Transcriptome Analysis Reveals Genes Involved in Thermogenesis in Two Cold-Exposed Sheep Breeds
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. RNA-Seq Analysis—RNA Extraction, Library Construction and Sequencing
2.3. Gene Expression Analysis
2.4. Differentially Expressed Gene Analysis
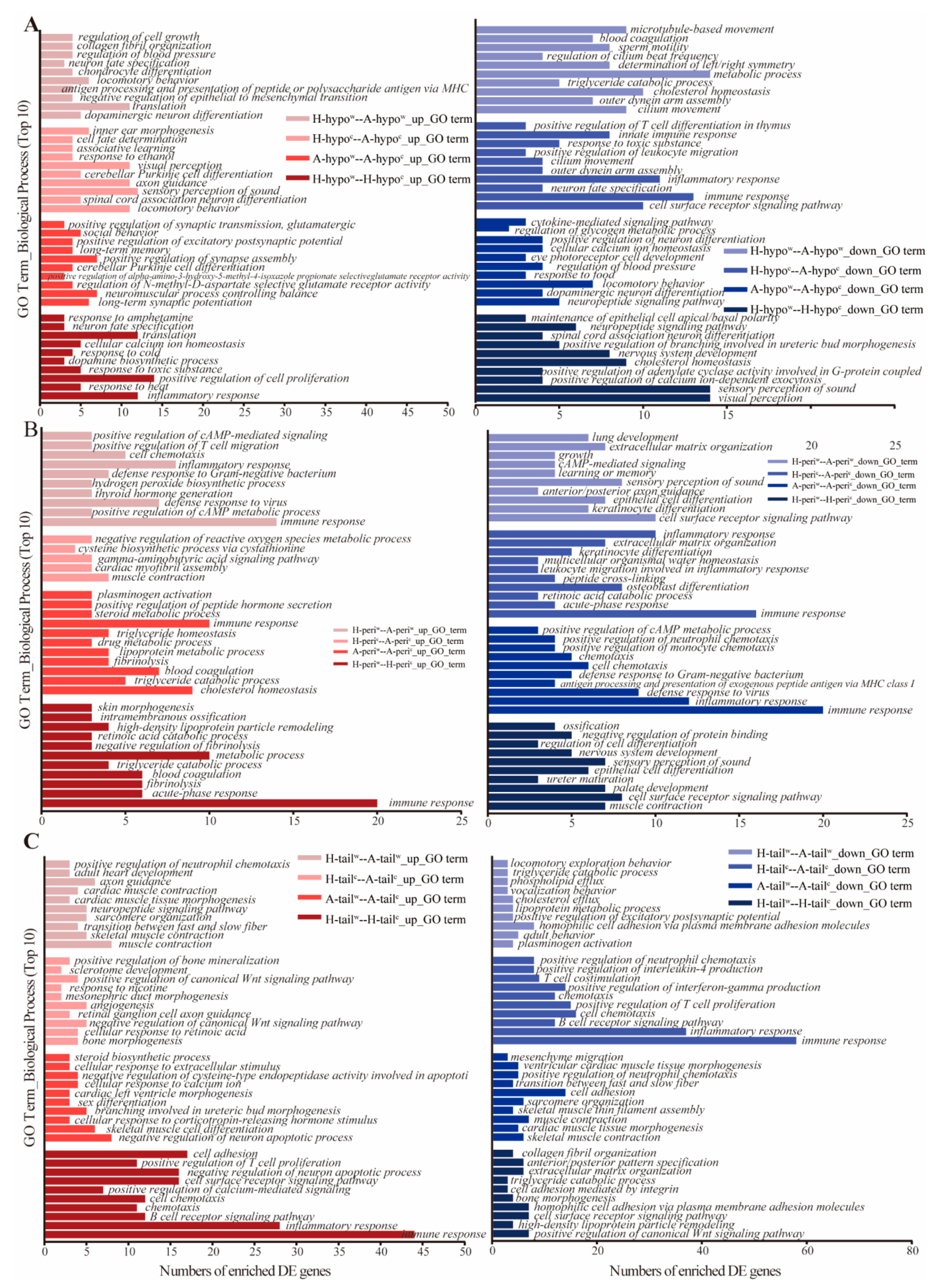
2.5. Function Enrichment and Analyses
2.6. RNA-Seq Validation by Quantitative Real-Time PCR
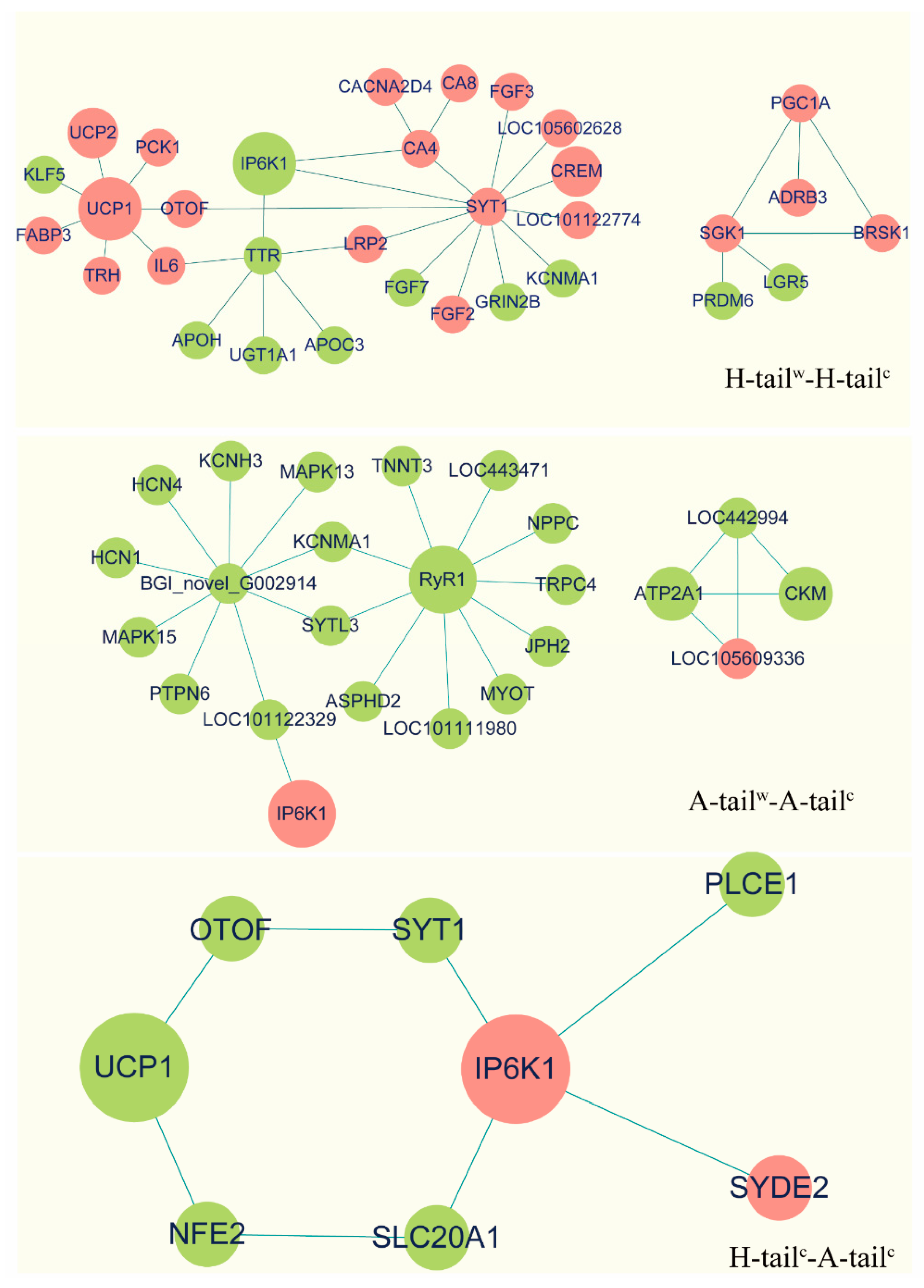
2.7. Protein-Protein Interaction Network Analyses
2.8. Statistical Analyses
2.9. Availability of Data and Materials
2.10. Ethics Approval and Consent to Participate
3. Results
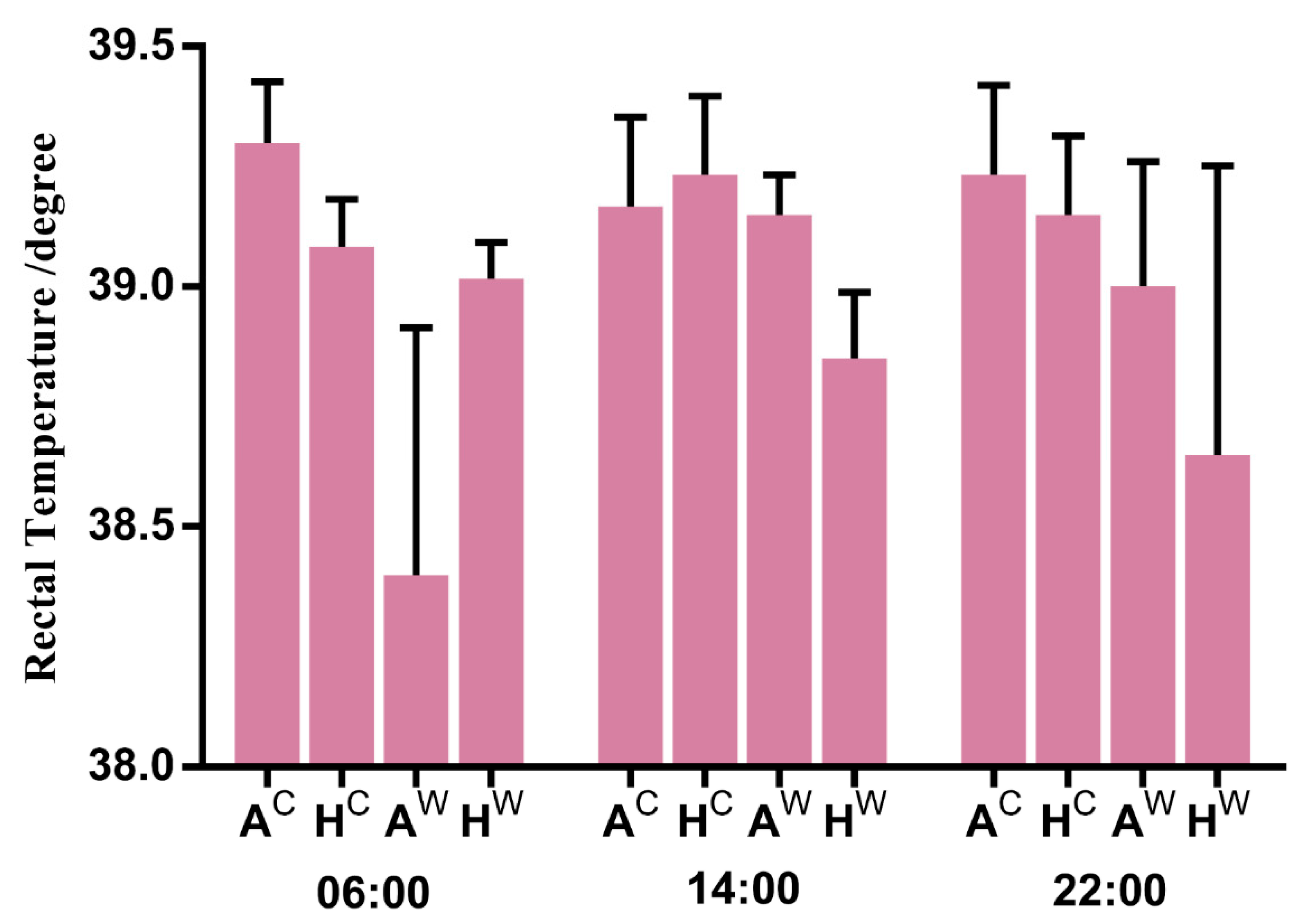
3.1. Sequencing and Mapping
3.2. Gene Annotation
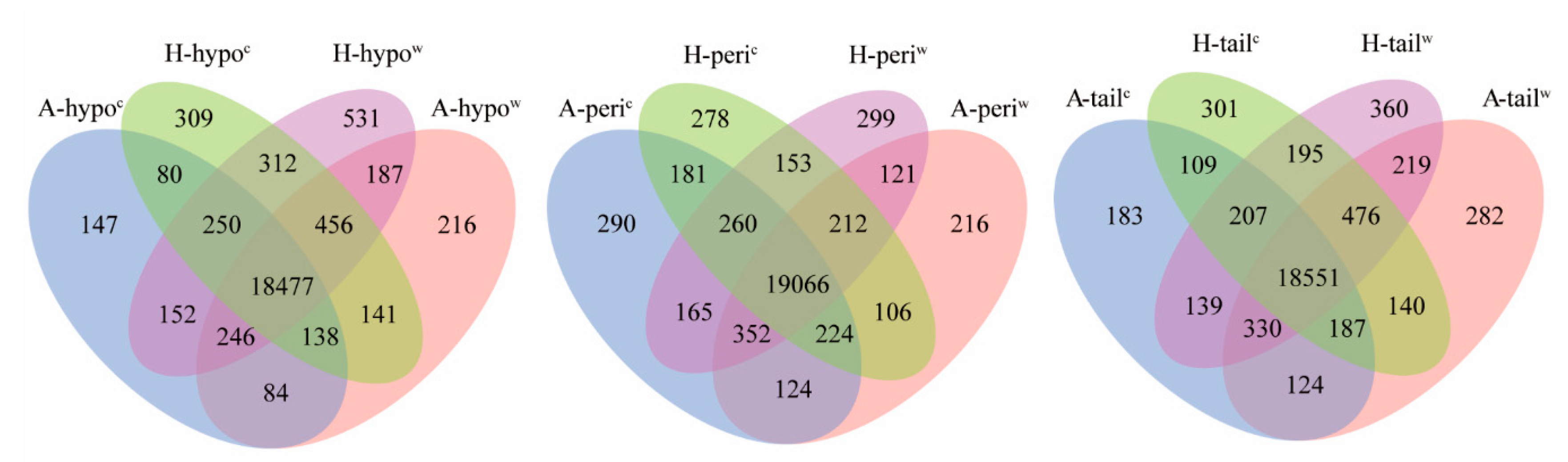
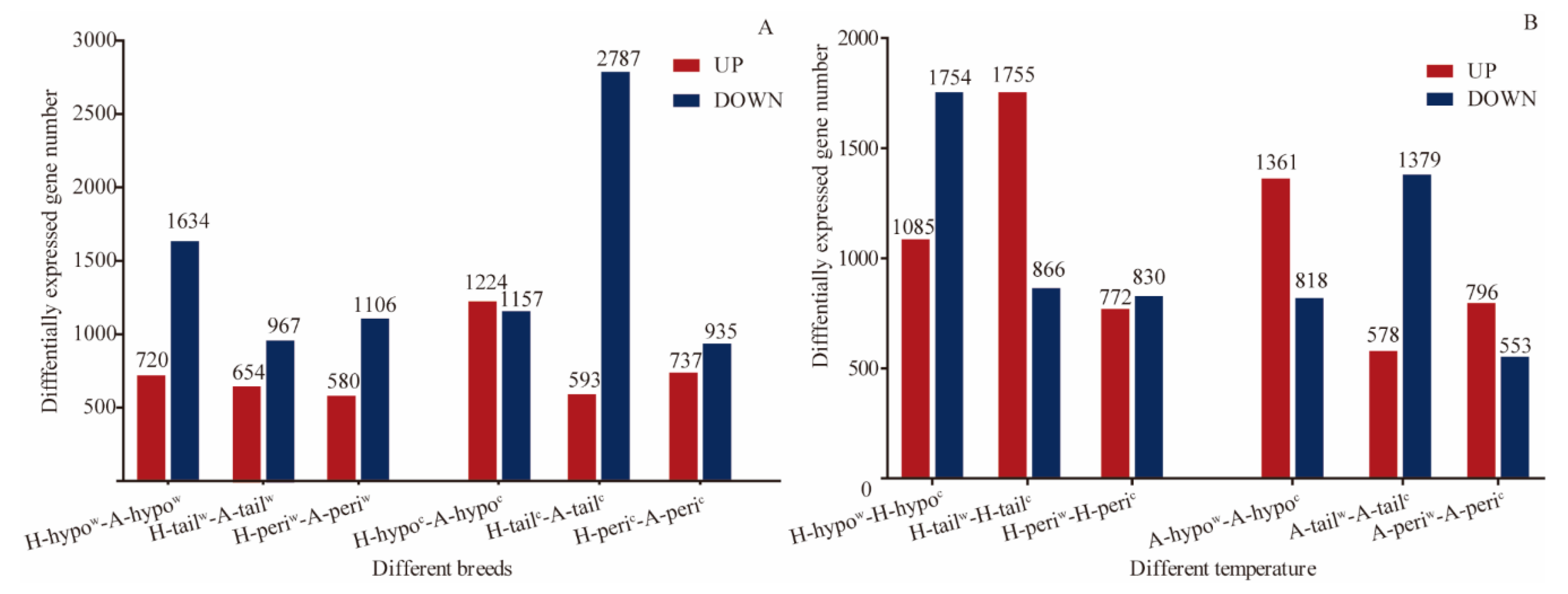
3.3. Analysis of the DEGs
3.4. Validation of RNA-Seq Results by RT-qPCR
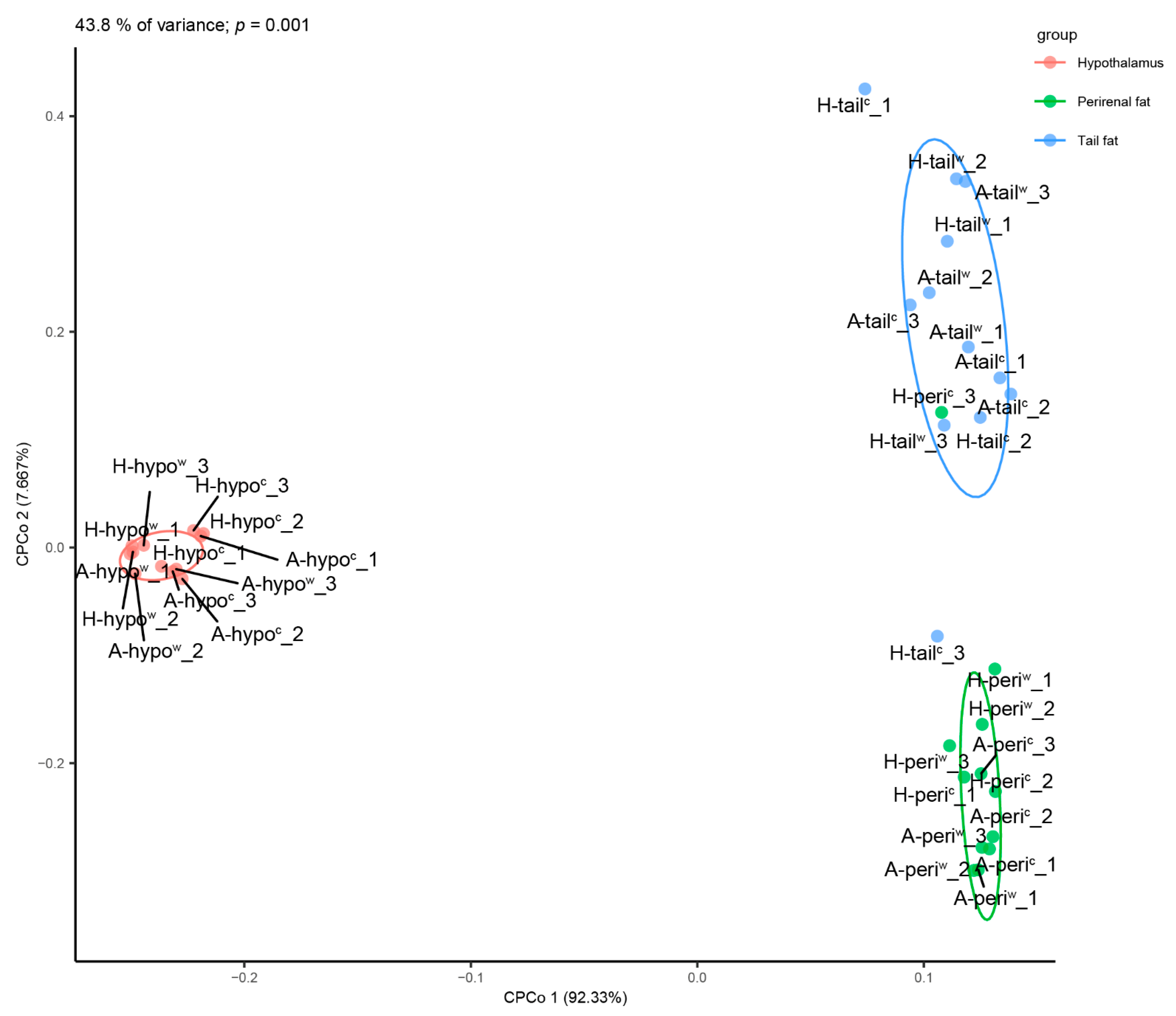
3.5. Protein-Protein Interaction (PPI) Analyses
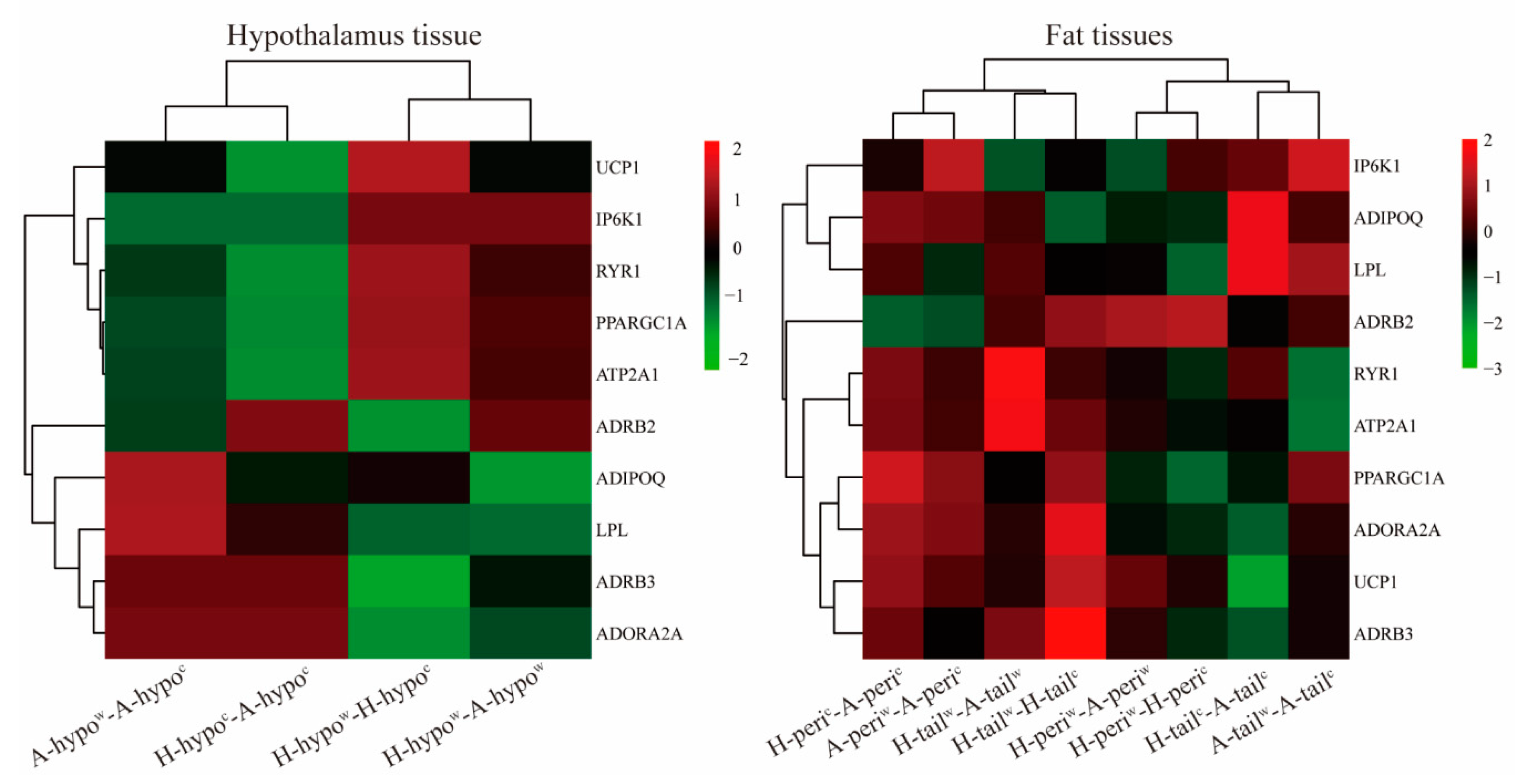
3.6. Candidate Genes Analyses
4. Discussion
4.1. Pathways Identified in Low Temperature Challenges—cAMP and Calcium Signaling Pathways
4.2. Analysis of the Genes Participating in Calcium and cAMP Signaling Pathways
4.3. Analysis of the Genes Participating in the UCP1 Related Thermogenesis Pathway
4.4. Other Genes in Low Temperature Challenges
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Stott, A.W.; Slee, J. The effect of environmental temperature during pregnancy on thermoregulation in the newborn lamb. Anim. Sci. 1985, 41, 341–347. [Google Scholar] [CrossRef]
- Alexander, G.; Williams, D. Shivering and non-shivering thermogenesis during summit metabolism in young lambs. J. Physiol. 1968, 198, 251–276. [Google Scholar] [CrossRef] [PubMed]
- Clarke, L.; Heasman, L.; Firth, K.; E Symonds, M. Influence of feeding and ambient temperature on thermoregulation in newborn lambs. Exp. Physiol. 1997, 82, 1029–1040. [Google Scholar] [CrossRef] [PubMed]
- Kazak, L.; Chouchani, E.T.; Jedrychowski, M.P.; Erickson, B.K.; Shinoda, K.; Cohen, P.; Vetrivelan, R.; Lu, G.Z.; Laznik-Bogoslavski, D.; Hasenfuss, S.C.; et al. A Creatine-Driven Substrate Cycle Enhances Energy Expenditure and Thermogenesis in Beige Fat. Cell 2015, 163, 643–655. [Google Scholar] [CrossRef] [PubMed]
