Traces of Late Bronze and Early Iron Age Mongolian Horse Mitochondrial Lineages in Modern Populations
Abstract
1. Introduction
2. Materials and Methods
2.1. Information about the Samples
2.2. Ancient-Mitogenome Sequencing
2.3. Sequence Data Analysis
2.4. Phylogenetic Analysis
2.5. Data Availability
3. Results
3.1. Authenticity of Ancient DNA Data
3.2. Characteristics of the Ancient Mongolian Horse Mitochondrial Genomes
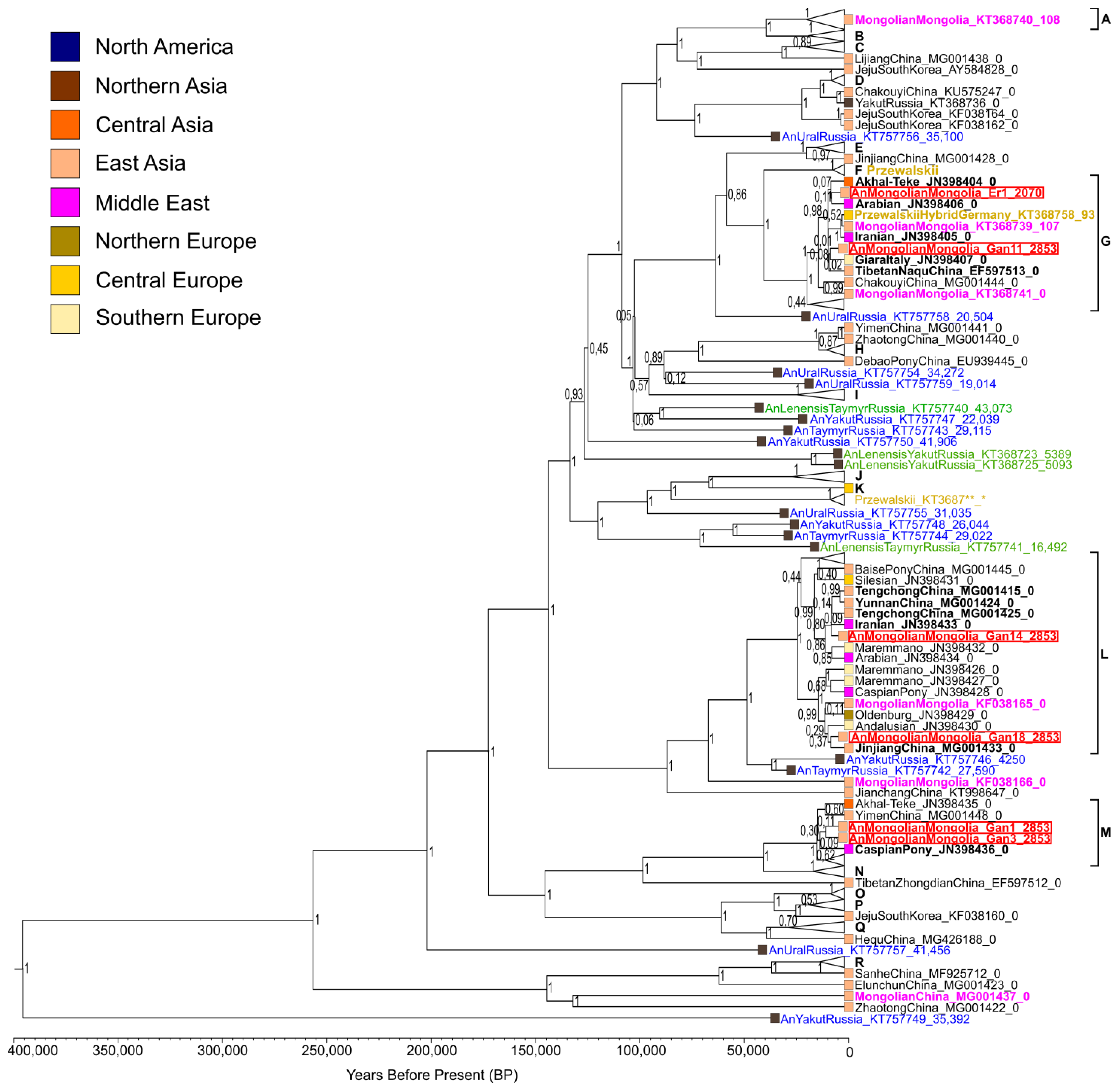
3.3. Phylogenetic Reconstructions
3.4. Time of Haplotype Divergence
4. Discussion
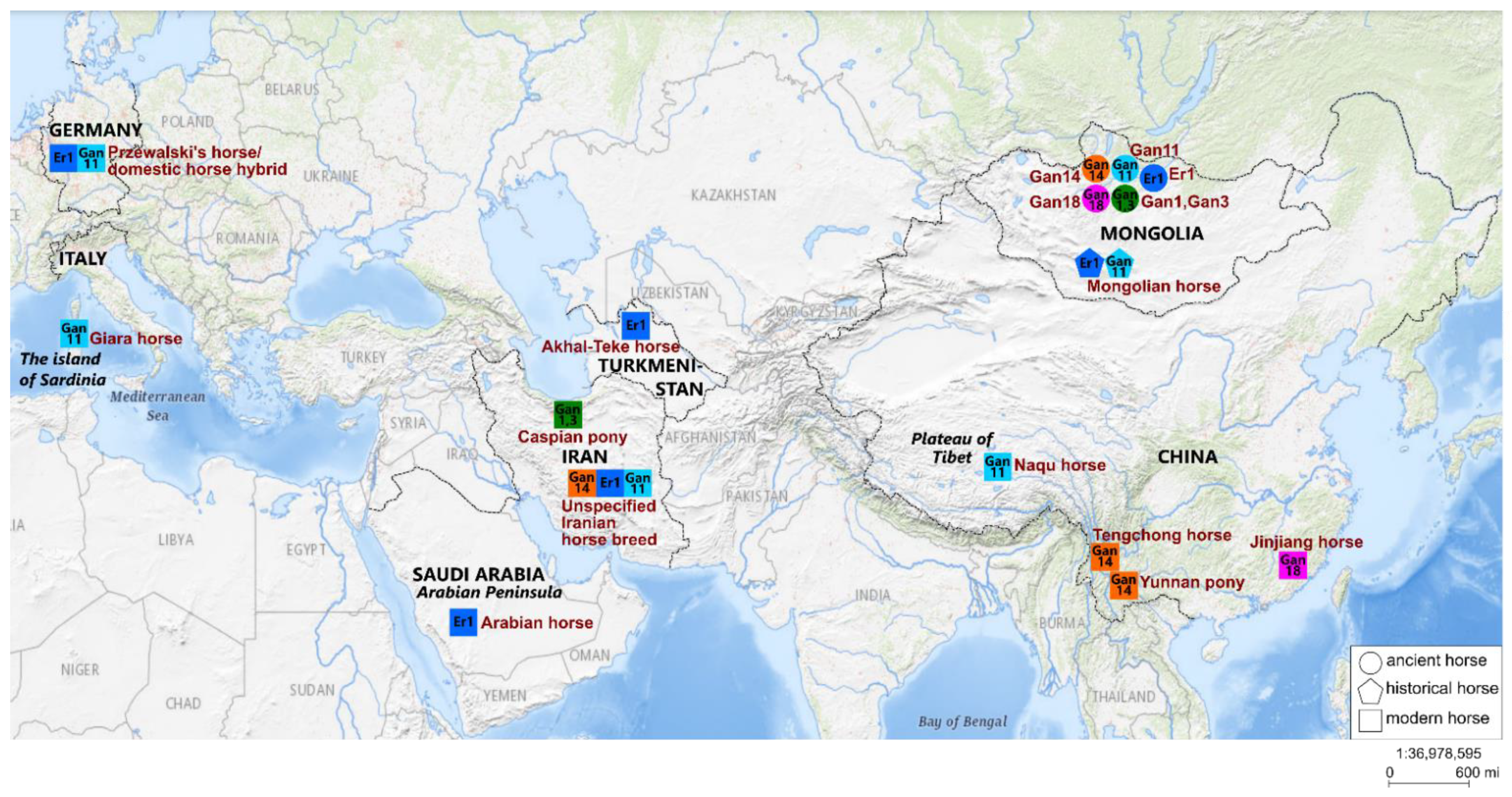
4.1. Phylogeographic Relationships
4.2. Cultural Context
4.3. Population Dynamics of the Mongolian Horse Breed
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Petersen, J.L.; Mickelson, J.R.; Cothran, E.G.; Andersson, L.S.; Axelsson, J.; Bailey, E.; Bannasch, D.; Binns, M.M.; Borges, A.S.; Brama, P.; et al. Genetic Diversity in the Modern Horse Illustrated from Genome-Wide SNP Data. PLoS ONE 2013, 8, e54997. [Google Scholar] [CrossRef]
- Hendricks, B.L. International Encyclopedia of Horse Breeds; The University of Oklahoma Press, Publishing Division of the University: Norman, OK, USA, 1995. [Google Scholar]
- Nozawa, K.; Shotake, T.; Ito, S.; Kawamoto, Y. Phylogenetic Relationships among Japanese Native and Alien Horses Estimated by Protein Polymorphisms. J. Equine Sci. 1998, 9, 53–69. [Google Scholar] [CrossRef]
- Fages, A.; Hanghøj, K.; Khan, N.; Gaunitz, C.; Seguin-Orlando, A.; Leonardi, M.; McCrory Constantz, C.; Gamba, C.; Al-Rasheid, K.A.S.; Albizuri, S.; et al. Tracking Five Millennia of Horse Management with Extensive Ancient Genome Time Series. Cell 2019, 177, 1419–1435.e31. [Google Scholar] [CrossRef] [PubMed]
- Gaunitz, C.; Fages, A.; Hanghøj, K.; Albrechtsen, A.; Khan, N.; Schubert, M.; Seguin-Orlando, A.; Owens, I.J.; Felkel, S.; Bignon-Lau, O.; et al. Ancient genomes revisit the ancestry of domestic and Przewalski’s horses. Science 2018, 360, 111–114. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.-I.; Yang, Y.-H.; Lee, S.-S.; Park, C.; Ma, R.; Bouzat, J.L.; Lewin, H.A. Phylogenetic relationships of Cheju horses to other horse breeds as determined by mtDNA D-loop sequence polymorphism. Anim. Genet. 1999, 30, 102–108. [Google Scholar] [CrossRef]
- Cho, G.J. Genetic Relationship among the Korean Native and Alien Horses Estimated by Microsatellite Polymorphism. Asian-Australas. J. Anim. Sci. 2006, 19, 784–788. [Google Scholar] [CrossRef]
