Abstract
MYO7A gene encodes unconventional myosin VIIA, which, when mutated, causes a phenotypic spectrum ranging from recessive hearing loss DFNB2 to deaf-blindness, Usher Type 1B (USH1B). MYO7A mutations are reported in nine DFNB2 families to date, none from sub-Saharan Africa.In DNA, from a cohort of 94 individuals representing 92 families from the Limpopo province of South Africa, eight MYO7A variations were detected among 10 individuals. Family studies identified homozygous and compound heterozygous mutations in 17 individuals out of 32 available family members. Four mutations were novel, p.Gly329Asp, p.Arg373His, p.Tyr1780Ser, and p.Pro2126Leufs*5. Two variations, p.Ser617Pro and p.Thr381Met, previously listed as of uncertain significance (ClinVar), were confirmed to be pathogenic. The identified mutations are predicted to interfere with the conformational properties of myosin VIIA through interruption or abrogation of multiple interactions between the mutant and neighbouring residues. Specifically, p.Pro2126Leufs*5, is predicted to abolish the critical site for the interactions between the tail and the motor domain essential for the autoregulation, leaving a non-functional, unregulated protein that causes hearing loss. We have identified MYO7A as a possible key deafness gene among indigenous sub-Saharan Africans. The spectrum of MYO7A mutations in this South African population points to DFNB2 as a specific entity that may occur in a homozygous or in a compound heterozygous state.
1. Introduction
MYO7A encodes an unconventional myosin, myosin VIIA, which, when mutated, causes a phenotypic spectrum ranging from recessive nonsyndromic hearing loss (DFNB2) to syndromic deaf-blindness, Usher Type 1B (USH1B). Studies in transfected mouse hair cells demonstrated that DNFB2 mutant myosin VIIA protein localizes correctly to the hair cell stereocilia, yet the USH1B myosin VIIA proteins do not, confirming that, in contrast to USH1B-associated alleles, the DNFB2-associated alleles retain some residual function and that DFNB2 and USH1B are different ends of the same disease spectrum. Furthermore, elucidation of the three distinct forms of Usher Syndrome Type 1 demonstrated that USH1B, USH1C, and USH1D are distinct genetic disorders caused by mutations in three different genes, MYO7A, CHD23, and USH1C, respectively [1,2,3,4,5]. DFNB2 is a recessive form of prelingual sensorineural hearing loss that may, on rare occasions, occur with vestibular dysfunction and vertigo.
1.1. Background Information
Tip links and the mechanoelectrical transduction apparatus of the cochlea hair cells.
The hearing mechanism at the cochlear level depends on the integrity of both the physiologic conditions and the mechanical conditions in the cochlea. Of these factors, the most vital is the state of the hair cells and their stereocilia. Inner ear hair cell stereocilia exhibit links between the tips and sides of adjacent stereocilia [6,7,8]. These links are part of the mechanoelectrical transduction (MET) apparatus of the sensory cells of the inner ear [9,10,11]. The links also ensure that the stereocilia are mechanically integrated into bundles that can withstand the repeated stress caused by shearing movements that occur during sound conduction [6,7,8,12].
Myosin VIIA belongs to the myosin superfamily, a large and diverse family of over twenty proteins whose members are involved in a number of cellular pathways [13,14]. In humans, myosins are grouped into twelve classes. Myosin sub-class VIIA is expressed in the inner ear. Specifically, myosin VIIA is a vital component of the tripartite complex in the hair cell stereocilia where it helps to maintain the mechanical tension across the cadherin links [12,15,16] and to transport the protein complex to the tips [17,18,19]. Myosin VIIA forms tripartite complexes involving two modular adaptor proteins, SANS and Harmonin, that anchor the cytoplasmic domains of cadherins to the actin cytoskeleton [12,17,20,21]. Both harmonin and myosin VIIA localize along the length of stereocilia, but the three Usher proteins are localized at the upper-tip link density (UTLD), where they associate with the tip link and regulate the function of MET [22,23,24,25]. In the assembly (Figure 1), the cytoplasmic tail of CDH23 binds to the PDZ domain scaffolding protein harmonin (USH1C), which in turn forms a tripartite complex with the ankyrin-repeat SANS adaptor (USH1G), and which in turn binds to the tail domain of myosin VIIA [17,26]. The cooperation of these three proteins is essential because the earliest connections between the growing stereocilia are critical for shaping the hair bundle as a coherent unit. Failure of this interaction leads to disorganized stereocilia and to hearing loss.
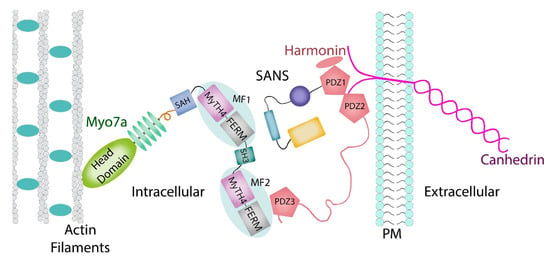
Figure 1.
