From Laboratory towards Industrial Operation: Biomarkers for Acidophilic Metabolic Activity in Bioleaching Systems
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains and Media
2.2. Bioleaching Column Test
2.3. Industrial Bioleaching Heap
2.4. Candidate Genes Selection and Primer Design
2.5. RNA Isolation and Transcriptional Analysis
3. Results
3.1. Cell Harvesting, DNA, and RNA Extraction
3.2. Biomarkers for CO2 Availability in Acidithiobacillus spp.
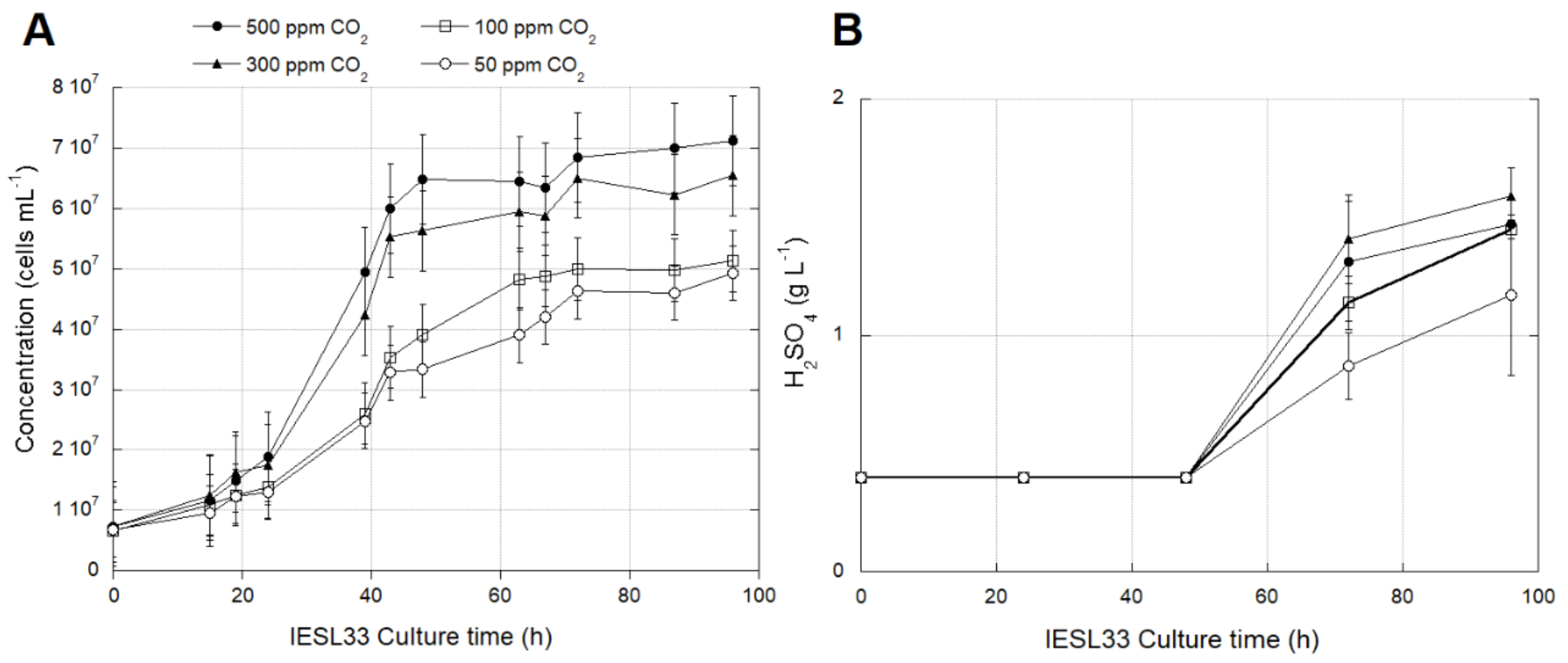
3.2.1. Effects of CO2 Availability on Growth and Activity of At. thiooxidans Strain IESL-33
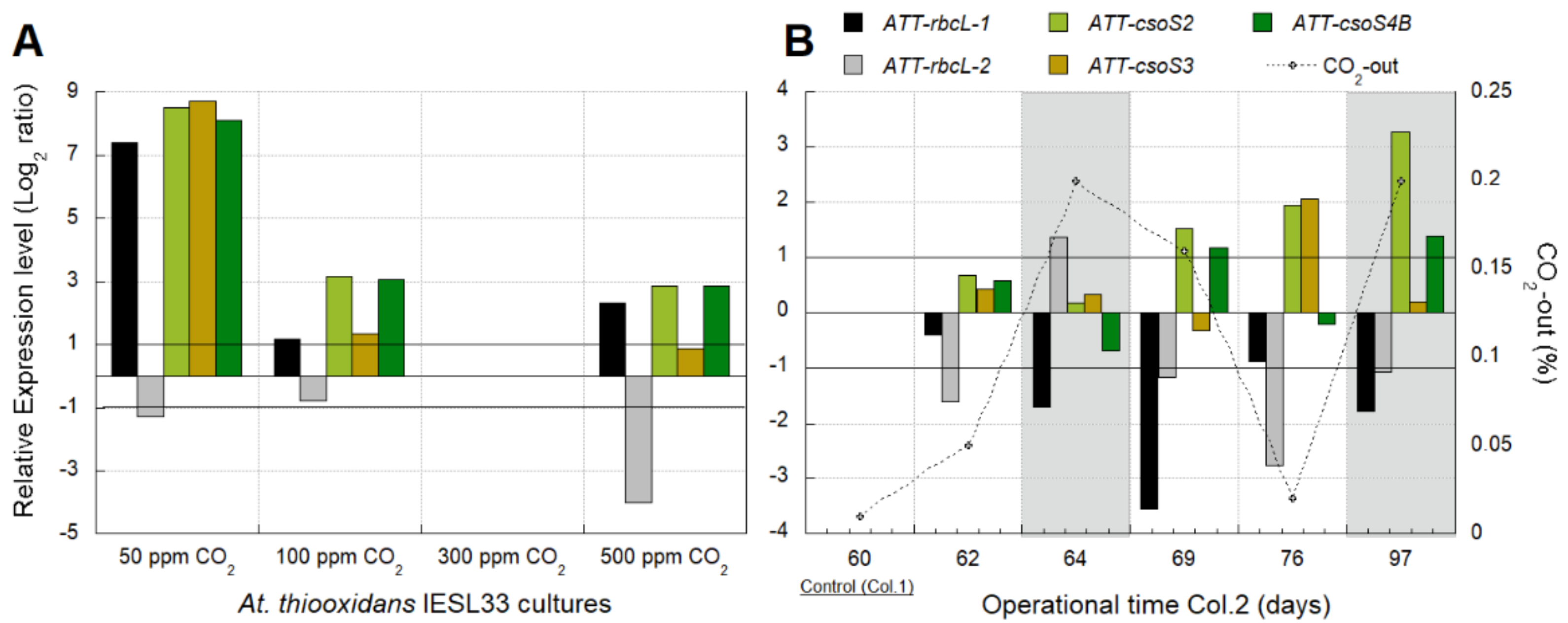
3.2.2. Identification of Transcriptomic Markers Associated with CO2 Availability
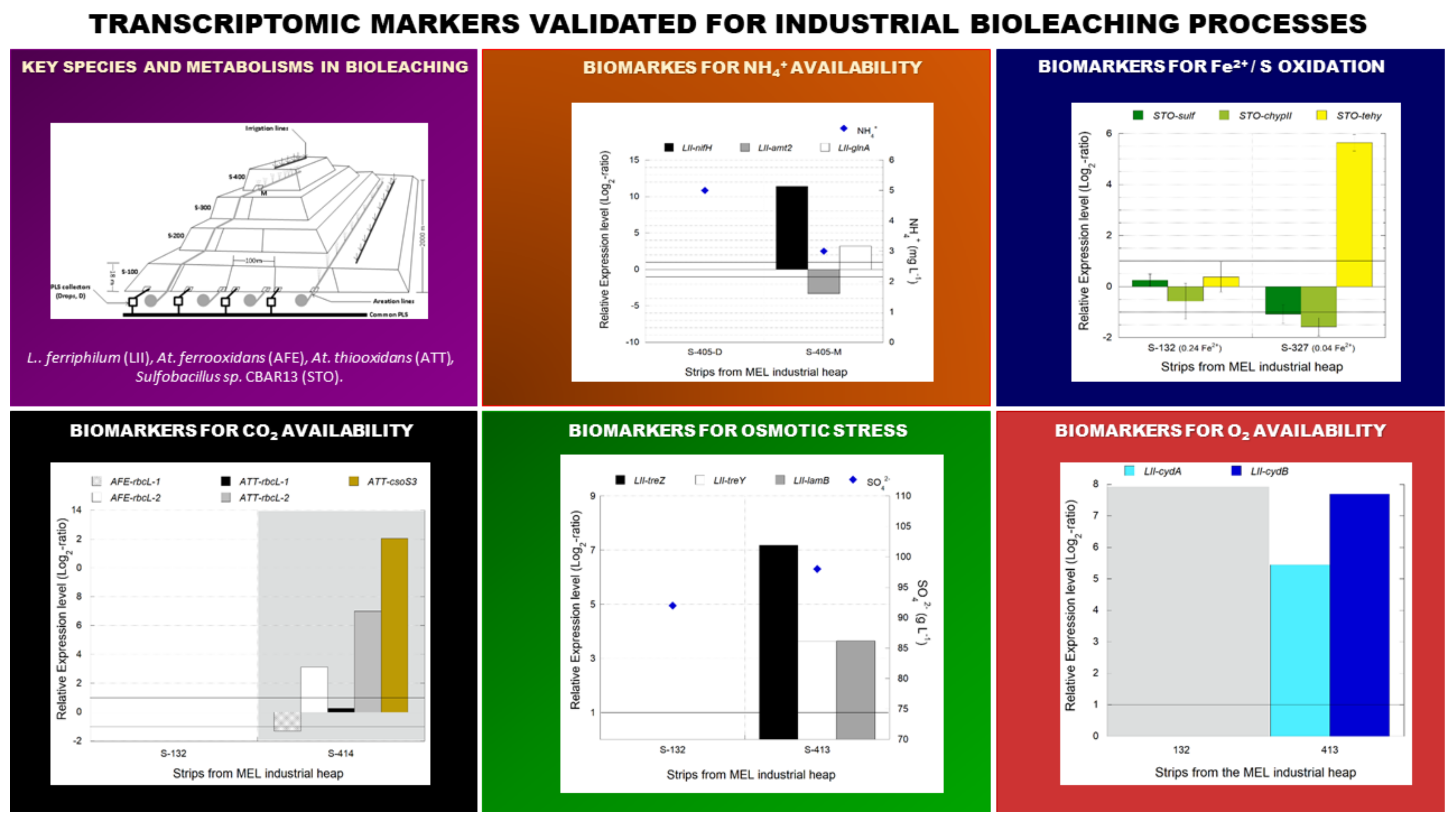
3.2.3. Industrial Validation of Transcriptomic Markers Associated with CO2 Fixation
3.3. Biomarkers for N2 Availability in L. ferriphilum
3.3.1. Identification of Transcriptomic Markers Associated with N2 Availability
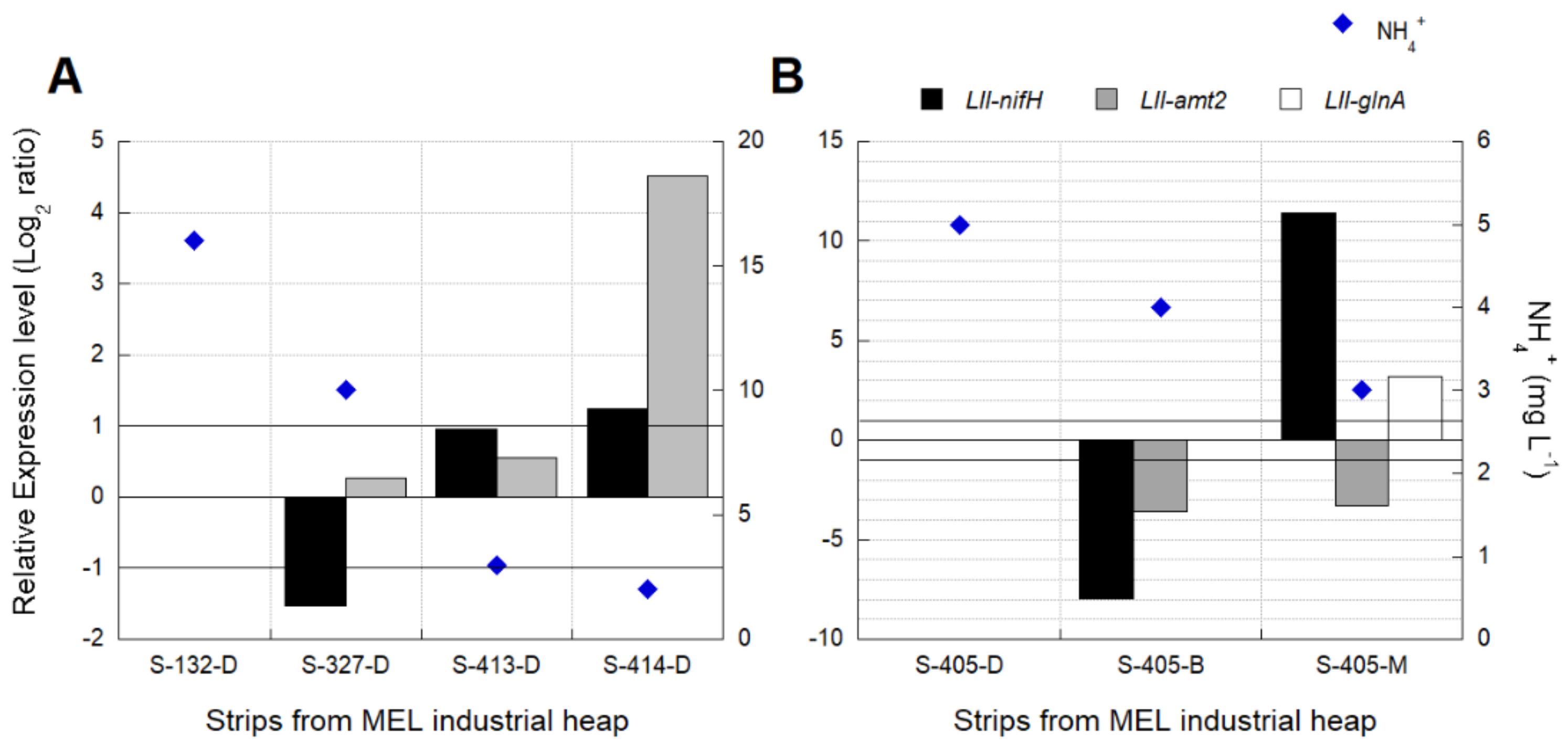
3.3.2. Industrial Validation of Transcriptomic Markers Associated with N2 Metabolism
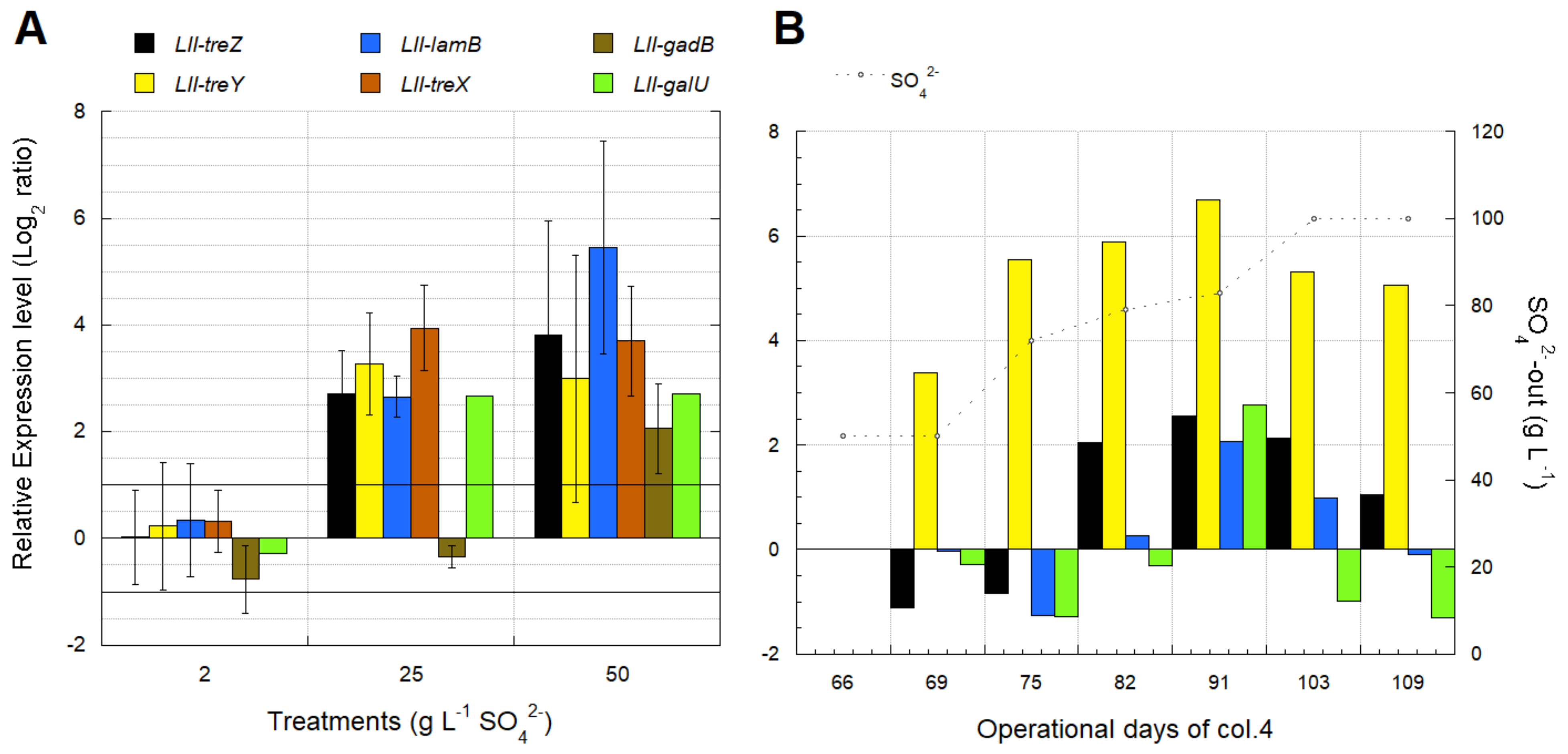
3.4. Biomarkers for Osmotic Stress in L. ferriphilum
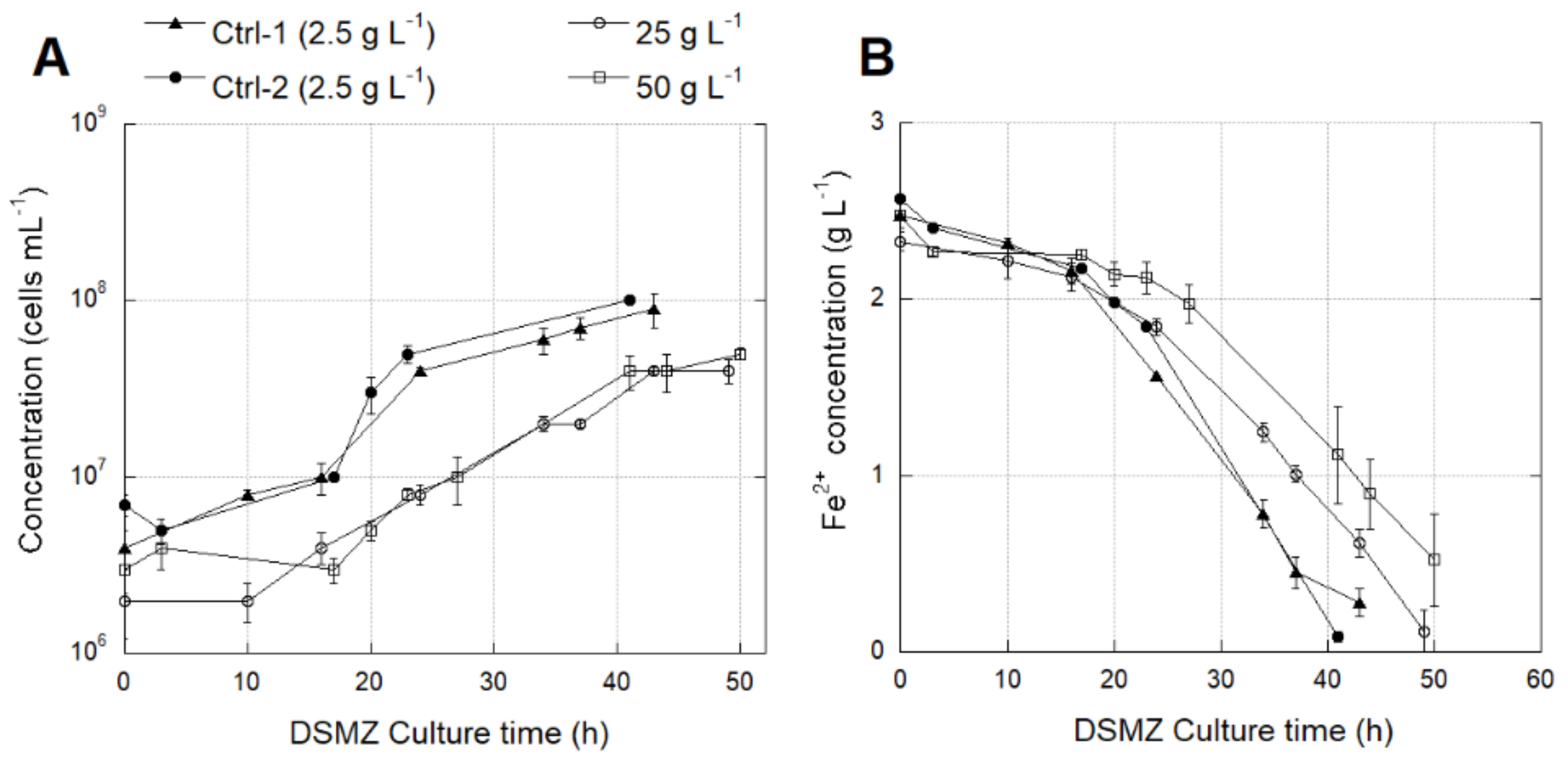
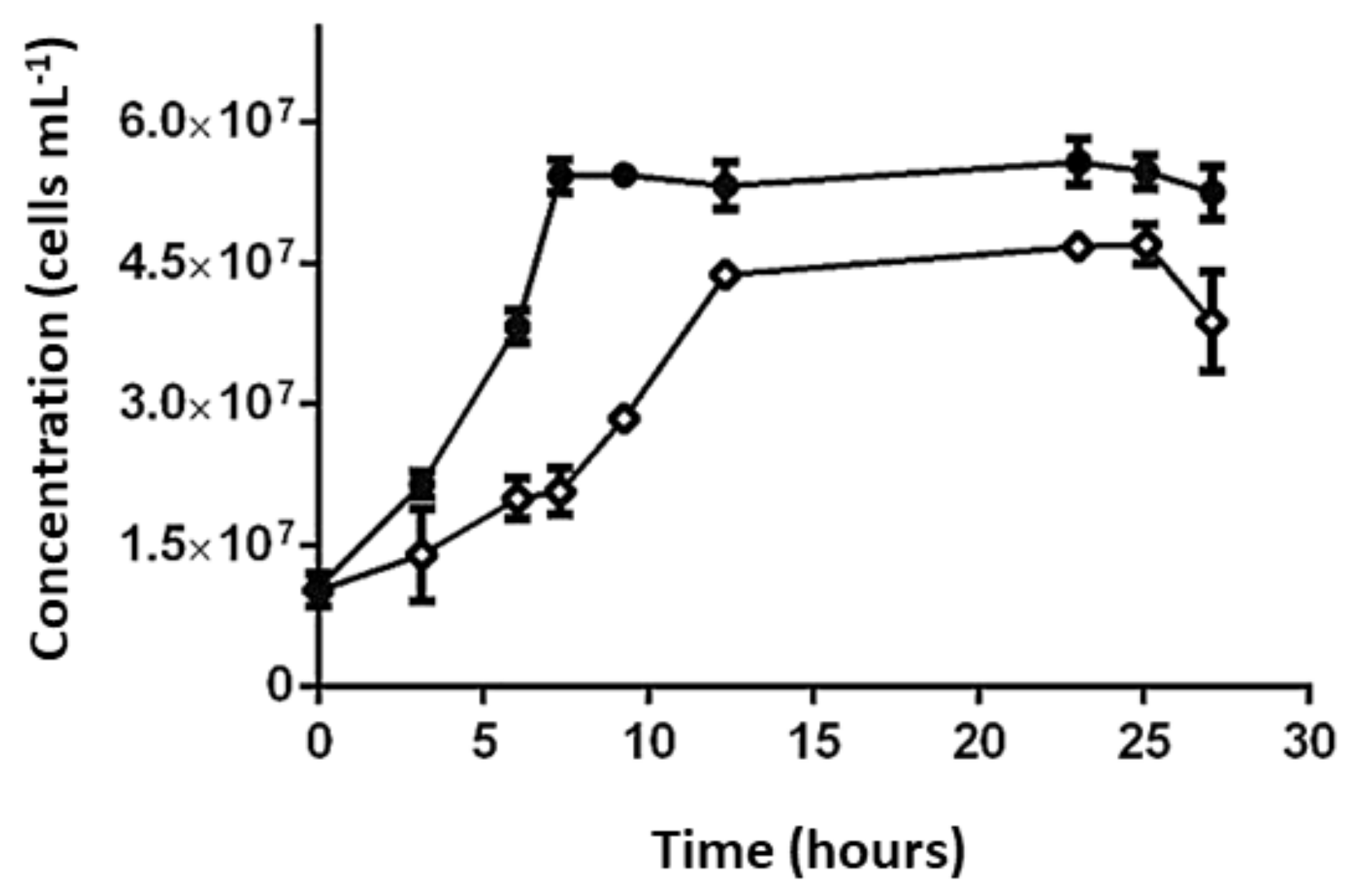
3.4.1. Effect of SO42− Concentration on Growth and Activity of L. ferriphilum Strains DSMZ 14647 and IESL-25
3.4.2. Identification of Transcriptomic Markers Associated with Osmotic Stress
3.4.3. Industrial Validation of Transcriptomic Markers Associated with Osmotic Stress
3.5. Biomarkers for Iron and Sulfur Oxidation in Sulfobacillus sp. CBAR13
3.5.1. Effect of the Energy Source on Growth of Sulfobacillus sp.
3.5.2. Identification of Transcriptomic Markers Associated with Ferrous and Sulfur Oxidation
3.5.3. Industrial Validation of Transcriptomic Markers Associated with Ferrous and Sulfur Oxidation
4. Discussion
4.1. Carbon Fixation
4.2. Nitrogen Metabolism
4.3. Osmotic Stress
4.4. Ferrous and Sulfur Oxidation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fundación-Chile. From Copper to Innovation: Technological Roadmap 2015–2035; A Impresores: Santiago, Chile, 2016; p. 387. ISBN 978-956-8200-30-5. [Google Scholar]
- Velasquez-Yevenes, L.; Torres, D.; Toro, N. Leaching of chalcopyrite ore agglomerated with high chloride concentration and high curing periods. Hydrometallurgy 2018, 181, 215–220. [Google Scholar] [CrossRef]
- Rautenbach, G. Heap Leaching of Copper. U.S. Patent No. 10041143, 7 August 2018. [Google Scholar]
- Kelly, G.; Aguilera, S.; Chico, V. Transition from traditional leaching to chloride leaching—Costs and impacts on infrastructure. In Proceedings of the Hydroprocess, Santiago, Chile, 26–28 October 2020. [Google Scholar]
- Hajdu-Rahkama, R.; Ahoranta, S.; Lakaniemi, A.M.; Puhakka, J.A. Effects of elevated pressures on the activity of acidophilic bioleaching microorganisms. Biochem. Eng. J. 2019, 150, 107286. [Google Scholar] [CrossRef]
- Acosta, M.; Galleguillos, P.; Ghorbani, Y.; Tapia, P.; Contador, Y.; Velasquez, A.; Espoz, C.; Pinilla, C.; Demergasso, C. Variation in microbial community from predominantly mesophilic to thermotolerant and moderately thermophilic species in an industrial copper heap bioleaching operation. Hydrometallurgy 2014, 150, 281–289. [Google Scholar] [CrossRef]