- Tozaki, T.; Takezaki, N.; Hasegawa, T.; Ishida, N.; Kurosawa, M.; Tomita, M.; Saitou, N.; Mukoyama, H. Microsatellite variation in Japanese and Asian horses and their phylogenetic relationship using a European horse outgroup. J. Hered. 2003, 94, 374–380. [Google Scholar] [CrossRef]
- Kovalev, A.A.; Erdenebaatar, D.; Iderkhangai, T.-O. Discovery of new Bronze Age culture in the South of Mongolia. In Proceedings of the Ancient Cultures of Mongolia and Baikal Siberia, Ulaanbaatar, Mongolia, 5–9 September 2012; National University of Mongolia: Ulaanbaatar, Mongolia, 2012; pp. 175–182. [Google Scholar]
- Kovalev, A.A.; Erdenebaatar, D.; Rukavishnikova, I.V. A ritual complex with deer stones at Uushigiin Uvur/Ulaan Uushig, Mongolia: Composition and construction stages (based on the 2013 excavations). Archaeol. Ethnol. Anthropol. Eurasia 2016, 44, 82–92. (In Russian) [Google Scholar] [CrossRef]
- Youn, M.; Kim, J.C.; Kim, H.K.; Tumen, D.; Navaan, D.; Erdene, M. Dating the Tavan Tolgoi Site, Mongolia: Burials of the Nobility from Genghis Khan’s Era. Radiocarbon 2007, 49, 685–691. [Google Scholar] [CrossRef]
- Cieslak, M.; Pruvost, M.; Benecke, N.; Hofreiter, M.; Morales, A.; Reissmann, M.; Ludwig, A. Origin and History of Mitochondrial DNA Lineages in Domestic Horses. PLoS ONE 2010, 5, e15311. [Google Scholar] [CrossRef] [PubMed]
- Pilipenko, A.S.; Romaschenko, A.G.; Molodin, V.I.; Parzinger, H.; Kobzev, V.F. Mitochondrial DNA studies of the Pazyryk people (4th to 3rd centuries BC) from northwestern Mongolia. Archaeol. Anthropol. Sci. 2010, 2, 231–236. [Google Scholar] [CrossRef]
- Tishkin, A.A. “Deer” stones of Mongolia and adjacent territories as one of the indicators of the archaic nomadic empire (to the formulation of the question). In Proceedings of the V (XXI) All-Russian Archaeological Congress, Belokurikha, Russia, 1–8 October 2017; Altai State University: Barnaul, Russia, 2017; p. 1026. [Google Scholar]
- Iderkhangai, T.-O.; Mijiddorj, E.; Orgilbayar, S.; Galbadrakh, B.; Erdene, J.; Өnөrbayar, B.; Enkhmagnay, G. 2015 Archaeological Rescue Excavation Report on the Egiin Gol Hydroelectric Dam, Khutag Undur Sum, Bulgan Aimag; Ulaanbaatar State University: Ulaanbaatar, Mongolia, 2015; pp. 67–150. [Google Scholar]
- Taylor, W. Horse demography and use in Bronze Age Mongolia. Quat. Int. 2017, 436, 270–282. [Google Scholar] [CrossRef]