- Sykes, A.R.; Griffiths, R.G.; Slee, J. Influence of breed, birth weight and weather on the body temperature of newborn lambs. Anim. Sci. 1976, 22, 395–402. [Google Scholar] [CrossRef]
- Slee, J.; Simpson, S.P.; Woolliams, J.A. Metabolic rate responses to cold and to exogenous noradrenaline in newborn Scottish Blackface lambs genetically selected for high or low resistance to cold. Anim. Sci. 1987, 45, 69–74. [Google Scholar] [CrossRef]
- Slee, J.; Stott, A.W. Genetic selection for cold resistance in Scottish Blackface lambs. Anim. Sci. 1986, 43, 397–404. [Google Scholar] [CrossRef]
- Li, Y.; Li, Y.B.; Liu, C.J. Changes in lipid oxidation and fatty acids in Altay sheep fat during a long time of low temperature storage. J. Oleo Sci. 2017, 66, 321–327. [Google Scholar] [CrossRef]
- Li, J.; Zhu, X.; Ma, L.; Xu, H.; Cao, X.; Luo, R.; Chen, H.; Sun, X.; Cai, Y.; Lan, X. Detection of a new 20-bp insertion/deletion (indel) within sheep PRND gene using mathematical expectation (ME) method. Prion 2017, 11, 143–150. [Google Scholar] [CrossRef]
- Chen, Y.; Chen, Y.; Shi, C.; Huang, Z.; Zhang, Y.; Li, S.; Li, Y.; Ye, J.; Yu, C.; Li, Z.; et al. SOAPnuke: A MapReduce acceleration-supported software for integrated quality control and preprocessing of high-throughput sequencing data. Gigascience 2018, 7, 1–6. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef]
- Buja, A.; Eyuboglu, N. Remarks on Parallel Analysis. Multivar. Behav. Res. 1992, 27, 509–540. [Google Scholar] [CrossRef]
- Linting, M.; Van Os, B.J.; Meulman, J.J. Statistical Significance of the Contribution of Variables to the PCA solution: An Alternative Permutation Strategy. Psychometrika 2011, 76, 440–460. [Google Scholar] [CrossRef]
- Wang, L.; Feng, Z.; Wang, X.; Wang, X.; Zhang, X. DEGseq: An R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 2009, 26, 136–138. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate—A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B-Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene Ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Ogata, H.; Goto, S.; Sato, K.; Fujibuchi, W.; Bono, H.; Kanehisa, M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 1999, 27, 29–34. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Qiao, L.; Yoo, H.S.; Bosco, C.; Lee, B.; Feng, G.-S.; Schaack, J.; Chi, N.-W.; Shao, J. Adiponectin reduces thermogenesis by inhibiting brown adipose tissue activation in mice. Diabetologia 2014, 57, 1027–1036. [Google Scholar] [CrossRef]
- Laperrousaz, E.; Denis, R.G.; Kassis, N.; Contreras, C.; López, M.; Luquet, S.; Cruciani-Guglielmacci, C.; Magnan, C. Lipoprotein Lipase Expression in Hypothalamus Is Involved in the Central Regulation of Thermogenesis and the Response to Cold Exposure. Front. Endocrinol. 2018, 9, 103. [Google Scholar] [CrossRef]
- Wang, H.; Eckel, R.H. Lipoprotein lipase: From gene to obesity. Am. J. Physiol. Metab. 2009, 297, E271–E288. [Google Scholar] [CrossRef]
- Oliver, S.R.; Anderson, K.J.; Hunstiger, M.M.; Andrews, M.T. Turning down the heat: Down-regulation of sarcolipin in a hibernating mammal. Neurosci. Lett. 2019, 696, 13–19. [Google Scholar] [CrossRef] [PubMed]
- Kashan, N.; Azar, G.M.; Afzalzadeh, A.; Salehi, A. Growth performance and carcass quality of fattening lambs from fat-tailed and tailed sheep breeds. Small Rumin. Res. 2005, 60, 267–271. [Google Scholar] [CrossRef]
- Attia, N.; Bocquierb, F.; Khaldi, G. Performance of the fat-tailed Barbarine sheep in its environment: Adaptive capacity to alternation of underfeeding and re-feeding periods. A review. Anim. Res. 2004, 53, 165–176. [Google Scholar] [CrossRef]
- Gan, S.Z.W.; Shen, M.; Liang, Y.; Yang, J.; Gao, L.; Liu, S.; Wang, X. Poly-morphism detection and analysis of the position (59327581) on X chromosome among sheep breeds of three different tail types. J. Shihezi Univ. (Nat. Sci.) 2013, 31, 587–591. [Google Scholar]
- Liu, X.; Li, Q.; Lin, Q.; Sun, R. Uncoupling protein1 mRNA, mitochondrial GTP-binding, and T4 5′-deiodinase of brown adipose tissue in euthermic Daurian ground squirrel during cold exposure. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2001, 128, 827–835. [Google Scholar] [CrossRef]