- Cautivo, D.; Soto, P.; Zepeda, V.; Galleguillos, P.; Velásquez, A.; Pinilla, C.; Demergasso, C. Estimation of ionic load effect on the oxidizing activity of the microbial population in the heap bioleaching process at Escondida Mine. Adv. Mater. Res. 2013, 825, 219–222. [Google Scholar] [CrossRef]
- Galleguillos, P.A.; Music, V.; Acosta, M.; Salazar, C.N.; Quatrini, R.; Shmaryahu, A.; Holmes, D.S.; Velásquez, A.; Espoz, C.; Pinilla, C.; et al. Temporal Dynamics of Genes Involved in Metabolic Pathways of C and N of L. ferriphilum, in the Industrial Bioleaching Process of Escondida Mine, Chile. Adv. Mater. Res. 2013, 825, 162–165. [Google Scholar] [CrossRef]
- Marin, S.; Acosta, M.; Galleguillos, P.; Chibwana, C.; Strauss, H.; Demergasso, C. Is the growth of microorganisms limited by carbon availability during chalcopyrite bioleaching? Hydrometallurgy 2017, 168, 13–20. [Google Scholar] [CrossRef]
- Zepeda, V.; Cautivo, D.; Galleguillos, P.; Salazar, C.; Velásquez, A.; Pinilla, C.; Demergasso, C. Effect of increased acid concentration on the microbial population inhabiting an industrial heap bioleaching plant. Adv. Mater. Res. 2013, 825, 348–351. [Google Scholar] [CrossRef]
- Cortes, M.; Marin, S.; Galleguillos, P.; Cautivo, D.; Demergasso, C. Validation of Genetic Markers Associated to Oxygen Availability in Low-Grade Copper Bioleaching Systems: An Industrial Application. Front. Microbiol. 2019, 10, 1841. [Google Scholar] [CrossRef]
- Remonsellez, F.; Galleguillos, F.; Moreno-Paz, M.; Parro, V.; Acosta, M.; Demergasso, C. Dynamic of active microorganisms inhabiting a bioleaching industrial heap of low-grade copper sulfide ore monitored by real-time PCR and oligonucleotide prokaryotic acidophile microarray. Microb. Biotechnol. 2009, 2, 613–624. [Google Scholar] [CrossRef]
- Roberto, F.F. Copper Heap Bioleach Microbiology—Progress and Challenges. Solid State Phenom. 2017, 262, 250–254. [Google Scholar] [CrossRef]
- Watling, H.R.; Watkin, E.L.J.; Ralph, D.E. The resilience and versatility of acidophiles that contribute to the bio-assisted extraction of metals from mineral sulphides. Environ. Technol. 2011, 31, 915–933. [Google Scholar] [CrossRef]
- Demergasso, C.; Galleguillos, F.; Soto, P.; Serón, M.; Iturriaga, V. Microbial succession during a heap bioleaching cycle of low grade copper sulfides: Does this knowledge mean a real input for industrial process design and control? Hydrometallurgy 2010, 104, 382–390. [Google Scholar] [CrossRef]
- Demergasso, C.; Galleguillos, P.; Escudero, L.; Zepeda, V.; Castillo, D.; Casamayor, E. Molecular characterization of microbial populations in a low-grade copper ore bioleaching test heap. Hydrometallurgy 2005, 80, 241–253. [Google Scholar] [CrossRef]
- Roberto, F.F. 16S-rRNA gene-targeted amplicon sequence analysis of an enargite-dominant bioleach demonstration in Peru. Hydrometallurgy 2018, 180, 271–276. [Google Scholar] [CrossRef]
- Ramirez-Flandes, S.; Gonzalez, B.; Ulloa, O. Redox traits characterize the organization of global microbial communities. Proc. Natl. Acad. Sci. USA 2019, 116, 3630–3635. [Google Scholar] [CrossRef] [PubMed]
- Escalas, A.; Hale, L.; Voordeckers, J.W.; Yang, Y.F.; Firestone, M.K.; Alvarez-Cohen, L.; Zhou, J.Z. Microbial functional diversity: From concepts to applications. Ecol. Evol. 2019, 9, 12000–12016. [Google Scholar] [CrossRef]
- Techtmann, S.M.; Hazen, T.C. Metagenomic applications in environmental monitoring and bioremediation. J. Ind. Microbiol. Biotechnol. 2016, 43, 1345–1354. [Google Scholar] [CrossRef] [PubMed]