- Taylor, W.T.T.; Jargalan, B.; Lowry, K.B.; Clark, J.; Tuvshinjargal, T.; Bayarsaikhan, J. A Bayesian chronology for early domestic horse use in the Eastern Steppe. J. Archaeol. Sci. 2017, 81, 49–58. [Google Scholar] [CrossRef]
- Kradin, N.N. The Xiongnu Empire, 2nd ed.; “The Publishing Group Logos” Ltd.: Moscow, Russia, 2001; ISBN 5-94010-124-0. [Google Scholar]
- Garutt, V.E.; Yuriev, K.B. Paleofauna of the Ivolginsky settlement according to archaeological excavations of 1949–1956. Archaeol. Festschr. 1959, 80–82. [Google Scholar]
- Rudenko, S.I. The Culture of the Huns and the Noin-Ula Burial Mounds; Academy of Sciences of the USSR: Moscow/Leningrad, Russia, 1962. [Google Scholar]
- Yang, D.Y.; Eng, B.; Waye, J.S.; Dudar, J.C.; Saunders, S.R. Improved DNA extraction from ancient bones using silica-based spin columns. Am. J. Phys. Anthropol. 1998, 105, 539–543. [Google Scholar] [CrossRef]
- Sanderson, C.; Radley, K.; Mayton, L. Ethylenediaminetetraacetic Acid in Ammonium Hydroxide for Reducing Decalcification Time. Biotech. Histochem. 1995, 70, 12–18. [Google Scholar] [CrossRef] [PubMed]
- Maricic, T.; Whitten, M.; Pääbo, S. Multiplexed DNA Sequence Capture of Mitochondrial Genomes Using PCR Products. PLoS ONE 2010, 5, e14004. [Google Scholar] [CrossRef]
- Vorobieva, N.V.; Makunin, A.I.; Druzhkova, A.S.; Kusliy, M.A.; Trifonov, V.A.; Popova, K.O.; Polosmak, N.V.; Molodin, V.I.; Vasiliev, S.K.; Shunkov, M.V.; et al. High genetic diversity of ancient horses from the Ukok Plateau. PLoS ONE 2020, 15, e0241997. [Google Scholar] [CrossRef] [PubMed]
- Schubert, M.; Jónsson, H.; Chang, D.; Der Sarkissian, C.; Ermini, L.; Ginolhac, A.; Albrechtsen, A.; Dupanloup, I.; Foucal, A.; Petersen, B.; et al. Prehistoric genomes reveal the genetic foundation and cost of horse domestication. Proc. Natl. Acad. Sci. USA 2014, 111, E5661–E5669. [Google Scholar] [CrossRef] [PubMed]
- Schubert, M.; Lindgreen, S.; Orlando, L. AdapterRemoval v2: Rapid adapter trimming, identification, and read merging. BMC Res. Notes 2016, 9, 88. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Jónsson, H.; Ginolhac, A.; Schubert, M.; Johnson, P.L.F.; Orlando, L. mapDamage2.0: Fast approximate Bayesian estimates of ancient DNA damage parameters. Bioinformatics 2013, 29, 1682–1684. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Misawa, K.; Kuma, K.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef] [PubMed]
- Lanfear, R.; Calcott, B.; Ho, S.Y.W.; Guindon, S. PartitionFinder: Combined Selection of Partitioning Schemes and Substitution Models for Phylogenetic Analyses. Mol. Biol. Evol. 2012, 29, 1695–1701. [Google Scholar] [CrossRef] [PubMed]
