Genome-Wide Identification of Genes Involved in Acid Stress Resistance of Salmonella Derby
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Plasmids, and Cell Lines
2.2. Genome Sequencing, Assembly and Annotation of the S. Derby 14T
2.3. Transposon Insertion Library Construction
2.4. Library Construction, Sequencing and Bioinformatics Analysis
2.5. Construction of Gene Deletion Mutant Strains
2.6. Quantitative Real Time PCR
2.7. The Growth Characteristics of WT and Deletion Mutant Strains
2.8. Survival Analysis
2.9. Adhesion and Invasion Assay of MC 38 Cells
2.10. Data Availability
3. Results
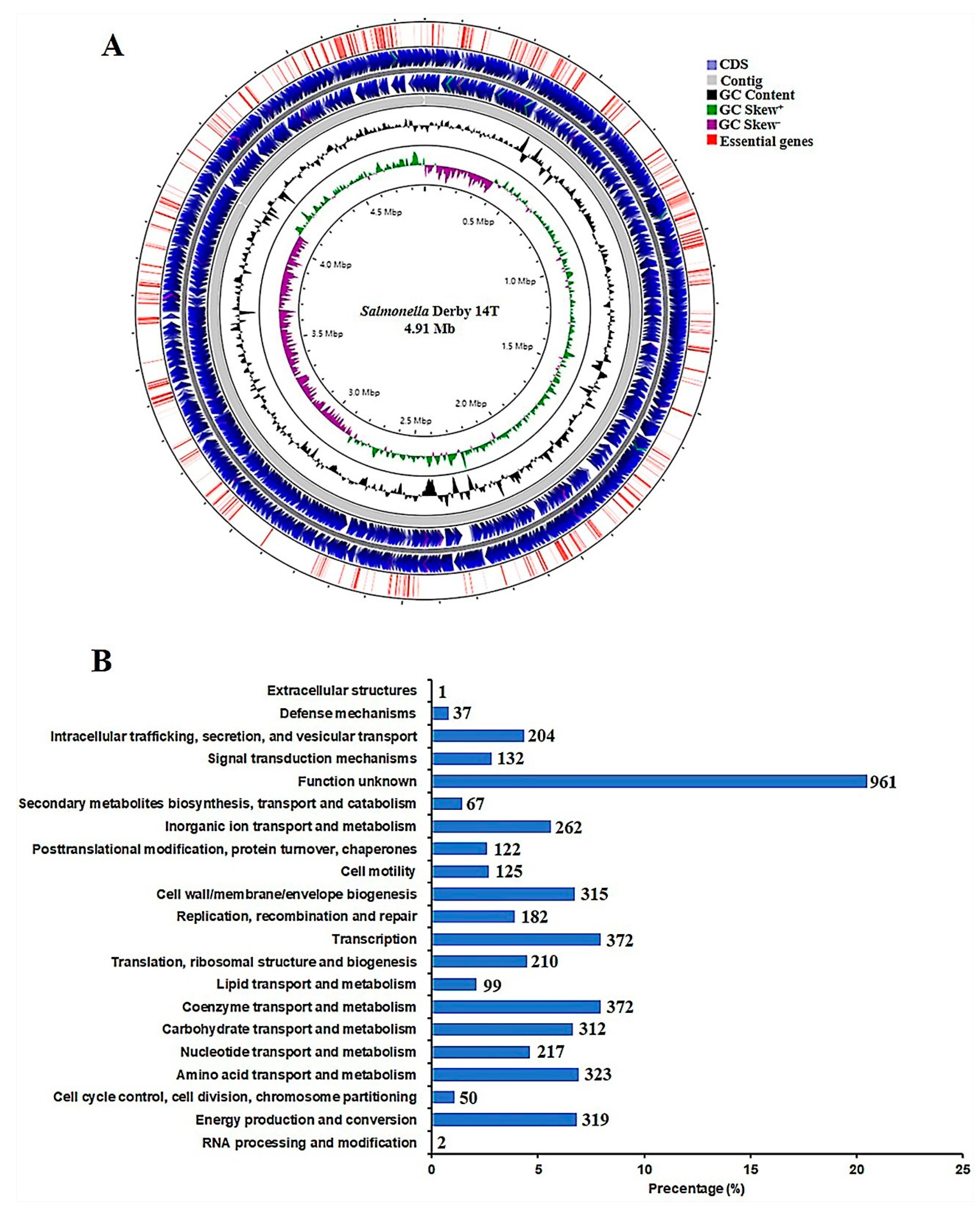
3.1. Overview of the S. Derby 14T Genome
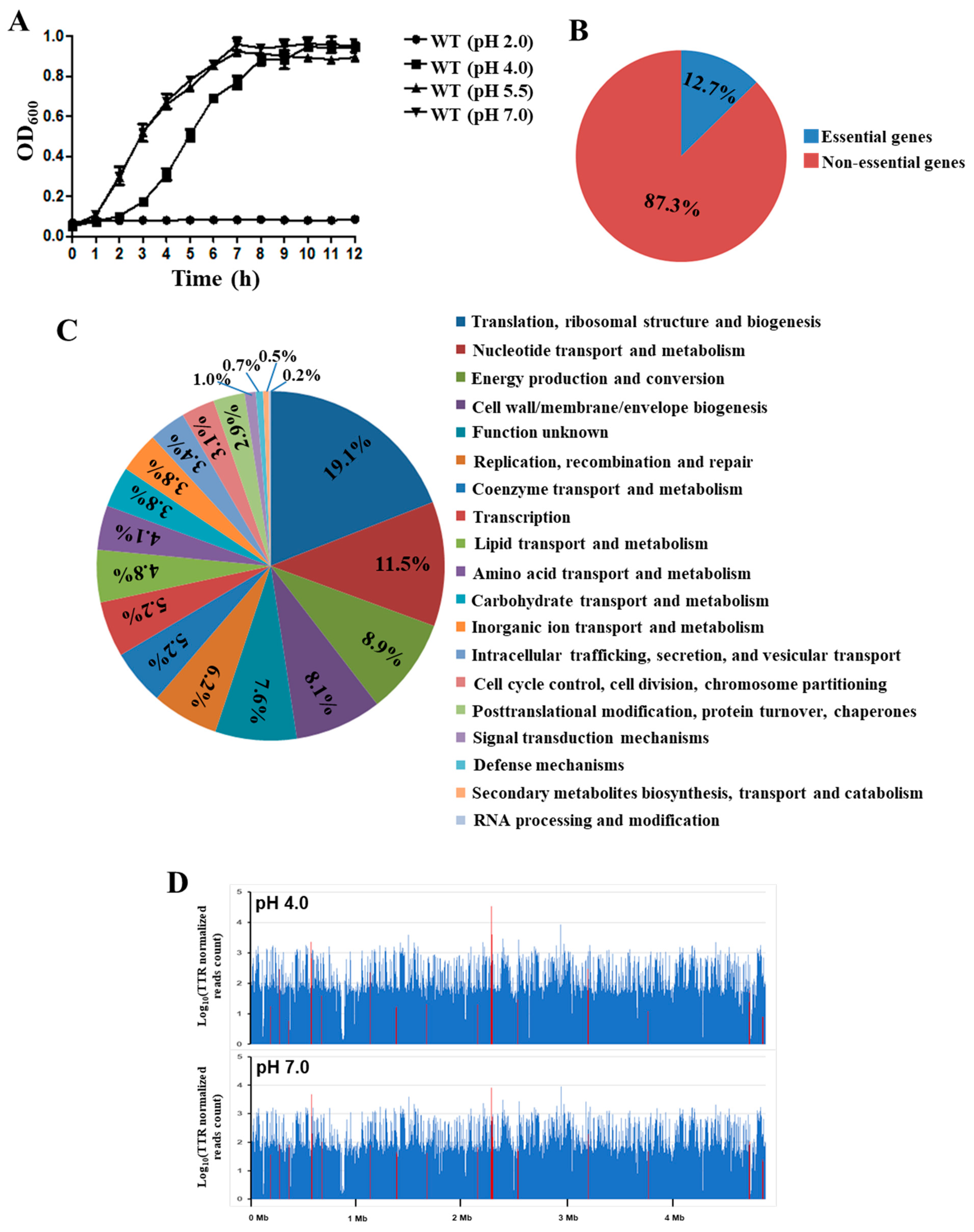
3.2. Identification of Genes Involved in Acid Stress Resistance by Tn-seq Analysis
3.3. Validation of the Tn-seq Identified Genes with qRT-PCR
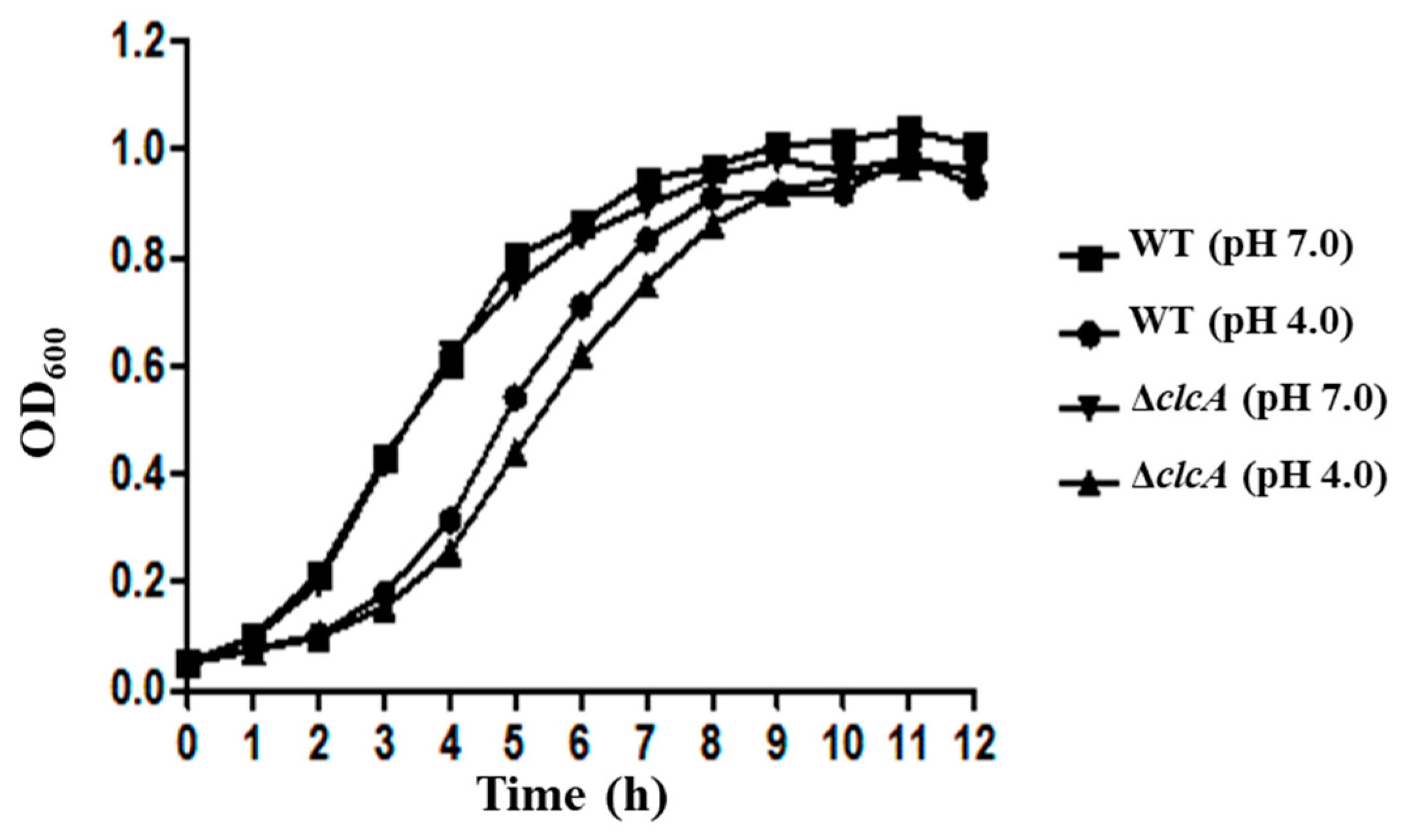
3.4. ClcA Was Responsible for Acid Stress Resistance
3.5. CpxAR Was Responsible for the Acid Stress Resistance and Virulence of S. Derby
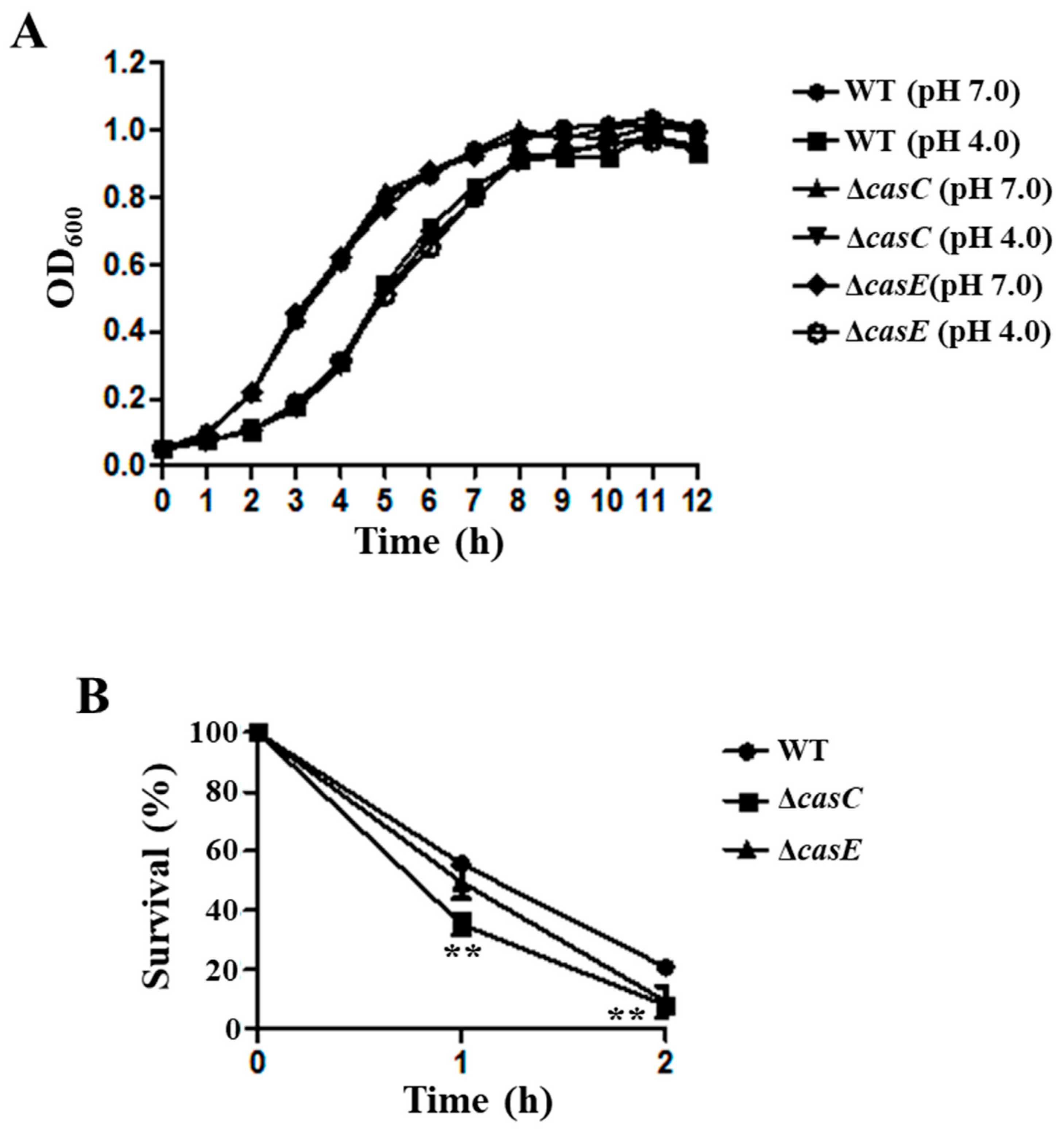
3.6. CasC/CasE Were Involved in the Acid Stress Resistance
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Simon, S.; Trost, E.; Bender, J.; Fuchs, S.; Flieger, A. Evaluation of WGS based approaches for investigating a food-borne outbreak caused by Salmonella enterica serovar Derby in Germany. Food Microbiol. 2018, 71, 46–54. [Google Scholar] [CrossRef] [PubMed]
- Sévellec, Y.; Granier, S.A.; Hello, S.L.; Weill, F.X.; Cadel-Six, S. Source Attribution Study of Sporadic Salmonella Derby Cases in France. Front. Microbiol. 2020, 11, 889. [Google Scholar] [CrossRef]
- Sévellec, Y.; Felten, A.; Radomski, N.; Granier, S.A.; Hello, S.L.; Petrovska, L.; Mistou, M.Y.; Cadel-Six, S. Genetic Diversity of Salmonella Derby from the Poultry Sector in Europe. Pathogens 2019, 8, 46. [Google Scholar] [CrossRef]
- European Food Safety Authority and European Centre for Disease Prevention and Control. The European Union One Health 2018 Zoonoses Report. EFSA J. 2019, 17, 5956. [Google Scholar] [CrossRef]
- Yann, S.; Marie-Léone, V.; Granier, S.A.; Renaud, L.; Carole, F.; Simon, L.H.; Michel-Yves, M.; Sabrina, C.S. Polyphyletic Nature of Salmonella enterica Serotype Derby and Lineage-Specific Host-Association Revealed by Genome-Wide Analysis. Front. Microbiol. 2018, 9, 891. [Google Scholar] [CrossRef]
- Zheng, H.; Hu, Y.; Li, Q.; Tao, J.; Cai, Y.; Wang, Y.; Li, J.; Zhou, Z.; Pan, Z.; Jiao, X. Subtyping Salmonella enterica serovar Derby with multilocus sequence typing (MLST) and clustered regularly interspaced short palindromic repeats (CRISPRs). Food Control 2017, 73, 474–484. [Google Scholar] [CrossRef]