- Buetti-Dinh, A.; Herold, M.; Christel, S.; El Hajjami, M.E.; Delogu, F.; Ilie, O.; Bellenberg, S.; Wilmes, P.; Poetsch, A.; Sand, W.; et al. Reverse engineering directed gene regulatory networks from transcriptomics and proteomics data of biomining bacterial communities with approximate Bayesian computation and steady-state signalling simulations. BMC Bioinform. 2020, 21, 23. [Google Scholar] [CrossRef] [PubMed]
- Esparza, M.; Jedlicki, E.; Gonzalez, C.; Dopson, M.; Holmes, D.S. Effect of CO2 Concentration on Uptake and Assimilation of Inorganic Carbon in the Extreme Acidophile Acidithiobacillus ferrooxidans. Front. Microbiol. 2019, 10, 603. [Google Scholar] [CrossRef] [PubMed]
- Demergasso, C.; Veliz, R.; Galleguillos, P.; Marin, S.; Acosta, M.; Zepeda, V.; Zeballos, J.; Henriquez, F.; Pizarro, R.; Sekios-Calfa, J. Decision support system for bioleaching processes. Hydrometallurgy 2018, 181, 113–122. [Google Scholar] [CrossRef]
- Véliz, R.; Demergasso, C.; Galleguillos, P.A.; Marín, S.; Acosta, M. Molecular microbial indicators for industrial bioleaching operations and its implementation in a Decision support system. In Proceedings of the International Biohydrometallurgy Symposium, Fukuoka, Japan, 21–23 October 2019. [Google Scholar]
- Sutton, R.T.; Pincock, D.; Baumgart, D.C.; Sadowski, D.C.; Fedorak, R.N.; Kroeker, K.I. An overview of clinical decision support systems: Benefits, risks, and strategies for success. NPJ Digit. Med. 2020, 3, 17. [Google Scholar] [CrossRef]
- Galleguillos, P. Biodiversity and Stress Response of Extremophilic Prokaryotes Isolated from the Escondida Copper Mine, Chile. Ph.D. Thesis, Bangor University, Gwynedd, UK, 2011. [Google Scholar]
- Norris, P.R.; Clark, D.A.; Owen, J.P.; Waterhouse, S. Characteristics of Sulfobacillus acidophilus sp. nov. and other moderately thermophilic mineral-sulphide-oxidizing bacteria. Microbiology 1996, 142, 775–783. [Google Scholar] [CrossRef] [PubMed]
- Bryan, C.G.; Davis-Belmar, C.S.; van Wyk, N.; Fraser, M.K.; Dew, D.; Rautenbach, G.F.; Harrison, S.T.L. The effect of CO2 availability on the growth, iron oxidation and CO2-fixation rates of pure cultures of Leptospirillum ferriphilum and Acidithiobacillus ferrooxidans. Biotechnol. Bioeng. 2012, 109, 1693–1703. [Google Scholar] [CrossRef] [PubMed]
- Davis-Belmar, C.S.; Nicolle, J.L.C.; Norris, P.R. Ferrous iron oxidation and leaching of copper ore with halotolerant bacteria in ore columns. Hydrometallurgy 2008, 94, 144–147. [Google Scholar] [CrossRef]
- Paipa, C.; Mateo, M.; Godoy, I.; Poblete, E.; Toral, M.; Vargas, T. Comparative study of alternative methods for the simultaneous determination of Fe+3 and Fe+2 in leaching solutions and in acid mine drainages. Miner. Eng. 2005, 18, 1116–1119. [Google Scholar] [CrossRef]
- Weisberg, S. Applied Linear Regression, 4th ed.; Barnett, V.S.H., Hunter, J.S., Kendall, D.G., Teugels, J.L., Eds.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2014; p. 368. [Google Scholar]
- Demergasso, C.; Iturriaga, V.; Galleguillos, P.A.; Davis, S. Effect of SX on microbial populations inhabiting in solutions of a heap bioleaching process. In Proceedings of the 19th International Biohydrometallurgy Symposium, Biohydrometallurgy: Biotech Key to Unlock Mineral Resource Value, Changsha, China, 18–22 September 2011; p. 1015. [Google Scholar]