- Suchard, M.A.; Lemey, P.; Baele, G.; Ayres, D.L.; Drummond, A.J.; Rambaut, A. Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol. 2018, 4, vey016. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, M.A.R.; Suchard, M.A. Bayesian analysis of elapsed times in continuous-time Markov chains. Can. J. Stat. 2008, 36, 355–368. [Google Scholar] [CrossRef]
- Drummond, A.J.; Nicholls, G.K.; Rodrigo, A.G.; Solomon, W. Estimating mutation parameters, population history and genealogy simultaneously from temporally spaced sequence data. Genetics 2002, 161, 1307–1320. [Google Scholar]
- Kingman, J.F.C. The coalescent. Stoch. Process. Appl. 1982, 13, 235–248. [Google Scholar] [CrossRef]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior Summarization in Bayesian Phylogenetics Using Tracer 1.7. Syst. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef]
- Drummond, A.J.; Rambaut, A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 2007, 7, 1–8. [Google Scholar] [CrossRef]
- Sawyer, S.; Krause, J.; Guschanski, K.; Savolainen, V.; Pääbo, S. Temporal Patterns of Nucleotide Misincorporations and DNA Fragmentation in Ancient DNA. PLoS ONE 2012, 7, e34131. [Google Scholar] [CrossRef] [PubMed]
- Achilli, A.; Olivieri, A.; Soares, P.; Lancioni, H.; Kashani, B.H.; Perego, U.A.; Nergadze, S.G.; Carossa, V.; Santagostino, M.; Capomaccio, S.; et al. Mitochondrial genomes from modern horses reveal the major haplogroups that underwent domestication. Proc. Natl. Acad. Sci. USA 2012, 109, 2449–2454. [Google Scholar] [CrossRef] [PubMed]
- Orlando, L.; Ginolhac, A.; Zhang, G.; Froese, D.; Albrechtsen, A.; Stiller, M.; Schubert, M.; Cappellini, E.; Petersen, B.; Moltke, I.; et al. Recalibrating Equus evolution using the genome sequence of an early Middle Pleistocene horse. Nature 2013, 499, 74–78. [Google Scholar] [CrossRef] [PubMed]
- Librado, P.; Der Sarkissian, C.; Ermini, L.; Schubert, M.; Jónsson, H.; Albrechtsen, A.; Fumagalli, M.; Yang, M.A.; Gamba, C.; Seguin-Orlando, A.; et al. Tracking the origins of Yakutian horses and the genetic basis for their fast adaptation to subarctic environments. Proc. Natl. Acad. Sci. USA 2015, 112, E6889–E6897. [Google Scholar] [CrossRef] [PubMed]
- Boeskorov, G.G.; Potapova, O.R.; Protopopov, A.V.; Plotnikov, V.V.; Maschenko, E.N.; Shchelchkova, M.V.; Petrova, E.A.; Kowalczyk, R.; van der Plicht, J.; Tikhonov, A.N. A study of a frozen mummy of a wild horse from the Holocene of Yakutia, East Siberia, Russia. Mammal. Res. 2018, 63, 307–314. [Google Scholar] [CrossRef]