- Diemert, S.; Yan, T. Municipal wastewater surveillance reveals high community disease burden of rarely reported and possibly subclinical Salmonella Derby strain. Appl. Environ. Microbiol. 2020, 86, e00814–e00820. [Google Scholar] [CrossRef] [PubMed]
- Hayward, M.R.; Jansen, V.; Woodward, M.J. Comparative genomics of Salmonella enterica serovars Derby and Mbandaka, two prevalent serovars associated with different livestock species in the UK. BMC Genom. 2013, 14, 365. [Google Scholar] [CrossRef] [PubMed]
- Cao, L.; Wang, J.; Sun, L.; Kong, Z.; Wu, Q.; Wang, Z. Transcriptional analysis reveals the relativity of acid tolerance and antimicrobial peptide resistance of Salmonella. Microb. Pathog. 2019, 136, 103701. [Google Scholar] [CrossRef]
- Howden, C.W.; Hunt, R.H.; Howden, C.W.; Hunt, R.H. Relationship between gastric secretion and infection. Gut 1987, 28, 96–107. [Google Scholar] [CrossRef]
- Dykhuizen, R.S.; Frazer, R.; Duncan, C.; Smith, C.C.; Golden, M.; Benjamin, N.; Leifert, C. Antimicrobial effect of acidified nitrite on gut pathogens: Importance of dietary nitrate in host defense. Antimicrob. Agents Chemother. 1996, 40, 1422–1425. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.L. The role of gastric acid in preventing foodborne disease and how bacteria overcome acid conditions. J. Food Prot. 2003, 66, 1292–1303. [Google Scholar] [CrossRef] [PubMed]
- Denbesten, L. Gastrointestinal Disease: Pathophysiology, Diagnosis, Management. Ann. Surg. 1984, 200, S0025–S6196. [Google Scholar]
- Bird, T.; Hall, M.R.; Schade, R.O. Gastric histology and its relation to anaemia in the elderly. Gerontology 1977, 23, 309–321. [Google Scholar] [CrossRef]
- Cevallos-Almeida, M.; Martin, L.; Houdayer, C.; Rose, V.; Guionnet, J.M.; Paboeuf, F.; Denis, M.; Kerouanton, A. Experimental infection of pigs by Salmonella Derby, S. Typhimurium and monophasic variant of S. Typhimurium: Comparison of colonization and serology. Vet. Microbiol. 2019, 231, 147–153. [Google Scholar] [CrossRef]
- Rathman, M.; Sjaastad, M.D.; Falkow, S. Acidification of phagosomes containing Salmonella Typhimurium in murine macrophages. Infect. Immun. 1996, 64, 2765–2773. [Google Scholar] [CrossRef] [PubMed]
- Alpuche, A.C.M.; Swanson, J.A.; Loomis, W.P.; Miller, S.I. Salmonella Typhimurium activates virulence gene transcription within acidified macrophage phagosomes. Proc. Natl. Acad. Sci. USA 1992. [Google Scholar] [CrossRef]
- Choi, J.; Groisman, E.A. Acidic pH sensing in the bacterial cytoplasm is required for Salmonella virulence. Mol. Microbiol. 2016, 101, 1024–1038. [Google Scholar] [CrossRef] [PubMed]
- Lang, C.; Zhang, Y.; Mao, Y.; Yang, X.; Wang, X.; Luo, X.; Dong, P.; Zhu, L. Acid tolerance response of Salmonella during simulated chilled beef storage and its regulatory mechanism based on the PhoP/Q system. Food Microbiol. 2021, 95, 103716. [Google Scholar] [CrossRef]