- Makaula, D.X.; Huddy, R.J.; Fagan-Endres, M.A.; Harrison, S.T.L. Using isothermal microcalorimetry to measure the metabolic activity of the mineral-associated microbial community in bioleaching. Miner. Eng. 2017, 106, 33–38. [Google Scholar] [CrossRef]
- Zepeda, V.J.; Galleguillos, F.; Urtuvia, V.; Molina, J.; Demergasso, C. Comparison between the bacterial populations from solutions and minerals in 1 m test columns and the industrial low grade copper sulphide bioleaching process in the Escondida Mine, Chile. Adv. Mater. Res. 2009, 71–73, 63–66. [Google Scholar] [CrossRef]
- Bellenberg, S.; Barthen, R.; Boretska, M.; Zhang, R.; Sand, W.; Vera, M. Manipulation of pyrite colonization and leaching by iron-oxidizing Acidithiobacillus species. Appl. Microbiol. Biotechnol. 2015, 99, 1435–1449. [Google Scholar] [CrossRef]
- Demergasso, C.; Shmaryahu, A.; Quatrini, R.; Galleguillos, P.; Acosta Grinok, M.; Holmes, D. DNA Microarray for the Quantitative Analysis of Gene Expression from Microorganisms Inhabiting Liquid and Solid Samples of Industrial Bioleaching Processes; Method for Analyzing Gene Expression Using the Microarray. Chile Patent No. 58023, 23 September 2019. [Google Scholar]
- Acosta, M.; Galleguillos, P.; Guajardo, M.; Demergasso, C. Microbial survey on industrial bioleaching heap by high-throughput 16S sequencing and metagenomics analysis. Solid State Phenom. 2017, 262, 219–223. [Google Scholar] [CrossRef]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid annotations using subsystems technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T.L. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 2012, 13, 134. [Google Scholar] [CrossRef] [PubMed]
- Svec, D.; Tichopad, A.; Novosadova, V.; Pfaffl, M.W.; Kubista, M. How good is a PCR efficiency estimate: Recommendations for precise and robust qPCR efficiency assessments. Biomol. Detect. Quantif. 2015, 3, 9–16. [Google Scholar] [CrossRef]
- Pfaffl, M.W.; Horgan, G.W.; Dempfle, L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real time PCR. Nucleic Acid Res. 2002, 30, e36. [Google Scholar] [CrossRef]
- Andersen, C.; Jensen, J.; Orntoft, T. Normalization of Real-Time quantitative transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cáncer data sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef] [PubMed]
- Klein, M.G.; Zwart, P.; Bagby, S.C.; Cai, F.; Chisholm, S.W.; Heinhorst, S.; Cannon, G.C.; Kerfeld, C.A. Identification and structural analysis of a novel carboxysome shell protein with implications for metabolite transport. J. Mol. Biol. 2009, 392, 319–333. [Google Scholar] [CrossRef] [PubMed]
- Petersen, J.; Minnaar, S.; du Plessis, C. Carbon dioxide and oxygen consumption during the bioleaching of a copper ore in a large isothermal column. Hydrometallurgy 2010, 104, 356–362. [Google Scholar] [CrossRef]
- Acevedo, F.; Gentina, J.; García, N. CO2 supply in the biooxidation of an enargite-pyrite gold concentrate. Biotechnol. Lett. 1998, 20, 257–259. [Google Scholar]
- Kurosawa, H.; Konno, Y.; Nakamura, K.; Amano, Y. Estimation of the CO2 fixation ability of Thiobacillus thiooxidans JCM 7814. J. Ferment. Bioeng. 1993, 75, 71–72. [Google Scholar] [CrossRef]
- Toyoda, K.; Yoshizawa, Y.; Arai, H.; Ishii, M.; Igarashi, Y. The role of two CbbRs in the transcriptional regulation of three ribulose-1,5-bisphosphate carboxylase/oxygenase genes in Hydrogenovibrio marinus strain MH-110. Microbiol. SGM 2005, 151, 3615–3625. [Google Scholar] [CrossRef] [PubMed]