- Cosgrove, E.J.; Sadeghi, R.; Schlamp, F.; Holl, H.M.; Moradi-Shahrbabak, M.; Miraei-Ashtiani, S.R.; Abdalla, S.; Shykind, B.; Troedsson, M.; Stefaniuk-Szmukier, M.; et al. Genome Diversity and the Origin of the Arabian Horse. Sci. Rep. 2020, 10, 9702. [Google Scholar] [CrossRef]
- Szontagh, A.; Bán, B.; Bodó, I.; Cothran, E.G.; Hecker, W.; Józsa, C.; Major, Á. Genetic diversity of the Akhal-Teke horse breed in Turkmenistan based on microsatellite analysis. In Conservation Genetics of Endangered Horse Breeds; Bodo, I., Alderson, L., Langlois, B., Eds.; Wageningen Academic Publishers: Bled, Slovenia, 2005; pp. 123–128. [Google Scholar]
- Kovalevskaya, V.B. Ancestors of the Oriental Horse in Eurasia: Origin and Distribution. Archaeol. Ethnol. Anthropol. Eurasia 2020, 48, 129–139. [Google Scholar] [CrossRef]
- Yang, L.; Kong, X.; Yang, S.; Dong, X.; Yang, J.; Gou, X.; Zhang, H. Haplotype diversity in mitochondrial DNA reveals the multiple origins of Tibetan horse. PLoS ONE 2018, 13, e0201564. [Google Scholar] [CrossRef] [PubMed]
- Du, D.; Zhao, C.; Zhang, H.; Han, G. Genetic Diversity of Tibetan Horse and its Relationships with Mongolian Horse and Ningqiang Pony Assessed by Microsatellite Polymorphism. Asian J. Anim. Vet. Adv. 2011, 6, 564–571. [Google Scholar] [CrossRef][Green Version]
- Morelli, L.; Useli, A.; Sanna, D.; Barbato, M.; Contu, D.; Pala, M.; Cancedda, M.; Francalacci, P. Mitochondrial DNA lineages of Italian Giara and Sarcidano horses. Genet. Mol. Res. 2014, 13, 8241–8257. [Google Scholar] [CrossRef] [PubMed]
- Ling, Y.; Ma, Y.; Guan, W.; Cheng, Y.; Wang, Y.; Han, J.; Jin, D.; Mang, L.; Mahmut, H. Identification of Y Chromosome Genetic Variations in Chinese Indigenous Horse Breeds. J. Hered. 2010, 101, 639–643. [Google Scholar] [CrossRef] [PubMed]
- Ling, Y.H.; Ma, Y.H.; Guan, W.J.; Cheng, Y.J.; Wang, Y.P.; Han, J.L.; Mang, L.; Zhao, Q.J.; He, X.H.; Pu, Y.B.; et al. Evaluation of the genetic diversity and population structure of Chinese indigenous horse breeds using 27 microsatellite markers. Anim. Genet. 2011, 42, 56–65. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Zhu, Q.; Liu, S.; Zhao, C.; Wu, C. The origin of Chinese domestic horses revealed with novel mtDNA variants. Anim. Sci. J. 2017, 88, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Wang, S.; Zeng, G.; Guo, J.; Guo, M.; Dong, X.; Hua, G.; Liu, Y.; Wang, M.; Ling, Y.; et al. The Origin of a Coastal Indigenous Horse Breed in China Revealed by Genome-Wide SNP Data. Genes 2019, 10, 241. [Google Scholar] [CrossRef]