- Bearson, B.L.; Wilson, L.; Foster, J.W. A low pH-inducible, PhoPQ-dependent acid tolerance response protects Salmonella Typhimurium against inorganic acid stress. J. Bacteriol. 1998, 180, 2409–2417. [Google Scholar] [CrossRef] [PubMed]
- Quinn, H.J.; Cameron, A.D.; Dorman, C.J. Bacterial regulon evolution: Distinct responses and roles for the identical OmpR proteins of Salmonella Typhimurium and Escherichia coli in the acid stress response. PLoS Genet. 2014, 10, e1004215. [Google Scholar] [CrossRef]
- Muller, C.; Bang, I.-S.; Velayudhan, J.; Karlinsey, J.; Papenfort, K. Acid stress activation of the σE stress response in Salmonella enterica serovar Typhimurium. Mol. Microbiol. 2009, 71, 1228–1238. [Google Scholar] [CrossRef] [PubMed]
- Audia, J.P.; Foster, J.W. Acid Shock Accumulation of Sigma S in Salmonella enterica Involves Increased Translation, Not Regulated Degradation. J. Mol. Microbiol. Biotechnol. 2003, 5, 17–28. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Ordonez, A.A.; Begley, M.; Prieto, M.; Messens, W.; Hill, C. Salmonella spp. survival strategies within the host gastrointestinal tract. Microbiology 2011, 157, 3268–3281. [Google Scholar] [CrossRef]
- Ren, J.; Sang, Y.; Ni, J.; Tao, J.; Lu, J.; Zhao, M.; Yao, Y.F. Acetylation Regulates Survival of Salmonella enterica Serovar Typhimurium under Acid Stress. Appl. Environ. Microbiol. 2015, 81, 5675–5682. [Google Scholar] [CrossRef] [PubMed]
- Ryan, D.; Pati, N.B.; Ojha, U.K.; Padhi, C.; Suar, M. Global Transcriptome and Mutagenic Analyses of the Acid Tolerance Response of Salmonella enterica Serovar Typhimurium. Appl. Environ. Microbiol. 2015, 81, 8054–8065. [Google Scholar] [CrossRef] [PubMed]
- Kenney, L.J. The role of acid stress in Salmonella pathogenesis. Curr. Opin. Microbiol. 2019, 47, 45–51. [Google Scholar] [CrossRef]
- Lee, S.A.; Dunne, J.; Febery, E.; Wilcock, P.; Mottram, T.; Bedford, M.R. Superdosing phytase reduces real-time gastric pH in broilers and weaned piglets. Br. Poult Sci. 2018, 59, 330–339. [Google Scholar] [CrossRef]
- Aluthge, N.D.; van Sambeek, D.M.; Carney-Hinkle, E.E.; Li, Y.S.S.; Fernando, S.C.; Burkey, T.E. BOARD INVITED REVIEW: The pig microbiota and the potential for harnessing the power of the microbiome to improve growth and health. J. Anim. Sci. 2019, 97, 3741–3757. [Google Scholar] [CrossRef] [PubMed]
- Chesson, A. 6—supplemeary enzymes to improve the utilization of pig and poultry diets. Recent Adv. Anim. Nutr. 1987, 71–89. [Google Scholar] [CrossRef]
- Wetmore, K.M.; Price, M.N.; Waters, R.J.; Lamson, J.S.; He, J.; Hoover, C.A.; Blow, M.J.; Bristow, J.; Butland, G.; Arkin, A.P.; et al. Rapid quantification of mutant fitness in diverse bacteria by sequencing randomly bar-coded transposons. MBio 2015, 6, e00306–e00315. [Google Scholar] [CrossRef]