- Bieber, L.L.; Pettersson, B.; Lindberg, O. Studies on Norepinephrine-Induced Efflux of Free Fatty Acid from Hamster Brown-Adipose-Tissue Cells. JBIC J. Biol. Inorg. Chem. 1975, 58, 375–381. [Google Scholar] [CrossRef]
- Bukowiecki, L.; Folléa, N.; Lupien, J.; Paradis, A. Metabolic relationships between lipolysis and respiration in rat brown adipocytes. The role of long chain fatty acids as regulators of mitochondrial respiration and feedback inhibitors of lipolysis. J. Biol. Chem. 1981, 256, 12840–12848. [Google Scholar] [CrossRef]
- Zhou, J.; Ji, K.; Liu, H.; Zhang, Y.; Degen, A.A.; Jiao, D.; Wang, W.; Xie, Z.; Wang, X.; Zhou, P.; et al. Effect of air temperature on growth performance, apparent digestibilities, rumen fermentation and serum metabolites in Altay and Hu lambs. J. Anim. Physiol. Anim. Nutr. 2020, 104, 1024–1033. [Google Scholar] [CrossRef]
- Sano, H.; Sawada, H.; Takenami, A.; Oda, S.; Al-Mamun, M. Effects of dietary energy intake and cold exposure on kinetics of plasma glucose metabolism in sheep. J. Anim. Physiol. Anim. Nutr. 2007, 91, 1–5. [Google Scholar] [CrossRef]
- Stefanidis, A.; Wiedmann, N.M.; Tyagi, S.; Allen, A.M.; Watt, M.J.; Oldfield, B.J. Insights into the neurochemical signature of the Innervation of Beige Fat. Mol. Metab. 2018, 11, 47–58. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Demena, R.; Anedda, A.; Cadenas, S.; Obregon, M.-J. TSH effects on thermogenesis in rat brown adipocytes. Mol. Cell. Endocrinol. 2015, 404, 151–158. [Google Scholar] [CrossRef] [PubMed]
- Cannon, B.; Houstek, J.; Nedergaard, J. Brown Adipose Tissue: More Than an Effector of Thermogenesis? Ann. N. Y. Acad. Sci. 1998, 856, 171–187. [Google Scholar] [CrossRef]
- Daval, M.; Foufelle, F.; Ferré, P. Functions of AMP-activated protein kinase in adipose tissue. J. Physiol. 2006, 574, 55–62. [Google Scholar] [CrossRef] [PubMed]
- Chu, S.; Narayan, V.P.; Sung, M.-K.; Park, T. Piperonal attenuates visceral adiposity in mice fed a high-fat diet: Potential involvement of the adenylate cyclase-protein kinase A dependent pathway. Mol. Nutr. Food Res. 2017, 61, 1601124. [Google Scholar] [CrossRef]
- Tong, T.; Shen, Y.; Lee, H.-W.; Yu, R.; Park, T. Adenylyl cyclase 3 haploinsufficiency confers susceptibility to diet-induced obesity and insulin resistance in mice. Sci. Rep. 2016, 6, 34179. [Google Scholar] [CrossRef]
