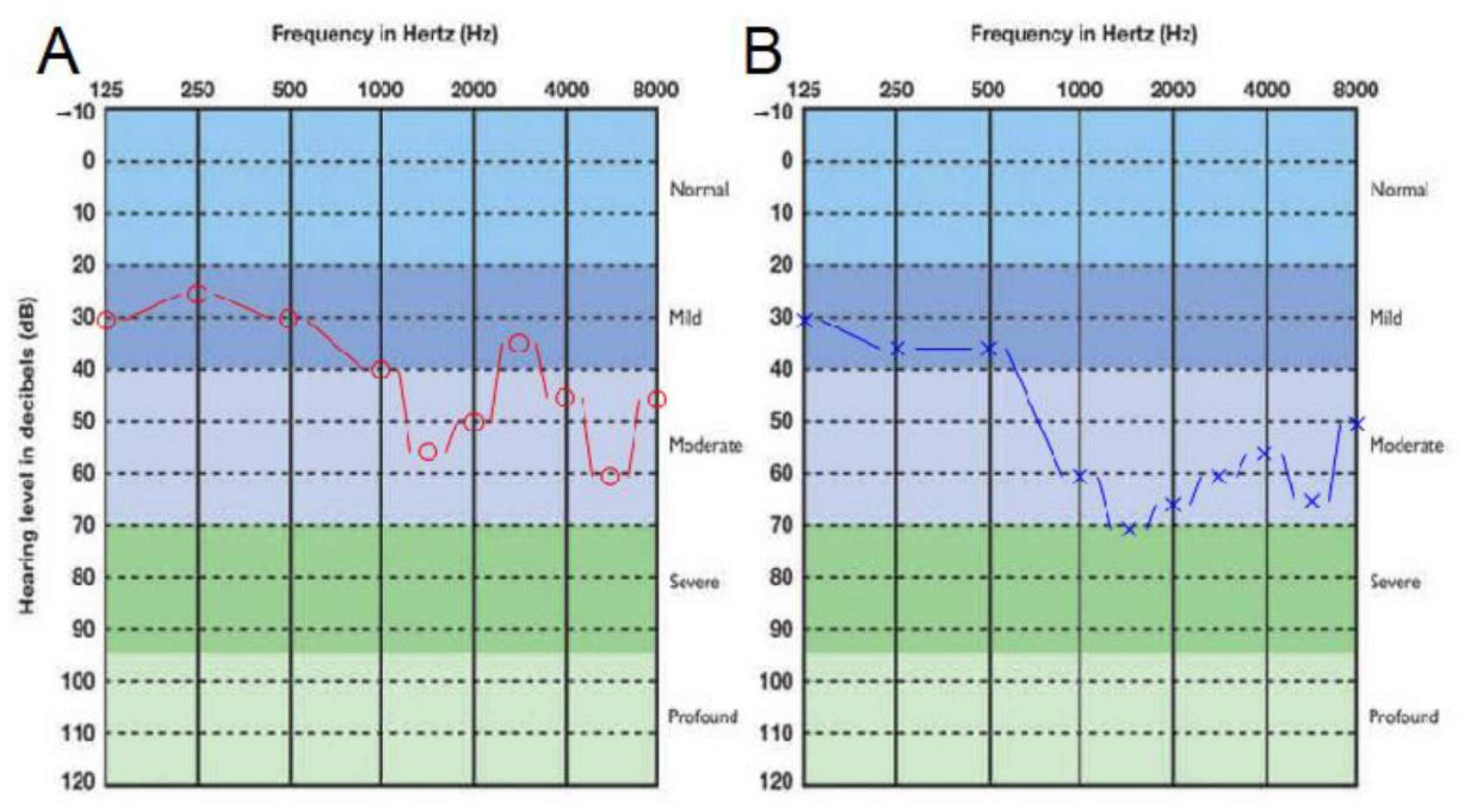
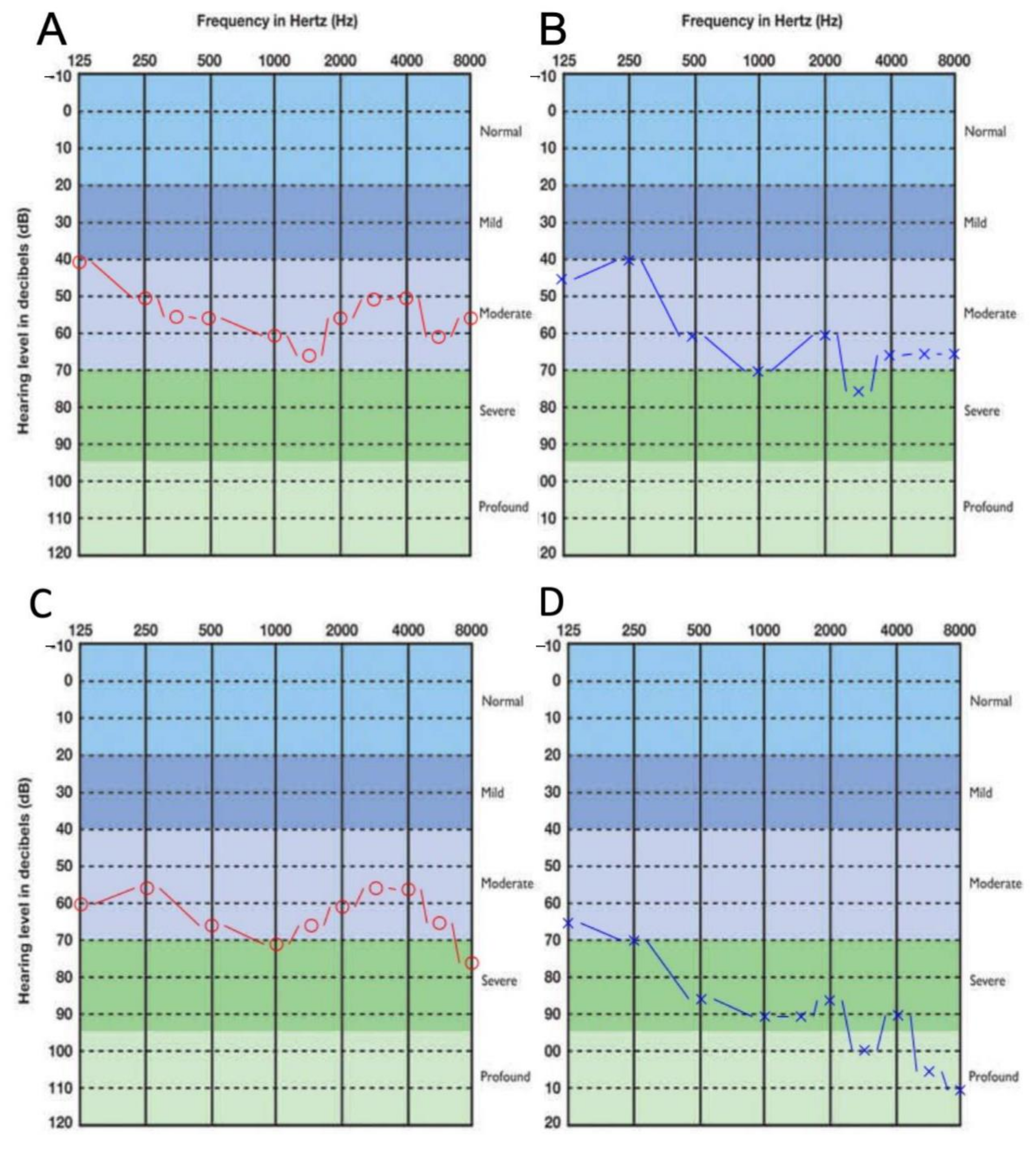
3.1. Clinical Presentation
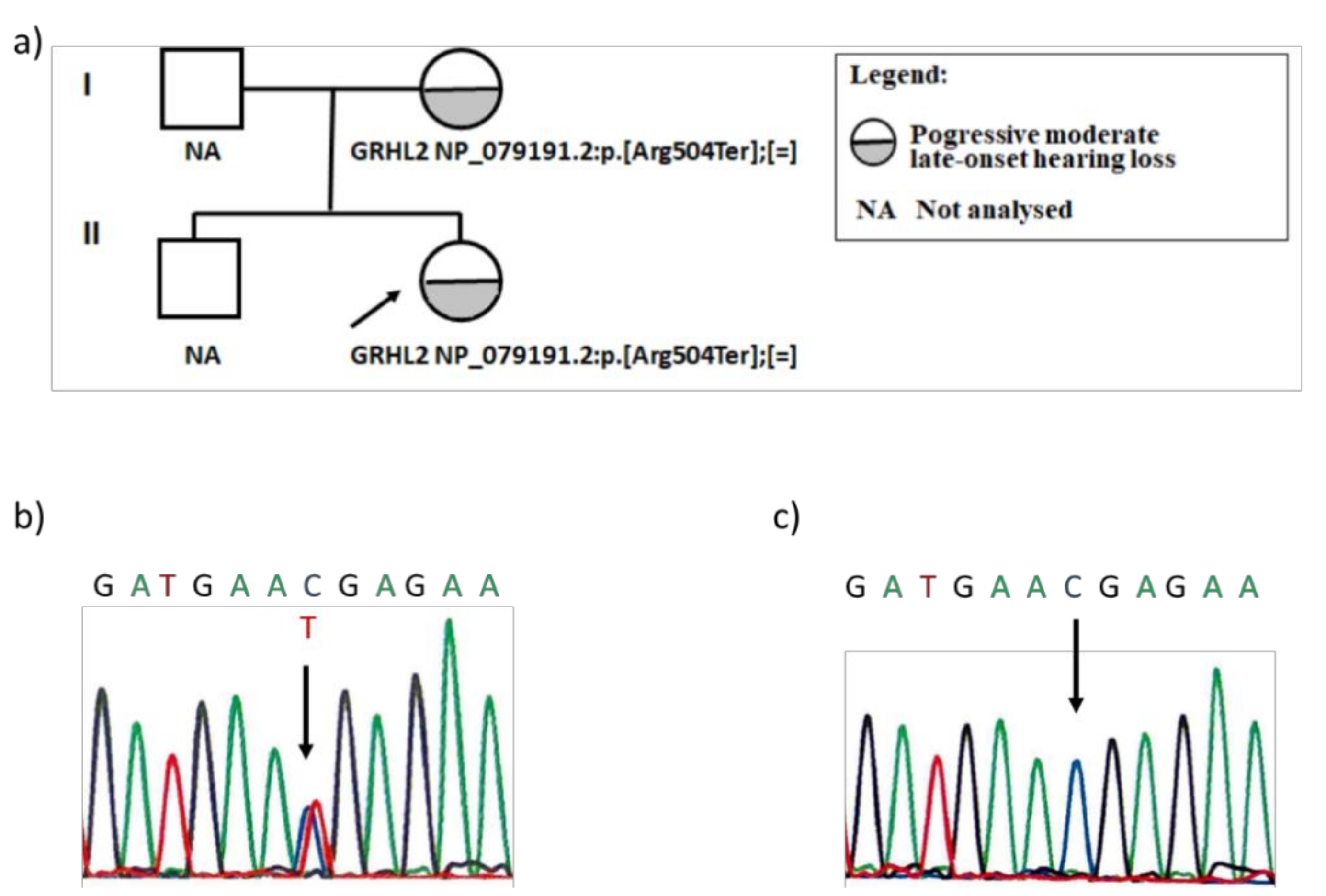
3.2. Genetic Analysis
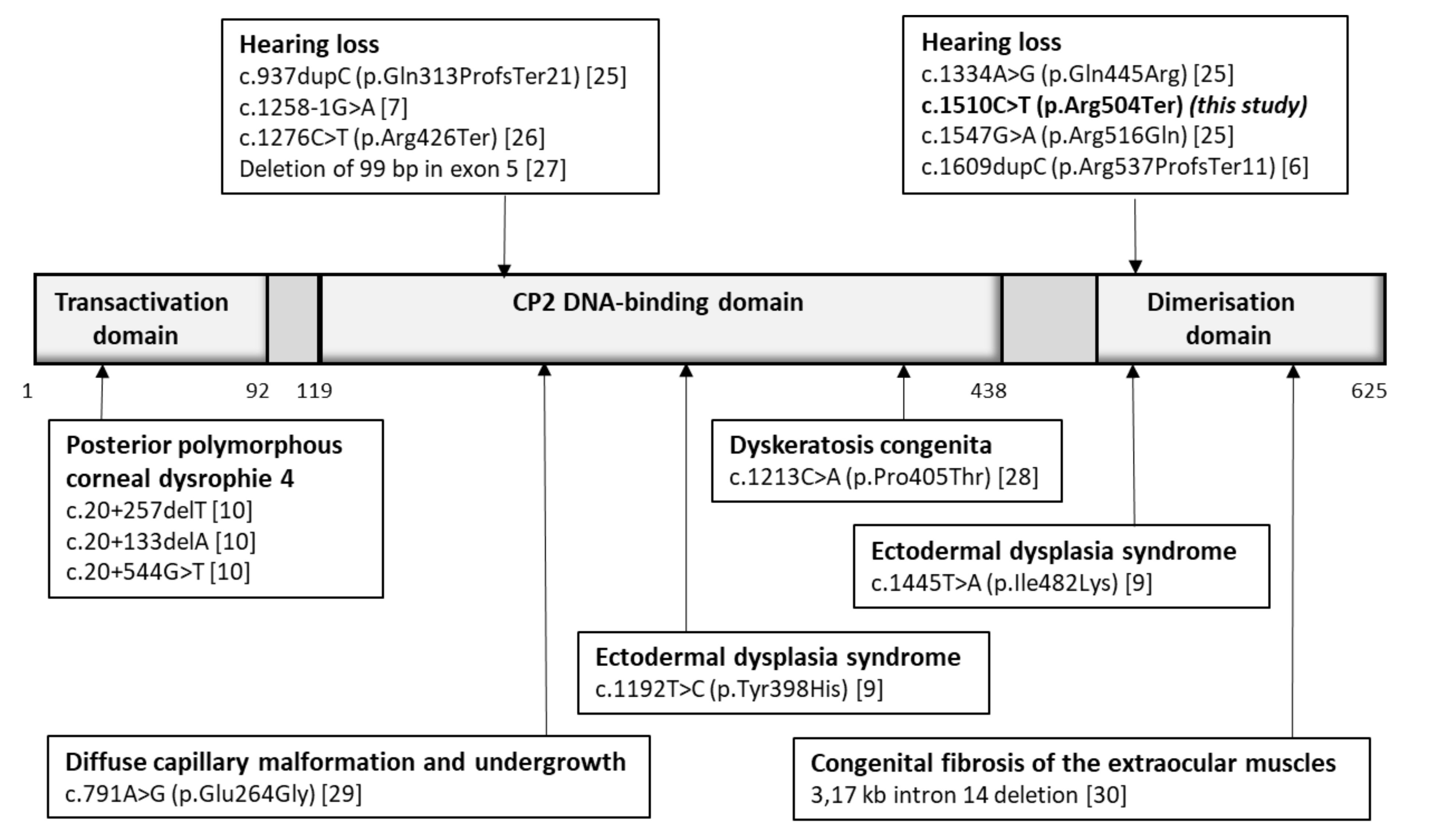
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chang, K.W. Genetics of Hearing Loss-Nonsyndromic. Otolaryngol. Clin. N. Am. 2015, 48, 1063–1072. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Xu, L.; Zhang, S.; Xu, Y. Epidemiological and Genetic Studies of Congenital Profound Deafness in the General Population of Sichuan, China. Am. J. Med. Genet. 1994, 2, 192–195. [Google Scholar] [CrossRef] [PubMed]
- Parker, M.; Bitner-Glindzicz, M. Genetic investigations in childhood deafness. Arch. Dis. Child. 2015, 100, 271–278. [Google Scholar] [CrossRef]