- Liang, W.L.; Wang, S.X.; Yu, F.G.; Zhang, L.J.; Qi, G.M.; Liu, Y.Q.; Gao, S.Y.; Kan, B. Construction and evaluation of a safe, live, oral Vibrio cholerae vaccine candidate, IEM108. Infect. Immun. 2003, 71, 5498–5504. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wang, S.Y.; Lauritz, J.; Jass, J.; Milton, D.L. A ToxR homolog from Vibrio anguillarum serotype O1 regulates its own production, bile resistance, and biofilm formation. J. Bacteriol. 2002, 184, 1630–1639. [Google Scholar] [CrossRef] [PubMed]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Huerta-Cepas, J.; Szklarczyk, D.; Forslund, K.; Cook, H.; Heller, D.; Walter, M.C.; Rattei, T.; Mende, D.R.; Sunagawa, S.; Kuhn, M.; et al. eggNOG 4.5: A hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res. 2016, 44, D286–D293. [Google Scholar] [CrossRef]
- Grant, J.R.; Stothard, P. The CGView Server: A comparative genomics tool for circular genomes. Nucleic Acids Res. 2008, 36, W181–W184. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Billings, G.; Hubbard, T.P.; Park, J.S.; Yin Leung, K.; Liu, Q.; Davis, B.M.; Zhang, Y.; Wang, Q.; Waldor, M.K. Time-Resolved Transposon Insertion Sequencing Reveals Genome-Wide Fitness Dynamics during Infection. MBio 2017, 8, e01581–e01617. [Google Scholar] [CrossRef] [PubMed]
- DeJesus, M.A.; Ambadipudi, C.; Baker, R.; Sassetti, C.; Ioerger, T.R. TRANSIT--A Software Tool for Himar1 TnSeq Analysis. PLoS Comput. Biol. 2015, 11, e1004401. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Gu, D.; Huang, T.; Cao, L.; Zhu, X.; Zhou, Y.; Wang, K.; Kang, X.; Meng, C.; Jiao, X.; et al. Essential role of Salmonella Enteritidis DNA adenine methylase in modulating inflammasome activation. BMC Microbiol. 2020, 20, 226. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Zhao, Z.; Tong, W.; Ding, Y.; Liu, B.; Shi, Y.; Wang, J.; Sun, S.; Liu, M.; Wang, Y.; et al. An acid-tolerance response system protecting exponentially growing Escherichia coli. Nat. Commun. 2020, 11, 1496. [Google Scholar] [CrossRef] [PubMed]
- Nunez-Hernandez, C.; Tierrez, A.; Ortega, A.D.; Pucciarelli, M.G.; Godoy, M.; Eisman, B.; Casadesus, J.; Garcia-del Portillo, F. Genome expression analysis of nonproliferating intracellular Salmonella enterica serovar Typhimurium unravels an acid pH-dependent PhoP-PhoQ response essential for dormancy. Infect. Immun. 2013, 81, 154–165. [Google Scholar] [CrossRef] [PubMed]
- Eguchi, Y.; Ishii, E.; Hata, K.; Utsumi, R. Regulation of acid resistance by connectors of two-component signal transduction systems in Escherichia coli. J. Bacteriol. 2011, 193, 1222–1228. [Google Scholar] [CrossRef]