- Vansal, S.S. β3-Adrenergic Receptor Agonists and Other Potential Anti-obesity Agents. Am. J. Pharm. Educ. 2004, 68. [Google Scholar] [CrossRef]
- Marhl, M.; Haberichter, T.; Brumen, M.; Heinrich, R. Complex calcium oscillations and the role of mitochondria and cytosolic proteins. Biosystems 2000, 57, 75–86. [Google Scholar] [CrossRef]
- Grubelnik, V.; Larsen, A.Z.; Kummer, U.; Olsen, L.F.; Marhl, M. Mitochondria regulate the amplitude of simple and complex calcium oscillations. Biophys. Chem. 2001, 94, 59–74. [Google Scholar] [CrossRef]
- Hu, R.; He, M.-L.; Hu, H.; Yuan, B.-X.; Zang, W.-J.; Lau, C.-P.; Tse, H.-F.; Li, G.-R. Characterization of calcium signaling pathways in human preadipocytes. J. Cell. Physiol. 2009, 220, 765–770. [Google Scholar] [CrossRef]
- Andrienko, T.; Kuznetsov, A.V.; Kaambre, T.; Usson, Y.; Orosco, A.; Appaix, F.; Tiivel, T.; Sikk, P.; Vendelin, M.; Margreiter, R.; et al. Metabolic consequences of functional complexes of mitochondria, myofibrils and sarcoplasmic reticulum in muscle cells. J. Exp. Biol. 2003, 206, 2059–2072. [Google Scholar] [CrossRef]
- Shkryl, V.M.; Shirokova, N. Transfer and Tunneling of Ca2+ from Sarcoplasmic Reticulum to Mitochondria in Skeletal Muscle. J. Biol. Chem. 2006, 281, 1547–1554. [Google Scholar] [CrossRef]
- De Meis, L.; Oliveira, G.M.; Arruda, A.P.; Santos, R.; Da Costa, R.M.; Benchimol, M. The Thermogenic Activity of Rat Brown Adipose Tissue and Rabbit White Muscle Ca2+-ATPase. IUBMB Life 2005, 57, 337–345. [Google Scholar] [CrossRef]
- Gnad, T.; Scheibler, S.; Von Kügelgen, I.; Scheele, C.; Kilić, A.; Glöde, A.; Hoffmann, L.S.; Reverte-Salisa, L.; Horn, P.; Mutlu, S.; et al. Adenosine activates brown adipose tissue and recruits beige adipocytes via A2A receptors. Nat. Cell Biol. 2014, 516, 395–399. [Google Scholar] [CrossRef] [PubMed]
- Leaver, E.V.; Pappone, P.A. β-Adrenergic potentiation of endoplasmic reticulum Ca2+ release in brown fat cells. Am. J. Physiol. Physiol. 2002, 282, C1016–C1024. [Google Scholar] [CrossRef] [PubMed]