- Badger, M.R.; Bek, E.J. Multiple Rubisco forms in proteobacteria: Their functional significance in relation to CO2 acquisition by the CBB cycle. J. Exp. Bot. 2008, 59, 1525–1541. [Google Scholar] [CrossRef] [PubMed]
- Valdes, J.; Cardenas, J.P.; Quatrini, R.; Esparza, M.; Osorio, H.; Duarte, F.; Lefimil, C.; Sepulveda, R.; Jedlicki, E.; Holmes, D.S. Comparative genomics begins to unravel the ecophysiology of bioleaching. Hydrometallurgy 2010, 104, 471–476. [Google Scholar] [CrossRef]
- Ahoranta, S.H.; Peltola, M.K.; Lakaniemi, A.M.; Puhakka, J.A. Enhancing the activity of iron-oxidising bacteria: A case study with process liquors from heap bioleaching of a complex sulphide ore. Hydrometallurgy 2017, 167, 163–172. [Google Scholar] [CrossRef]
- Goltsman, D.S.; Dasari, M.; Thomas, B.C.; Shah, M.B.; VerBerkmoes, N.C.; Hettich, R.L.; Banfield, J.F. New group in the Leptospirillum clade: Cultivation-independent community genomics, proteomics, and transcriptomics of the new species “Leptospirillum group IV UBA BS”. Appl. Environ. Microbiol. 2013, 79, 5384–5393. [Google Scholar] [CrossRef] [PubMed]
- Goltsman, D.S.; Denef, V.J.; Singer, S.W.; VerBerkmoes, N.C.; Lefsrud, M.; Mueller, R.S.; Dick, G.J.; Sun, C.L.; Wheeler, K.E.; Zemla, A.; et al. Community genomic and proteomic analyses of chemoautotrophic iron-oxidizing “Leptospirillum rubarum” (Group II) and “Leptospirillum ferrodiazotrophum” (Group III) bacteria in acid mine drainage biofilms. Appl. Environ. Microbiol. 2009, 75, 4599–4615. [Google Scholar] [CrossRef] [PubMed]
- Issotta, F.; Galleguillos, P.A.; Moya-Beltran, A.; Davis-Belmar, C.S.; Rautenbach, G.; Covarrubias, P.C.; Acosta, M.; Ossandon, F.J.; Contador, Y.; Holmes, D.S.; et al. Draft genome sequence of chloride-tolerant Leptospirillum ferriphilum Sp-Cl from industrial bioleaching operations in northern Chile. Stand. Genom. Sci. 2016, 11, 19. [Google Scholar] [CrossRef]
- Galleguillos, P.; Demergasso, C.S.; Johnson, D.B.; Quatrini, R.; Holmes, D.S.; Hallberg, K.B. Identification and analysis of diazotrophy in strains of Leptospirillum ferriphilum from heap bioleaching operations. In Proceedings of the Biohydrometallurgy: Biotech Key to Unlock Mineral Resources Value of the 19th International Biohydrometallurgy Symposium, Changsha, China, 18–22 September 2011; p. 1015. [Google Scholar]
- Csonka, L.N. Physiological and genetic responses of bacteria to osmotic-stress. Microbiol. Rev. 1989, 53, 121–147. [Google Scholar] [CrossRef] [PubMed]
- Galleguillos, P.A.; Grail, B.M.; Hallberg, K.B.; Demergasso, C.S.; Johnson, D.B. Identification of trehalose as a compatible solute in different species of acidophilic bacteria. J. Microbiol. 2018, 56, 727–733. [Google Scholar] [CrossRef]
- Cai, X.; Seitl, I.; Mu, W.M.; Zhang, T.; Stressler, T.; Fischer, L.; Jiang, B. Biotechnical production of trehalose through the trehalose synthase pathway: Current status and future prospects. Appl. Microbiol. Biotechnol. 2018, 102, 2965–2976. [Google Scholar] [CrossRef]
- Maruta, K.; Kubota, M.; Fukuda, S.; Kurimoto, M. Cloning and nucleotide sequence of a gene encoding a glycogen debranching enzyme in the trehalose operon from Arthrobacter sp Q36. Biochim. Biophys. Acta Protein Struct. Mol. Enzymol. 2000, 1476, 377–381. [Google Scholar] [CrossRef]