- Gu, D.; Guo, M.; Yang, M.; Zhang, Y.; Zhou, X.; Wang, Q. A sigmaE-Mediated Temperature Gauge Controls a Switch from LuxR-Mediated Virulence Gene Expression to Thermal Stress Adaptation in Vibrio alginolyticus. PLoS Pathog. 2016, 12, e1005645. [Google Scholar] [CrossRef]
- Tierney, A.R.; Rather, P.N. Roles of two-component regulatory systems in antibiotic resistance. Future Microbiol. 2019, 14, 533–552. [Google Scholar] [CrossRef]
- Lippa, A.M.; Goulian, M. Feedback inhibition in the PhoQ/PhoP signaling system by a membrane peptide. PLoS Genet. 2009, 5, e1000788. [Google Scholar] [CrossRef] [PubMed]
- Salazar, M.E.; Podgornaia, A.I.; Laub, M.T. The small membrane protein MgrB regulates PhoQ bifunctionality to control PhoP target gene expression dynamics. Mol. Microbiol. 2016, 102, 430–445. [Google Scholar] [CrossRef]
- Lin, Z.W.; Cai, X.; Chen, M.L.; Ye, L.; Wu, Y.; Wang, X.F.; Lv, Z.H.; Shang, Y.P.; Qu, D. Virulence and Stress Responses of Shigella flexneri Regulated by PhoP/PhoQ. Front. Microbiol. 2018, 8, 2689. [Google Scholar] [CrossRef]
- Accardi, A.; Miller, C. Secondary active transport mediated by a prokaryotic homologue of ClC Cl-channels. Nature 2004, 427, 803–807. [Google Scholar] [CrossRef]
- Iyer, R.; Iverson, T.M.; Accardi, A.; Miller, C. A biological role for prokaryotic ClC chloride channels. Nature 2002, 419, 715–718. [Google Scholar] [CrossRef]
- Yan, K.; Liu, T.; Duan, B.; Liu, F.; Cao, M.; Peng, W.; Dai, Q.; Chen, H.; Yuan, F.; Bei, W. The CpxAR Two-Component System Contributes to Growth, Stress Resistance, and Virulence of Actinobacillus pleuropneumoniae by Upregulating wecA Transcription. Front. Microbiol. 2020, 11, 1026. [Google Scholar] [CrossRef] [PubMed]
- Fujimoto, M.; Goto, R.; Haneda, T.; Okada, N.; Miki, T. Salmonella enterica Serovar Typhimurium CpxRA Two-Component System Contributes to Gut Colonization in Salmonella-Induced Colitis. Infect. Immun. 2018, 86, e00280–e00318. [Google Scholar] [CrossRef] [PubMed]
- De la Cruz, M.A.; Pérez-Morales, D.; Palacios, I.J.; Fernández-Mora, M.; Calva, E.; Bustamante, V.H. The two-component system CpxR/A represses the expression of Salmonella virulence genes by affecting the stability of the transcriptional regulator HilD. Front. Micriobiol. 2015, 6, 807. [Google Scholar] [CrossRef]
- Spinola, S.M.; Fortney, K.R.; Baker, B.; Janowicz, D.M.; Zwickl, B.; Katz, B.P.; Blick, R.J.; Munson, R.S., Jr. Activation of the CpxRA system by deletion of cpxA impairs the ability of Haemophilus ducreyi to infect humans. Infect. Immun. 2010, 78, 3898–3904. [Google Scholar] [CrossRef]
- Matter, L.B.; Ares, M.A.; Abundes-Gallegos, J.; Cedillo, M.L.; Yáñez, J.A.; Martínez-Laguna, Y.; De la Cruz, M.A.; Girón, J.A. The CpxRA stress response system regulates virulence features of avian pathogenic Escherichia coli. Environ. Microbiol. 2018, 20, 3363–3377. [Google Scholar] [CrossRef] [PubMed]