- Strosberg, A.D. STRUCTURE AND FUNCTION OF THE β3-ADRENERGIC RECEPTOR. Annu. Rev. Pharmacol. Toxicol. 1997, 37, 421–450. [Google Scholar] [CrossRef] [PubMed]
- Henry, B.A.; Pope, M.; Birtwistle, M.; Loughnan, R.; Alagal, R.; Fuller-Jackson, J.-P.; Perry, V.; Budge, H.; Clarke, I.J.; Symonds, M.E. Ontogeny and Thermogenic Role for Sternal Fat in Female Sheep. Endocrinology 2017, 158, 2212–2225. [Google Scholar] [CrossRef]
- Hampton, M.; Melvin, R.G.; Andrews, M.T. Transcriptomic Analysis of Brown Adipose Tissue across the Physiological Extremes of Natural Hibernation. PLoS ONE 2013, 8, e85157. [Google Scholar] [CrossRef] [PubMed]
- Orozco-Solis, R.; Aguilar-Arnal, L.; Murakami, M.; Peruquetti, R.; Ramadori, G.; Coppari, R.; Sassone-Corsi, P. The Circadian Clock in the Ventromedial Hypothalamus Controls Cyclic Energy Expenditure. Cell Metab. 2016, 23, 467–478. [Google Scholar] [CrossRef] [PubMed]
- Kurylowicz, A.; Jonas, M.; Lisik, W.; Jonas, M.; Wicik, Z.A.; Wierzbicki, Z.; Chmura, A.; Puzianowska-Kuznicka, M. Obesity is associated with a decrease in expression but not with the hypermethylation of thermogenesis-related genes in adipose tissues. J. Transl. Med. 2015, 13, 31. [Google Scholar] [CrossRef]
- De Meis, L. Brown Adipose Tissue Ca2+-ATPase: Uncoupled ATP hydrolysis and thermogenic activity. J. Biol. Chem. 2003, 278, 41856–41861. [Google Scholar] [CrossRef] [PubMed]
- De Meis, L.; Arruda, A.P.; Da Costa, R.M.; Benchimol, M. Identification of a Ca2+-ATPase in Brown Adipose Tissue Mitochondria: Regulation of thermogenesis by ATP and Ca2+. J. Biol. Chem. 2006, 281, 16384–16390. [Google Scholar] [CrossRef]
- Bal, N.C.; Sahoo, S.K.; Maurya, S.K.; Periasamy, M. The Role of Sarcolipin in Muscle Non-shivering Thermogenesis. Front. Physiol. 2018, 9, 1217. [Google Scholar] [CrossRef]
- Bal, N.C.; Singh, S.; Reis, F.C.G.; Maurya, S.K.; Pani, S.; Rowland, L.A.; Periasamy, M. Both brown adipose tissue and skeletal muscle thermogenesis processes are activated during mild to severe cold adaptation in mice. J. Biol. Chem. 2017, 292, 16616–16625. [Google Scholar] [CrossRef]
- Yao, C.-K.; Liu, Y.-T.; Lee, I.-C.; Wang, Y.-T.; Wu, P.-Y. A Ca2+ channel differentially regulates Clathrin-mediated and activity-dependent bulk endocytosis. PLoS Biol. 2017, 15, e2000931. [Google Scholar] [CrossRef] [PubMed]