Assembly of the myosin 7a-SANS-harmonin complex at a cellular level.
1.2. Myosin VIIA Heavy Chain Structure and Function
Structurally (Figure 1), the well conserved myosin VIIA heavy chain contains domains that provide for its motor-and cargo-binding functions [27,28,29]. The head and neck are composed of a conserved N-terminal region containing sixty amino acids, a characteristic motor domain, and five isoleucine-glutamine (IQ) motifs that bind calmodulin [30,31] and/or specific light chains. Next is a stable single α–helix (SAH) [29] followed by a tandem repeat of myosin tail homology 4 (MyTH4) domains and partial four-point 1, ezrin, radixin, and moesin (FERM) domains that are separated by an SH3 subdomain. Myosin VIIA demonstrates actin activated ATPase activity [32,33].
The highly conserved motor domain contains ATP- and actin binding sites associated with nucleotide binding, hydrolysis and product release, essential for myosin enzyme motility, while the tail region mediates dimerization and binding to other proteins or cargo [27]. Furthermore, myosin VIIA motor activity plays a key role in the differentiation and organization of hair cell stereocilia [34]. The myosin MF domains bind to the cytoplasmic tails of signaling receptors and adhesion receptors and interact with adapter proteins [27,35,36,37,38] as well as play a major role in autoinhibition [39]. Studies across species show that FERM subdomains exhibit class-specific sequence variations yet retain a very high degree of conservation of the supramodule, and of the cloverleaf configuration which controls not only the three lobes of the FERM domain, but the rotational freedom of the MyTH4 as well. Indeed, though the FERM domain may bind to actin binding proteins, signaling receptors and adhesion molecules [35,36,37,38,39,40], it remains vital in the autoinhibition of mammalian Myo7a and Myo10 as well as in Drosophila Myo7a [28,39,40,41].
Biochemical experiments have shown that myosin VIIA has a compact structure that exhibits, in its monomeric form, intramolecular bending as well as a unique regulatory mechanism [39]. Furthermore, myosin VIIA adopts a bent conformation in the presence of ATP and is extended in the absence of ATP [39]. A portion of the tail interacts with the motor domain [39] in the bent conformation. Yang et al., 2009 [39] demonstrated that, upon release of the tail inhibition, the full length myosin VIIA can transport its cargo molecules, such as the USH1 proteins. Myosin VIIA has been shown to associate with large protein complexes, either transporting them to a particular site or stabilizing the cargo in a specific position [27,42]. In the neuroretina, myosin VIIA plays a key role in the distribution and migration of retinal pigment epithelial (RPE) melanosomes and phagosomes [43,44], and enables the regulation of opsin transport in retinal photoreceptors [45,46].
1.3. MYO7A Gene Expression and Phenotypes
MYO7A is expressed in cochlea neuroepithelia, vestibular neuroepithelia, retinal photoreceptor cells, and retinal pigment epithelium. In humans, MYO7A is expressed in both the pigment cells and in the photoreceptor cells in the retina. In mice, however, myo7a is not expressed in the photoreceptor cells but only in pigment epithelium. For this reason, the shaker-1 mouse has no retinal defects, only demonstrating hearing and balance disorders [47]. In humans, a primary rod and cone defect were shown to cause the syndrome depicting the retinal defects associated with the vestibuloauditory defects of USH1B [47]. The DFNB2 mutations do not give rise to retinal defects and blindness, even though they are found in the same gene as the USH1B mutations.
Non-syndromic sensorineural hearing loss was reported in 22 individuals from a large consanguineous family and mapped to chromosome 11q13 [48] through linkage analysis. It was subsequently identified to be due to homozygous mutations in the MYO7A gene [49]. Following on these findings, two Chinese families were identified with homozygous and heterozygous MYO7A mutations [50], extending the DFNB2 genotype from homozygous recessive mutations alone to include compound heterozygous mutations. Since then, characterization studies point to DFNB2 and Usher syndrome as representing a disease spectrum with variable expressivity. Molecular findings demonstrate that the DNFB2-associated alleles retain some residual function at the hair cell apices in contrast to USH1B-associated alleles [51].