- Padilla, L.; Morbach, S.; Kramer, R.; Agosin, E. Impact of heterologous expression of Escherichia coli UDP-glucose pyrophosphorylase on trehalose and glycogen synthesis in Corynebacterium glutamicum. Appl. Environ. Microbiol. 2004, 70, 3845–3854. [Google Scholar] [CrossRef] [PubMed]
- Carpinelli, J.; Kramer, R.; Agosin, E. Metabolic engineering of Corynebacterium glutamicum for trehalose overproduction: Role of the TreYZ trehalose biosynthetic pathway. Appl. Environ. Microbiol. 2006, 72, 1949–1955. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Chandra, G.; Chater, K.F.; Bornemann, S. Unexpected and widespread connections between bacterial glycogen and trehalose metabolism. Microbiology 2011, 157, 1565–1572. [Google Scholar] [CrossRef]
- Ruhal, R.; Kataria, R.; Choudhury, B. Trends in bacterial trehalose metabolism and significant nodes of metabolic pathway in the direction of trehalose accumulation. Microb. Biotechnol. 2013, 6, 493–502. [Google Scholar] [CrossRef]
- Feehily, C.; Karatzas, K.A.G. Role of glutamate metabolism in bacterial responses towards acid and other stresses. J. Appl. Microbiol. 2013, 114, 11–24. [Google Scholar] [CrossRef]
- Wang, R.M.; Li, N.; Zheng, K.; Hao, J.F. Enhancing acid tolerance of the probiotic bacterium Lactobacillus acidophilus NCFM with trehalose. FEMS Microbiol. Lett. 2018, 365, fny217. [Google Scholar] [CrossRef]
- Shiers, D.W.; Ralph, D.E.; Watling, H.R. A comparative study of substrate utilisation by Sulfobacillus species in mixed ferrous ion and tetrathionate growth medium. Hydrometallurgy 2010, 104, 363–369. [Google Scholar] [CrossRef]
- Guo, X.; Yin, H.; Liang, Y.; Hu, Q.; Zhou, X.; Xiao, Y.; Ma, L.; Zhang, X.; Qiu, G.; Liu, X. Comparative Genome Analysis Reveals Metabolic Versatility and Environmental Adaptations of Sulfobacillus thermosulfidooxidans Strain ST. PLoS ONE 2014, 9, e99417. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Liu, X.; Liang, Y.; Guo, X.; Xiao, Y.; Ma, L.; Miao, B.; Liu, H.; Peng, D.; Huang, W.; et al. Adaptive Evolution of Extreme Acidophile Sulfobacillus thermosulfidooxidans Potentially Driven by Horizontal Gene Transfer and Gene Loss. Appl. Environ. Microbiol. 2017, 83. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.B.; Zhang, H.J.; Zhou, W.G.; Wang, Y.G.; Zhou, H.B.; Chen, X.H. Sulfur Metabolism Pathways in Sulfobacillus acidophilus TPY, A Gram-Positive Moderate Thermoacidophile from a Hydrothermal Vent. Front. Microbiol. 2016, 7, 1861. [Google Scholar] [CrossRef] [PubMed]
- Panyushkina, A.E.; Babenko, V.V.; Nikitina, A.S.; Selezneva, O.V.; Tsaplina, I.A.; Letarova, M.A.; Kostryukova, E.S.; Letarov, A.V. Sulfobacillus thermotolerans: New insights into resistance and metabolic capacities of acidophilic chemolithotrophs. Sci. Rep. 2019, 9, 15069. [Google Scholar] [CrossRef] [PubMed]
- Acosta, M.; Galleguillos, P.; Marín, S.; Chibwana, C.; Strauss, H.; Demergasso, C. Blue-Copper Proteins: Expression of Coding Genes from Sulfobacillus spp. and Iron Oxidation in Column Bioleaching Tests. Adv. Mater. Res. 2015, 1130, 333–337. [Google Scholar] [CrossRef]
- Christel, S.; Herold, M.; Bellenberg, S.; Buetti-Dinh, A.; El Hajjami, M.; Pivkin, I.V.; Sand, W.; Wilmes, P.; Poetsch, A.; Vera, M.; et al. Weak Iron Oxidation by Sulfobacillus thermosulfidooxidans Maintains a Favorable Redox Potential for Chalcopyrite Bioleaching. Front. Microbiol. 2018, 9, 3059. [Google Scholar] [CrossRef] [PubMed]