- Cui, L.; Wang, X.; Huang, D.; Zhao, Y.; Feng, J.; Lu, Q.; Pu, Q.; Wang, Y.; Cheng, G.; Wu, M.; et al. CRISPR-cas3 of Salmonella Upregulates Bacterial Biofilm Formation and Virulence to Host Cells by Targeting Quorum-Sensing Systems. Pathogens 2020, 9, 53. [Google Scholar] [CrossRef] [PubMed]


| Strain or Plasmid | Relevant Characteristics | Reference |
|---|---|---|
| Escherichia coli | ||
| WM3064 | conjugal donor | [31] |
| SM10 λpir | Host for π requiring plasmids, conjugal donor | [32] |
| S. Derby | ||
| S. Derby 14T | Isolated from pork sample. Tcr | Laboratory collection |
| ΔcpxA | 14T-T8N3, in-frame deletion in cpxA, Tcr | This study |
| ΔcpxR | 14T-T8N3, in-frame deletion in cpxR, Tcr | This study |
| ΔclcA | 14T-T8N3, in-frame deletion in clcA, Tcr | This study |
| ΔcasC | 14T-T8N3, in-frame deletion in casC, Tcr | This study |
| ΔcasE | 14T-T8N3, in-frame deletion in casE, Tcr | This study |
| Plasmids | ||
| pKWM2 | Tn5 transposon vector library, OriT, R6K, Kanr | [31] |
| pDM4 | Suicide vector, pir dependent, R6K, SacBR, Cmr | [33] |
| Primer Name | Primer Sequence (5′ to 3′) | Target |
|---|---|---|
| cpxA-up-F | GAGCGGATAACAATTTGTGGAATCCCGGGAAACATTTAAGTCAGGAAGTGCTGGG | For cpxA deletion mutant |
| cpxA-up-R | TTCGACAATCGGATCGTTCGCAAGTTCAGCTTCTA | For cpxA deletion mutant |
| cpxA-down-F | CGAACGATCCGATTGTCGAAAGCGCCATGCAGCAG | For cpxA deletion mutant |
| cpxA-down-R | AGCGGAGTGTATATCAAGCTTATCGATACCATCTCTTGAGGAGCTTTGGGAGCGG | For cpxA deletion mutant |
| cpxA-out-F | GCTCCTCGAAATGGAAGGTTTTAAT | For cpxA deletion mutant |
| cpxA-out-R | GCTCGCTACAAGTGGGTGAAGAAGG | For cpxA deletion mutant |
| cpxR-up-F | GAGCGGATAACAATTTGTGGAATCCCGGGAAAAAAACTGAATGCCAGCGTTGAGG | For cpxR deletion mutant |
| cpxR-up-R | GTTAGAAATATCGATGCTGTCATCCAAAAGCTCAA | For cpxR deletion mutant |
| cpxR-down-F | ACAGCATCGATATTTCTAACCTGCGCCGCAAACTG | For cpxR deletion mutant |
| cpxR-down-R | AGCGGAGTGTATATCAAGCTTATCGATACCCGTCCTTCAGAGGTCACCAGTAATA | For cpxR deletion mutant |
| cpxR-out-F | TAACCAGCCGTCCATAGGTTTGATT | For cpxR deletion mutant |
| cpxR-out-R | CGACATCACCAGCAGGTCATTGATC | For cpxR deletion mutant |
| clcA-up-F | GAGCGGATAACAATTTGTGGAATCCCGGGAATATGCTTTGCCATCGACATCGTAC | For clcA deletion mutant |
| clcA-up-R | CTGGTAGTTAATTAACCGGCGAATCTGATCTCTGC | For clcA deletion mutant |
| clcA-down-F | GCCGGTTAATTAACTACCAGCTCATTTTGCCAATG | For clcA deletion mutant |
| clcA-down-R | AGCGGAGTGTATATCAAGCTTATCGATACCCCTTCGTTAACCTGATCGTCAAAGG | For clcA deletion mutant |
| clcA-out-F | AGACAGTCTGCGTGGCCGTGGTAGC | For clcA deletion mutant |
| clcA-out-R | TACGCTTATCGGGCCTGGAACATCT | For clcA deletion mutant |
| casC-up-F | GAGCGGATAACAATTTGTGGAATCCCGGGATGATAAGCAGCAAAAATTCGCCGCA | For casC deletion mutant |