- Seyedabadi, H.R.; Amirinia, S.; Bana, B.M.H.; Emrani, H. Parentage verification of Iranian Caspian horse using microsatellites markers. Iran. J. Biotechnol. 2006, 4, 260–264. [Google Scholar]
- Khazanov, A.M. Ecological limitations of nomadism in the Eurasian steppes and their social and cultural implications. Asian Afr. Stud. 1990, 24, 1–15. [Google Scholar]
- Taylor, W.; Fantoni, M.; Marchina, C.; Lepetz, S.; Bayarsaikhan, J.; Houle, J.-L.; Pham, V.; Fitzhugh, W. Horse sacrifice and butchery in Bronze Age Mongolia. J. Archaeol. Sci. Rep. 2020, 31, 102313. [Google Scholar] [CrossRef]
- Honeychurch, W. Inner Asia and the Spatial Politics of Empire; Springer: New York, NY, USA, 2015; ISBN 978-1-4939-1814-0. [Google Scholar]
- Kelekna, P. The Horse in Human History; Cambridge University Press: Cambridge, UK, 2009; ISBN 978-0-521-51659-4. [Google Scholar]
- Creel, H.G. The role of the horse in Chinese history. Am. Hist. Rev. 1965, 70, 647–672. [Google Scholar] [CrossRef]
- Panov, V.A. To the History of the Peoples of Central Asia, 2nd ed.; A. V. Dattan: Vladivostok, Russia, 1918. [Google Scholar]
- Schafer, E.H. The Golden Peaches of Samarkand; Nauka: Moscow, Russia, 1981. [Google Scholar]
- Polosmak, N.V. The light of distant Hellas. Sci. First Hand 2011, 30, 88–101. [Google Scholar]
- Erdenebaatar, D.; Iderkhangai, T.; Galbadrakh, B.; Minjiddorj, E.; Orgilbayar, S. Excavations of satellite burial 30, tomb 1 complex, Gol Mod 2 necropolis. In Proceedings of the Xiongnu archaeology: Multidisciplinary perspectives of the first steppe empire in Inner Asia; Brosseder, U., Miller, B.K., Eds.; Rheinische Friedrich-Wilhelms-Universitat: Ulaanbaatar, Mongolia, 2011; pp. 303–314. [Google Scholar]
- Prowse, T.L.; Barta, J.L.; von Hunnius, T.E.; Small, A.M. Stable isotope and mitochondrial DNA evidence for geographic origins on a Roman estate at Vagnari (Italy). J. Rom. Archaeol. 2010, 78, 175–198. [Google Scholar]
- Kusliy, M.A.; Druzhkova, A.S.; Popova, K.O.; Vorobieva, N.V.; Makunin, A.I.; Yurlova, A.A.; Tishkin, A.A.; Minyaev, S.S.; Trifonov, V.A.; Graphodatsky, A.S.; et al. Genotyping and coat colour detection of ancient horses from Buryatia. Tsitologiia 2016, 58, 304–308. [Google Scholar] [PubMed]