| Cultures and Columns Conditions | Identification Label | SO42− (g L−1) NH4+ (mg L−1) Feeding | Air Feeding |
|---|---|---|---|
| Sb. sp. CBAR13 cultures for markers identification associated with ferrous and sulfur oxidation. | SS | - | Atmospheric |
| DS | - | Atmospheric | |
| L. ferriphilum DSM 14647 cultures for markers identification associated with osmotic stress (SO42− impurity) | LII-DSM-2 | (2.5) | Atmospheric |
| LII-DSM-25 | (25) | Atmospheric | |
| LII-DSM-50 | (50) | Atmospheric | |
| L. ferriphilum IESL-25 cultures study of osmotic stress (SO42− impurity) | LII-IESL_2 | (2.5) | Atmospheric |
| LII-IESL_25 | (25) | Atmospheric | |
| LII-IESL_50 | (50) | Atmospheric | |
| At. thiooxidans IESL-33 cultures for markers identification associated with CO2 availability | ATH-50 | - | 50 ppm CO2-in |
| ATH-100 | - | 100 ppm CO2-in | |
| ATH-300 | - | 300 ppm CO2-in | |
| ATH-500 | - | 500 ppm CO2-in | |
| col.1: bioleaching column #1, control | OpD60 | - | 200 ppm CO2-out |
| col.2: bioleaching column #2 for markers identification associated with O2 and CO2 availability | (*) OpD62 | - | Air ON (500 ppm CO2-out) |
| OpD64 | - | Air OFF (2000 ppm CO2-out) | |
| (**) OpD69 | - | Air OFF (1600 ppm CO2-out) | |
| OpD76 | - | Air ON (200 ppm CO2-out) | |
| OpD97 | - | Air OFF (200 ppm CO2-out) | |
| col.3: bioleaching column #3 for markers identification associated with N2 and NH4+ availability (nutrients) | OpD62 | (0) | Atmospheric |
| OpD69 | (40) | Atmospheric | |
| OpD76 | (40) | Atmospheric | |
| OpD90 | (40) | Atmospheric | |
| OpD104 | (0) | Atmospheric | |
| col.4: bioleaching column #4 for markers identification associated with osmotic stress (SO42− impurity) | OpD67 | (50) | Atmospheric |
| OpD70 | (50) | Atmospheric | |
| OpD77 | (80) | Atmospheric | |
| OpD84 | (80) | Atmospheric | |
| OpD93 | (80) | Atmospheric | |
| OpD105 | (10) | Atmospheric | |
| OpD111 | (100) | Atmospheric |
| Sample | Day | Heap Lift | Total Fe (g L−1) | Fe2+ (g L−1) | H2SO4 (g L−1) | pH | Eh (mV) | NH4+ (mg L−1) | SO42− (g L−1) | Blowers Status |
|---|---|---|---|---|---|---|---|---|---|---|
| (1) S-405-D | 284 | 4th | 1.39 | - | 2.36 | 2.06 | 786 | 4.0 | 106 | L1 ON |
| (1) S-405-B | 284 | 4th | 0.84 | - | 1.57 | 2.28 | 798 | 5.0 | 96 | L1 ON |
| (1) S-405-M | 284 | 4th | 1.91 | - | 4.68 | 1.67 | 800 | 3.2 | 98 | L1 ON |
| (2) S410-W0-D | 377 | 4th | 1.46 | <0.01 | 2.69 | 2.05 | 790 | 4.0 | 113 | L1 and L4 ON |
| (2) S410-W0-M | 377 | 4th | 1.95 | 0.24 | 3.67 | 1.84 | 676 | 2.2 | 122 | L1 and L4 ON |
| (3) S410-W2-D | 391 | 4th | 1.53 | <0.01 | 3.01 | 2.05 | 765 | 1.8 | 104 | L1 ON L4 OFF |
| (3) S410-W2-M | 391 | 4th | 2.14 | 0.92 | 2.72 | 1.95 | 659 | 1.8 | 103 | L1 ON L4 OFF |
| (4) S410-W4-D | 405 | 4th | 1.59 | <0.01 | 2.09 | 2.14 | 826 | 5.4 | 104 | L1 ON L4 OFF |
| (4) S410-W4-M | 405 | 4th | 2.33 | 0.64 | 5.91 | 1.85 | 666 | 5.8 | 104 | L1 ON L4 OFF |
| (5) S-132-D | 63 | 1st | 1.73 | 0.24 | 2.07 | 2.00 | 708 | 16 | 92 | L1 ON |
| (5) S-327-D | 43 | 3th | 0.83 | 0.04 | 1.29 | 2.16 | 720 | 10 | 92 | L1 ON |
| (5) S-413-D | 394 | 4th | 1.82 | 0.15 | 2.88 | 1.97 | 707 | 3.0 | 97 | L1 and L4 ON |
| (5) S-414-D | 6 | 4th | 1.65 | 0.15 | 2.38 | 2.08 | 769 | 2.0 | 98 | L1 ON L4 OFF |
| Species | Gene | Gene Product | EC Number | Pathway |
|---|---|---|---|---|