- Van Camp, G.; Smith, R.J.H. Hereditary Hearing Loss Homepage. Available online: https://hereditaryhearingloss.org (accessed on 7 October 2020).
- Werth, M.; Walentin, K.; Aue, A.; Schönheit, J.; Wuebken, A.; Pode-Shakked, N.; Vilianovitch, L.; Erdmann, B.; Dekel, B.; Bader, M.; et al. The transcription factor grainyhead-like 2 regulates the molecular composition of the epithelial apical junctional complex. Development 2010, 137, 3835–3845. [Google Scholar] [CrossRef]
- Peters, L.M.; Anderson, D.W.; Griffith, A.J.; Grundfast, K.M.; San Agustin, T.B.; Madeo, A.C.; Friedman, T.B.; Morell, R.J. Mutation of a transcription factor, TFCP2L3, causes progressive autosomal dominant hearing loss, DFNA28. Hum. Mol. Genet. 2002, 1, 2877–2885. [Google Scholar] [CrossRef][Green Version]
- Vona, B.; Nanda, I.; Neuner, C.; Müller, T.; Haaf, T. Confirmation of GRHL2 as the gene for the DFNA28 locus. Am. J. Med. Genet. Part A 2013, 161, 2060–2065. [Google Scholar] [CrossRef]
- Van Laer, L.; Van Eyken, E.; Fransen, E.; Huyghe, J.R.; Topsakal, V.; Hendrickx, J.J.; Hannula, S.; Mäki-Torkko, E.; Jensen, M.; Demeester, K.; et al. The grainyhead like 2 gene (GRHL2), alias TFCP2L3, is associated with age-related hearing impairment. Hum. Mol. Genet. 2008, 17, 159–169. [Google Scholar] [CrossRef]
- Petrof, G.; Nanda, A.; Howden, J.; Takeichi, T.; McMillan, J.R.; Aristodemou, S.; Ozoemena, L.; Liu, L.; South, A.P.; Pourreyron, C.; et al. Mutations in GRHL2 result in an autosomal-recessive ectodermal dysplasia syndrome. Am. J. Hum. Genet. 2014, 95, 308–314. [Google Scholar] [CrossRef] [PubMed]
- Liskova, P.; Dudakova, L.; Evans, C.J.; Rojas Lopez, K.E.; Pontikos, N.; Athanasiou, D.; Jama, H.; Sach, J.; Skalicka, P.; Stranecky, V.; et al. Ectopic GRHL2 Expression Due to Non-coding Mutations Promotes Cell State Transition and Causes Posterior Polymorphous Corneal Dystrophy 4. Am. J. Hum. Genet. 2018, 102, 447–459. [Google Scholar] [CrossRef]
- Bcbio-Nextgen. Available online: https://github.com/bcbio/bcbio-nextgen (accessed on 7 October 2020).
- Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv 2013, arXiv:1303.3997. [Google Scholar]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The genome analysis toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Garrison, E.; Marth, G. Haplotype-based variant detection from short-read sequencing—Free bayes—Variant Calling—Longranger. arXiv 2012, arXiv:1207.3907. [Google Scholar]
- Kim, S.; Scheffler, K.; Halpern, A.L.; Bekritsky, M.A.; Noh, E.; Källberg, M.; Chen, X.; Kim, Y.; Beyter, D.; Krusche, P.; et al. Strelka2: Fast and accurate calling of germline and somatic variants. Nat. Methods 2018, 15, 591–594. [Google Scholar] [CrossRef] [PubMed]
- Lai, Z.; Markovets, A.; Ahdesmaki, M.; Chapman, B.; Hofmann, O.; Mcewen, R.; Johnson, J.; Dougherty, B.; Barrett, J.C.; Dry, J.R. VarDict: A novel and versatile variant caller for next-generation sequencing in cancer research. Nucleic Acids Res. 2016, 44, e108. [Google Scholar] [CrossRef] [PubMed]
- Desvignes, J.P.; Bartoli, M.; Delague, V.; Krahn, M.; Miltgen, M.; Béroud, C.; Salgado, D. VarAFT: A variant annotation and filtration system for human next generation sequencing data. Nucleic Acids Res. 2018, 46, W545–W553. [Google Scholar] [CrossRef]
- Urbančič, N.B.; Battelino, S.; Tesovnik, T.; Podkrajšek, K.T. The importance of early genetic diagnostics of hearing loss in children. Medicina 2020, 56, 471. [Google Scholar] [CrossRef]
- Human Gene Mutation Database. Available online: http://www.hgmd.org/ (accessed on 7 October 2020).
- Deafness Variation Database. Available online: http://deafnessvariationdatabase.org/ (accessed on 7 October 2020).
- Landrum, M.J.; Chitipiralla, S.; Brown, G.R.; Chen, C.; Gu, B.; Hart, J.; Hoffman, D.; Jang, W.; Kaur, K.; Liu, C.; et al. ClinVar: Improvements to accessing data. Nucleic Acids Res. 2020, 48, D835–D844. [Google Scholar] [CrossRef] [PubMed]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–424. [Google Scholar] [CrossRef] [PubMed]
- The Genome Aggregation Database. Available online: http://gnomad.broadinstitute.org// (accessed on 7 October 2020).