- Giontella, A.; Cardinali, I.; Lancioni, H.; Giovannini, S.; Pieramati, C.; Silvestrelli, M.; Sarti, F.M. Mitochondrial DNA Survey Reveals the Lack of Accuracy in Maremmano Horse Studbook Records. Animals 2020, 10, 839. [Google Scholar] [CrossRef] [PubMed]
| Sample Name | Number of Collapsed Reads | Number of Unique Mapped Collapsed Reads | Mitogenome Width of Coverage, % | Mitogenome Average Depth of Coverage | Ancient Library Average Fragment Size | Terminal Library Fragment Deamination, % |
|---|---|---|---|---|---|---|
| Er1 | 188,493 | 917 | 97.3 | 6.7 | 90 | 14.49 |
| Gan1 | 714,656 | 3143 | 99.1 | 18.6 | 60 | 24.31 |
| Gan3 | 743,175 | 6919 | 99.8 | 41.8 | 63 | 25.88 |
| Gan11 | 549,992 | 13,893 | 99.7 | 85.9 | 68 | 28.51 |
| Gan14 | 511,882 | 11,643 | 99.8 | 66.7 | 65 | 33.77 |
| Gan18 | 582,595 | 3322 | 99.5 | 18.5 | 59 | 25.26 |
| Node Name | Median Age Estimate | 95% HPD (Height Posterior Density) |
|---|---|---|
| Er1_Arabian_Akhal-Teke | 3885.2 | [2330.9; 6645.42] |
| Gan11_Giara_Naqu | 5056.25 | [3105.38; 7914.36] |
| Gan14_Iranian_Tengchong_Yunnan_Tengchong | 5687.42 | [3150.41; 9072.17] |
| Gan18_Jinjiang | 4432.33 | [2853.16; 7625.08] |
| Gan1_Gan3_Caspian | 9416.83 | [5911.57; 14,467.8] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kusliy, M.A.; Vorobieva, N.V.; Tishkin, A.A.; Makunin, A.I.; Druzhkova, A.S.; Trifonov, V.A.; Iderkhangai, T.-O.; Graphodatsky, A.S. Traces of Late Bronze and Early Iron Age Mongolian Horse Mitochondrial Lineages in Modern Populations. Genes 2021, 12, 412. https://doi.org/10.3390/genes12030412
Kusliy MA, Vorobieva NV, Tishkin AA, Makunin AI, Druzhkova AS, Trifonov VA, Iderkhangai T-O, Graphodatsky AS. Traces of Late Bronze and Early Iron Age Mongolian Horse Mitochondrial Lineages in Modern Populations. Genes. 2021; 12(3):412. https://doi.org/10.3390/genes12030412
Chicago/Turabian StyleKusliy, Mariya A., Nadezhda V. Vorobieva, Alexey A. Tishkin, Alexey I. Makunin, Anna S. Druzhkova, Vladimir A. Trifonov, Tumur-O. Iderkhangai, and Alexander S. Graphodatsky. 2021. "Traces of Late Bronze and Early Iron Age Mongolian Horse Mitochondrial Lineages in Modern Populations" Genes 12, no. 3: 412. https://doi.org/10.3390/genes12030412
APA StyleKusliy, M. A., Vorobieva, N. V., Tishkin, A. A., Makunin, A. I., Druzhkova, A. S., Trifonov, V. A., Iderkhangai, T.-O., & Graphodatsky, A. S. (2021). Traces of Late Bronze and Early Iron Age Mongolian Horse Mitochondrial Lineages in Modern Populations. Genes, 12(3), 412. https://doi.org/10.3390/genes12030412