| A. thiooxidans | rbcL1 | RubisCo type I | 4.1.1.39 | CO2 fixation |
| A. thiooxidans | rbcL2 | RubisCo type II | 4.1.1.39 | CO2 fixation |
| A. thiooxidans | csoS2 | Carboxysome shell proteins | - | CO2 fixation |
| A. thiooxidans | csoS3 | Carbonic anhydrase type ε | 4.2.1.1 | CO2 fixation |
| A. thiooxidans | csoS4 | Carboxysome shell proteins | - | CO2 fixation |
| A. thiooxidans | gyrA * | DNA gyrase subunit A | 5.99.1.3 | HKG |
| A. ferrooxidans | rbcL-1 | RubisCo type I | 4.1.1.39 | CO2 fixation |
| A. ferrooxidans | rbcL-2 | RubisCo type II | 4.1.1.39 | CO2 fixation |
| A. ferrooxidans | Alas * | DNA-binding transcription factor | - | HKG |
| L. ferriphilum | treZ | Malto-oligosyltrehalose trehalohydrolase | 3.2.1.141 | Trehalose synthetase |
| L. ferriphilum | treY | Malto-oligosyltrehalose synthase | 5.4.99.15 | Trehalose synthetase |
| L. ferriphilum | treX | Glycogen debranching enzyme | 3.2.1.68 | Trehalose synthetase |
| L. ferriphilum | lamB | Porine maltose | - | Trehalose synthetase |
| L. ferriphilum | galU | Glucose-1 -phosphate-UDP-pyrophosphorylase | 2.7.7.9 | Trehalose synthetase |
| L. ferriphilum | gadAB | Glutamate descarboxilase | 4.1.1.15 | Trehalose synthetase |
| L. ferriphilum | nifH | Nitrogenase | 1.18.6.1 | N2 fixation |
| L. ferriphilum | amt2 | Ammonium transporter | - | NH4+ transporter |
| L. ferriphilum | glnA | Glutamine synthetase | 6.3.1.2 | NH4+ asimilation |
| L. ferriphilum | glnB | Glutamine synthetase | 6.3.1.2 | NH4+ asimilation |
| L. ferriphilum | alas * | DNA-binding transcription factor | - | HKG |
| L. ferriphilum | 16S | Ribosomal RNA small subunit | - | HKG |
| Sb. sp. CBAR13 | carb | 2Fe-2S iron-sulfur cluster binding domain-containing protein | 1.2.7.4 | Fe2+ oxidation |
| Sb. sp. CBAR13 | cytbdl | Cytochrome bd ubiquinol oxidase subunit I | - | Fe2+oxidation |
| Sb. sp. CBAR13 | sulf | Hypothetical sulfocyanin | - | Fe2+oxidation |
| Sb. sp. CBAR13 | chypll | Hypothetical cbb3-type cytochrome c oxidase subunit I | - | Fe2+oxidation |
| Sb. sp. CBAR13 | fadp | NAD(P)/FAD-dependent oxidoreductase | - | SRC oxidation |
| Sb. sp. CBAR13 | tehy | Tetrathionate hydrolase | 3.12.1.B1 | SRC oxidation |
| Sb. sp. CBAR13 | gyrA * | DNA gyrase subunit A | 5.99.1.3 | HKG |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marín, S.; Cortés, M.; Acosta, M.; Delgado, K.; Escuti, C.; Ayma, D.; Demergasso, C. From Laboratory towards Industrial Operation: Biomarkers for Acidophilic Metabolic Activity in Bioleaching Systems. Genes 2021, 12, 474. https://doi.org/10.3390/genes12040474
Marín S, Cortés M, Acosta M, Delgado K, Escuti C, Ayma D, Demergasso C. From Laboratory towards Industrial Operation: Biomarkers for Acidophilic Metabolic Activity in Bioleaching Systems. Genes. 2021; 12(4):474. https://doi.org/10.3390/genes12040474
Chicago/Turabian StyleMarín, Sabrina, Mayra Cortés, Mauricio Acosta, Karla Delgado, Camila Escuti, Diego Ayma, and Cecilia Demergasso. 2021. "From Laboratory towards Industrial Operation: Biomarkers for Acidophilic Metabolic Activity in Bioleaching Systems" Genes 12, no. 4: 474. https://doi.org/10.3390/genes12040474
APA StyleMarín, S., Cortés, M., Acosta, M., Delgado, K., Escuti, C., Ayma, D., & Demergasso, C. (2021). From Laboratory towards Industrial Operation: Biomarkers for Acidophilic Metabolic Activity in Bioleaching Systems. Genes, 12(4), 474. https://doi.org/10.3390/genes12040474