| casC-up-R | CATTCATATTCAGGCTCTGAGAGGAGATACGCAGA | For casC deletion mutant |
| casC-down-F | TCAGAGCCTGAATATGAATGAGGTCTATGCACAGG | For casC deletion mutant |
| casC-down-R | AGCGGAGTGTATATCAAGCTTATCGATACCGCACAGCAAAAATTGGTAATGACGA | For casC deletion mutant |
| casC-out-F | CTGCTTGTCTGAGGGATTTCGCTCC | For casC deletion mutant |
| casC-out-R | AGCGGCAACGCCAGAGGGTGACTTT | For casC deletion mutant |
| casE-up-F | GAGCGGATAACAATTTGTGGAATCCCGGGACTTACACGTTGGCGCAGCTTCAGAC | For casE deletion mutant |
| casE-up-R | GCCAGACGTTTGCTGCCGGGAAAGAGATCCCATAA | For casE deletion mutant |
| casE-down-F | CCCGGCAGCAAACGTCTGGCGCAGGGGTACGGTAA | For casE deletion mutant |
| casE-down-R | AGCGGAGTGTATATCAAGCTTATCGATACCAGGAATAGACCCGCACTCCCGCCTC | For casE deletion mutant |
| casE-out-F | GGGATTATCACACGGTGCAGATGCC | For casE deletion mutant |
| casE-out-R | CGGCGATCTCAAATGCTTTGGGCAC | For casE deletion mutant |
| gyrB-q-F | TGATTGCGGTGGTTTCCGTA | qRT-PCR |
| gyrB-q-R | GACGACGATTTTCGCGTCAG | qRT-PCR |
| cpxA-q-F | AAGCTGAACTTGCGAACGAT | qRT-PCR |
| cpxA-q-R | CATCTCTACGCGGCCATATT | qRT-PCR |
| cpxR-q-F | TTGATGATGACCGAGAGCTG | qRT-PCR |
| cpxR-q-R | TACCGTTTTTCTTCGGCATC | qRT-PCR |
| clcA-q-F | CTCAGCAAATTGTGCGCTTA | qRT-PCR |
| clcA-q-R | CCAGTAACGCCGAAAGGATA | qRT-PCR |
| lysP-q-F | ACAACTGGGCGGTGACTATC | qRT-PCR |
| lysP-q-R | TACGCCAACGATGATGAAGA | qRT-PCR |
| phoQ-q-F | CTCGCCAAATGGGAAAATAA | qRT-PCR |
| phoQ-q-R | CATTCCGGTTGAATGCTTTT | qRT-PCR |
| phoP-q-F | TGCGCGTACTGGTTGTAGAG | qRT-PCR |
| phoP-q-R | TCATCCGGCAGACCTAAATC | qRT-PCR |
| degP-q-F | GTATGCCGCGTAATTTCCAG | qRT-PCR |
| degP-q-R | GAATTTACGCCCATCGCTAA | qRT-PCR |
| casE-q-F | TTATCGCCGCGAAGAGTTAC | qRT-PCR |
| casE-q-R | GGCTATCACCCTGCGTTTTA | qRT-PCR |
| mgrB-q-F | AATTTCGATGGGTCGTTCTC | qRT-PCR |
| mgrB-q-R | CGCAAATACCGCTGAAAAAT | qRT-PCR |
| crcB-q-F | CCCGGTGGATGCTAAGTATG | qRT-PCR |
| crcB-q-R | GGTCGTAATGAGCACTTTCCA | qRT-PCR |
| casC-q-F | AGAACATCGCCAACTGCTTT | qRT-PCR |
| casC-q-R | CAGTTCCGCTTCCTCTTCTG | qRT-PCR |
| Features | Value |
|---|---|
| Total reading base pairs (bp) | 4,914,080 |
| Scaffold number | 11 |
| N50 (bp) | 4,073,894 |
| GC content (%) | 52.03 |
| tRNAs | 84 |
| rRNAs | 22 |
| Protein-coding sequences | 4579 |
| Gene | Description | log2FC |
|---|---|---|
| T141_00187 | Sensor histidine kinase, CpxA | −1.08 |
| T141_02433 | Virulence transcriptional regulatory protein, PhoP | −1.07 |
| T141_02432 | Virulence sensor histidine kinase, PhoQ | −0.99 |
| T141_03065 | PhoP/PhoQ regulator, MgrB | −1.72 |
| T141_01611 | Sigma-E factor regulatory protein, RseB | −1.01 |
| T141_02070 | Lysine-specific permease, LysP | −1.73 |
| T142_00594 | Periplasmic serine endoprotease, DegP | −0.86 |
| T141_01333 | CRISPR system Cascade subunit, CasC | −1.41 |