- El-Merahbi, R.; Löffler, M.; Mayer, A.; Sumara, G. The roles of peripheral serotonin in metabolic homeostasis. FEBS Lett. 2015, 589, 1728–1734. [Google Scholar] [CrossRef]
- Sumara, G.; Sumara, O.; Kim, J.K.; Karsenty, G. Gut-Derived Serotonin Is a Multifunctional Determinant to Fasting Adaptation. Cell Metab. 2012, 16, 588–600. [Google Scholar] [CrossRef]
- Söhle, J.; Machuy, N.; Smailbegovic, E.; Holtzmann, U.; Grönniger, E.; Wenck, H.; Stäb, F.; Winnefeld, M. Identification of New Genes Involved in Human Adipogenesis and Fat Storage. PLoS ONE 2012, 7, e31193. [Google Scholar] [CrossRef]
- Harms, M.; Seale, P. Brown and beige fat: Development, function and therapeutic potential. Nat. Med. 2013, 19, 1252–1263. [Google Scholar] [CrossRef] [PubMed]
- Bostroem, P.; Wu, J.; Jedrychowski, M.P.; Korde, A.; Ye, L.; Lo, J.C.; Rasbach, K.A.; Bostroem, E.A.; Choi, J.H.; Long, J.Z.; et al. A PGC1-α-dependent myokine that drives brown-fat-like development of white fat and thermogenesis. Nat. Cell Biol. 2012, 481, 463–468. [Google Scholar] [CrossRef]
- Zhu, Q.; Ghoshal, S.; Rodrigues, A.; Gao, S.; Asterian, A.; Kamenecka, T.M.; Barrow, J.C.; Chakraborty, A. Adipocyte-specific deletion of Ip6k1 reduces diet-induced obesity by enhancing AMPK-mediated thermogenesis. J. Clin. Investig. 2016, 126, 4273–4288. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Q.; Ghoshal, S.; Tyagi, R.; Chakraborty, A. Global IP6K1 deletion enhances temperature modulated energy expenditure which reduces carbohydrate and fat induced weight gain. Mol. Metab. 2017, 6, 73–85. [Google Scholar] [CrossRef] [PubMed]
- Kubota, N.; Yano, W.; Kubota, T.; Yamauchi, T.; Itoh, S.; Kumagai, H.; Kozono, H.; Takamoto, I.; Okamoto, S.; Shiuchi, T.; et al. Adiponectin Stimulates AMP-Activated Protein Kinase in the Hypothalamus and Increases Food Intake. Cell Metab. 2007, 6, 55–68. [Google Scholar] [CrossRef]
- Duivenvoorden, I.; Teusink, B.; Rensen, P.C.; Romijn, J.A.; Havekes, L.M.; Voshol, P.J. Apolipoprotein C3 Deficiency Results in Diet-Induced Obesity and Aggravated Insulin Resistance in Mice. Diabetes 2005, 54, 664–671. [Google Scholar] [CrossRef] [PubMed]