- Zhang, X.; Liu, Y.; Zhang, L.; Yang, Z.; Yang, L.; Wang, X.; Jiang, C.X.; Wang, Q.; Xia, Y.; Chen, Y.; et al. Associations of genetic variations in EYA4, GRHL2 and DFNA5 with noise-induced hearing loss in Chinese population: A case-control study. Environ. Health 2015, 14, 1–7. [Google Scholar] [CrossRef]
- Iwasa, Y.I.; Nishio, S.Y.; Usami, S.I. Comprehensive genetic analysis of Japanese autosomal dominant sensorineural hearing loss patients. PLoS ONE 2016, 11, e0166781. [Google Scholar] [CrossRef]
- Wu, D.; Huang, W.; Xu, Z.; Li, S.; Zhang, J.; Chen, X.; Tang, Y.; Qiu, J.; Wang, Z.; Duan, X.; et al. Clinical and genetic study of 12 Chinese Han families with nonsyndromic deafness. Mol. Genet. Genom. Med. 2020, 8, e1177. [Google Scholar] [CrossRef] [PubMed]
- Ji, H.; Lu, J.; Wang, J.; Li, H.; Lin, X. Combined examination of sequence and copy number variations in human deafness genes improves diagnosis for cases of genetic deafness. BMC Ear Nose Throat Disord. 2014, 14, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Walne, A.J.; Collopy, L.; Cardoso, S.; Ellison, A.; Plagnol, V.; Albayrak, C.; Albayrak, D.; Kilic, S.S.; Patıroglu, T.; Akar, H.; et al. Marked overlap of four genetic syndromes with dyskeratosis congenita confounds clinical diagnosis. Haematologica 2016, 101, 1180–1189. [Google Scholar] [CrossRef] [PubMed]
- Cubiró, X.; Rozas-Muñoz, E.; Castel, P.; Roé Crespo, E.; Garcia-Melendo, C.; Puig, L.; Baselga, E. Clinical and genetic evaluation of six children with diffuse capillary malformation and undergrowth. Pediatr. Dermatol. 2020, 37, 833–838. [Google Scholar] [CrossRef]
- Abu-Amero, K.K.; Kondkar, A.A.; Khan, A.O. A microdeletion in the GRHL2 Gene in two unrelated patients with congenital fibrosis of the extra ocular muscles. BMC Res. Notes 2017, 10, 1–5. [Google Scholar] [CrossRef]
- Dworkin, S.; Darido, C.; Georgy, S.R.; Wilanowski, T.; Srivastava, S.; Ellett, F.; Pase, L.; Han, Y.; Meng, A.; Heath, J.K.; et al. Midbrain-hindbrain boundary patterning and morphogenesis are regulated by diverse grainy head-like 2-dependent pathways. Development 2012, 139, 525–536. [Google Scholar] [CrossRef] [PubMed]
- Sundararajan, V.; Pang, Q.Y.; Choolani, M.; Huang, R.Y.J. Spotlight on the Granules (Grainyhead-Like Proteins)—From an Evolutionary Conserved Controller of Epithelial Trait to Pioneering the Chromatin Landscape. Front. Mol. Biosci. 2020, 7, 213. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Feng, C.; Zhu, H.; Wu, S.; Jin, P.; Xu, T. Grainyhead-like 2 as a double-edged sword in development and cancer. Am. J. Transl. Res. 2020, 12, 310–331. [Google Scholar]
- Leth-Larsen, R.; Terp, M.G.; Christensen, A.G.; Elias, D.; Kühlwein, T.; Jensen, O.N.; Petersen, O.W.; Ditzel, H.J. Functional heterogeneity within the CD44 high human breast cancer stem cell-like compartment reveals a gene signature predictive of distant metastasis. Mol. Med. 2012, 8, 1109–1121. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Zhang, R.; Xie, C.; Gan, M.; Yao, S.; Yao, Y.; Jin, J.; Han, T.; Huang, Y.; Gong, Y.; et al. GRHL2 suppresses tumor metastasis via regulation of transcriptional activity of rhog in non-small cell lung cancer. Am. J. Transl. Res. 2017, 9, 4217–4226. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Trebusak Podkrajsek, K.; Tesovnik, T.; Bozanic Urbancic, N.; Battelino, S. Novel GRHL2 Gene Variant Associated with Hearing Loss: A Case Report and Review of the Literature. Genes 2021, 12, 484. https://doi.org/10.3390/genes12040484
Trebusak Podkrajsek K, Tesovnik T, Bozanic Urbancic N, Battelino S. Novel GRHL2 Gene Variant Associated with Hearing Loss: A Case Report and Review of the Literature. Genes. 2021; 12(4):484. https://doi.org/10.3390/genes12040484
Chicago/Turabian StyleTrebusak Podkrajsek, Katarina, Tine Tesovnik, Nina Bozanic Urbancic, and Saba Battelino. 2021. "Novel GRHL2 Gene Variant Associated with Hearing Loss: A Case Report and Review of the Literature" Genes 12, no. 4: 484. https://doi.org/10.3390/genes12040484
APA StyleTrebusak Podkrajsek, K., Tesovnik, T., Bozanic Urbancic, N., & Battelino, S. (2021). Novel GRHL2 Gene Variant Associated with Hearing Loss: A Case Report and Review of the Literature. Genes, 12(4), 484. https://doi.org/10.3390/genes12040484