| T141_01335 | CRISPR system Cascade subunit, CasE | −1.65 |
| T141_00366 | Low affinity potassium transport system protein, kup | −3.8 |
| T141_03609 | Putative fluoride ion transporter, CrcB | −2.03 |
| T142_00588 | H(+)/Cl(-) exchange transporter, ClcA | −1.7 |
| T142_00709 | Glutamate 5-kinase, ProB | −1.67 |
| T141_00659 | Endoglucanase, BcsZ | −1.42 |
| T142_00710 | γ-glutamyl phosphate reductase, ProA | −1.38 |
| T141_01331 | hypothetical protein | −1.15 |
| T141_00565 | hypothetical protein | −1.06 |
| T141_02199 | D-inositol-3-phosphate glycosyltransferase, MshA | 1.09 |
| T141_02191 | dTDP-4-dehydrorhamnose 3%2C5-epimerase, RfbC | 1.74 |
| T141_02189 | dTDP-4-dehydrorhamnose reductase, RfbD | 1.98 |
| T141_02193 | Glucose-1-phosphate cytidylyltransferase, RfbF | 2.29 |
| T141_02194 | CDP-glucose 4%2C6-dehydratase, RfbG | 2.02 |
| T141_02196 | CDP-abequose synthase, RfbJ | 2.64 |
| T141_02201 | Mannose-1-phosphate guanylyltransferase, RfbM | 1.23 |
| T141_02198 | Abequosyltransferase, RfbV | 2.37 |
| T141_01096 | Cell division protein, FtsP | 1.54 |
| T141_00272 | Sec-independent protein translocase protein, TatC | 1.81 |
| T141_02202 | Phosphoglucosamine mutase, GlmM | 1.9 |
| T141_02192 | CDP-6-deoxy-L-threo-D-glycero-4-hexulose-3-dehydrase reductase, AscD | 2.05 |
| T141_00569 | D-inositol-3-phosphate glycosyltransferase, MshA | 2.09 |
| T141_02195 | dTDP-4-dehydro-2%2C6-dideoxy-D-glucose 3-dehydratase, SpnO | 2.12 |
| T141_03062 | Peptidoglycan D%2CD-transpeptidase, FtsI | 2.41 |
| T141_02203 | hypothetical protein | 1.58 |
| T141_00570 | hypothetical protein | 1.7 |
| T141_02200 | hypothetical protein | 1.83 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gu, D.; Xue, H.; Yuan, X.; Yu, J.; Xu, X.; Huang, Y.; Li, M.; Zhai, X.; Pan, Z.; Zhang, Y.; et al. Genome-Wide Identification of Genes Involved in Acid Stress Resistance of Salmonella Derby. Genes 2021, 12, 476. https://doi.org/10.3390/genes12040476
Gu D, Xue H, Yuan X, Yu J, Xu X, Huang Y, Li M, Zhai X, Pan Z, Zhang Y, et al. Genome-Wide Identification of Genes Involved in Acid Stress Resistance of Salmonella Derby. Genes. 2021; 12(4):476. https://doi.org/10.3390/genes12040476
Chicago/Turabian StyleGu, Dan, Han Xue, Xiaohui Yuan, Jinyan Yu, Xiaomeng Xu, Yu Huang, Mingzhu Li, Xianyue Zhai, Zhiming Pan, Yunzeng Zhang, and et al. 2021. "Genome-Wide Identification of Genes Involved in Acid Stress Resistance of Salmonella Derby" Genes 12, no. 4: 476. https://doi.org/10.3390/genes12040476
APA StyleGu, D., Xue, H., Yuan, X., Yu, J., Xu, X., Huang, Y., Li, M., Zhai, X., Pan, Z., Zhang, Y., & Jiao, X. (2021). Genome-Wide Identification of Genes Involved in Acid Stress Resistance of Salmonella Derby. Genes, 12(4), 476. https://doi.org/10.3390/genes12040476