- Sahoo, A.; Paul, R.K.; Thirumurgan, P.; Sharma, S.; Kumawat, P.K.; De, K. Immunological and plasma antioxidant re-sponse following protection of newborn lambs from cold by umbrella-type housing and lamb-jacket in winter. Biol. Rhythm Res. 2019, 1–8. [Google Scholar] [CrossRef]
- Sejian, V.; Kumar, D.; Naqvi, S.M.K. Physiological rhythmicity in Malpura ewes to adapt to cold stress in a semi-arid tropical environment. Biol. Rhythm Res. 2017, 49, 215–225. [Google Scholar] [CrossRef]
| Gene Name | Primer Sequences (5′–3′) | Products Size (bp) | Tm (°C) |
|---|---|---|---|
| UCP1 | ACTTCGTGTCCGCTGTTGTTG | 104 | 61 |
| TGTGTACTGTCCTGGTGAAGAGTT | |||
| RYR1 | CAAGTGCTTCATCTGCGGTAT | 83 | 60 |
| TGTGTTCCTCCAGTGTGTGAGT | |||
| ADIPOQ | GAACAGTCCACAGGTCTAC | 190 | 61 |
| CCTTCCATTACCACTACATAAC | |||
| LPL | CGACAGGATTACAAGAGGAA | 100 | 60 |
| AGGAATGAGGTGGCAAGT | |||
| β-actin | AGCCTTCCTTCCTGGGCATGGA | 113 | 60 |
| GGACAGCACCGTGTTGGCGTAGA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiao, D.; Ji, K.; Liu, H.; Wang, W.; Wu, X.; Zhou, J.; Zhang, Y.; Zhou, H.; Hickford, J.G.H.; Degen, A.A.; et al. Transcriptome Analysis Reveals Genes Involved in Thermogenesis in Two Cold-Exposed Sheep Breeds. Genes 2021, 12, 375. https://doi.org/10.3390/genes12030375
Jiao D, Ji K, Liu H, Wang W, Wu X, Zhou J, Zhang Y, Zhou H, Hickford JGH, Degen AA, et al. Transcriptome Analysis Reveals Genes Involved in Thermogenesis in Two Cold-Exposed Sheep Breeds. Genes. 2021; 12(3):375. https://doi.org/10.3390/genes12030375
Chicago/Turabian StyleJiao, Dan, Kaixi Ji, Hu Liu, Wenqiang Wang, Xiukun Wu, Jianwei Zhou, Yunsheng Zhang, Huitong Zhou, Jon G. H. Hickford, Allan A. Degen, and et al. 2021. "Transcriptome Analysis Reveals Genes Involved in Thermogenesis in Two Cold-Exposed Sheep Breeds" Genes 12, no. 3: 375. https://doi.org/10.3390/genes12030375
APA StyleJiao, D., Ji, K., Liu, H., Wang, W., Wu, X., Zhou, J., Zhang, Y., Zhou, H., Hickford, J. G. H., Degen, A. A., & Yang, G. (2021). Transcriptome Analysis Reveals Genes Involved in Thermogenesis in Two Cold-Exposed Sheep Breeds. Genes, 12(3), 375. https://doi.org/10.3390/genes12030375