Confirmation of DFNB2 as a separate entity has been relatively slow, mainly because, initially, families from other population groups could not be identified. These negative findings led to calling into question the existence of DFNB2 altogether by the researchers who felt there was disparity in the ‘alleged causative mutations’ and the nonsyndromic phenotype [52]. Prior to the current study findings (Table 1), no DFNB2 families had been reported from sub-Saharan Africa. Over time, DFNB2 linked MYO7A mutations have now been identified in different population groups including Pakistani [19], Iranian [53], Middle Eastern populations including Jewish and Arab Palestinian populations [54], and Chinese [50,55,56] (Table 2). Mutations have been reported mainly among populations practicing close family marriages. However, DFNB2 has been suspected, either through linkage studies, or through characterization studies of MYO7A mutations, in a wider spectrum of population groups. These include Tunisian, Pakistani, Saudi Arabian, Iranian, Chinese, Japanese, Swedish, Spanish, Palestinian, and Moroccan population groups respectively [19,50,53,55,56,57,58,59,60,61]. Out of these, clearly characterized mutations in MYO7A have been confirmed in fourteen DFNB2 families to date (Table 2). We now report our findings demonstrating the existence of DFNB2, due to both homozygous and compound heterozygous MYO7A mutations, among sub-Saharan African populations from South Africa.

Table 1.
Summary of genotypes in the sub-Saharan South African DFNB2 families.

Table 2.
Reported DFNB2 families with characterized MYO7A mutations.
2. Materials and Methods
This study was approved by the Ethics Committee of the Faculty of Health Sciences, University of Pretoria, South Africa, Ethics Approval number 395/2014, and the Institutional Review Board of the University of Miami, Miller School of Medicine, Miami Fl, USA. A signed informed-consent form was obtained from each participant or, in the case of a minor, from the parents.
2.1. Subjects
We included in this study 94 GJB2 mutation-negative indigenous sub-Saharan African individuals representing 92 families from the Limpopo Province of South Africa. Of these, 23 individuals had a definite family history of hearing loss, 49 individuals had none and 22 individuals were uncertain. Since a three-generation pedigree was not available in some cases, we did not group multiplex families according to an inheritance pattern.
2.2. Clinical Evaluation
Clinical evaluation included a thorough physical examination and otoscopy in all cases. Vestibular assessment was performed according to the “The ten-minute examination of the dizzy patient” protocol [62]. Additional evaluations, including a high-resolution, thin-section computed tomography (CT) and magnetic resonance imaging (MRI) of the temporal bone, were performed when possible. DNA was extracted from peripheral blood leukocytes of probands according to the standard procedures. SNHL was established via the standard audiometry in a soundproofed room according to the current clinical standards. HL was congenital onset or prelingual onset with a severity ranging from severe to profound. Assessment for retinopathy was performed by ophthalmologists at the local hospitals where they were referred to at least three occasions for each individual. Fundoscopy as well as tests for visual acuity and visual fields were done during the assessments. ERG and fundus photos were not performed due to lack of the equipment at the local hospitals. The initial evaluation was done in childhood at school entry during middle school. The third evaluation was done more recently at the time of family recruitment for the current study. The clinical records obtained at all these assessments together with history taken during vestibular assessment were used to determine the presence or absence of visual defects.
2.3. Sequencing
Using the Agilent SureDesign online tool (Available online: https://earray.chem.agilent.com/suredesign/ (accessed on 23 November 2020)), a SureSelect custom kit (Agilent, Santa Clara, CA, USA) was designed to include all exons, 5′ UTRs and 3′ UTRs of 180 known and candidate deafness causing genes (Supplementary Table S1) [63]. This custom capture panel (MiamiOtoGenes), with a target size of approximately 1.158 MB encompassing 3494 regions, covers genes associated with both syndromic and non-syndromic forms of HL. The targeted sequencing was processed at the Hussman Institute for Human Genomics (HIHG) Sequencing core, University of Miami. The Agilent’s SureSelect Target Enrichment (Agilent, Santa Clara, CA, USA) of coding exons and flanking intronic sequences in-solution hybridization capture system was used following the manufacturer’s standard protocol. Adapter sequences for the Illumina HiSeq 2000 were ligated, and the enriched DNA samples were prepared using the standard methods for the HiSeq 2000 instrument (Illumina). Through the sample preparation, average insert size was 180 bp and paired end reads were used. Regions with lower coverage were not subjected to additional sequencing.
2.4. Bioinformatics Analysis
The Illumina CASAVA v1.8 pipeline was used to assemble 99 bp sequence reads. Burrows–Wheeler Aligner (BWA) was applied for alignment of sequence reads to the human reference genome (hg19) [64], and variants were called using FreeBayes [65]. Genesis 2.0 (Available online: https://www.genesis-app.com/ (accessed on 23 November 2020)) was then used for variant filtering based on quality/score read depth and minor allele frequency (MAF thresholds of 0.005 for ARNSHL and 0.0005 for ADNSHL variants) as reported in dbSNP141, the National Heart, Lung, and Blood Institute Exome Sequencing Project Exome Variant Server, Seattle, WA Project (Exome Variant Server 2012), Exome Aggregation Consortium (ExAC) browser (Available online: http://exac.broadinstitute.org/ (accessed on 23 November 2020)), gnomAD, the 1000 Genome Project Database, and our internal database of >3000 samples from European, Asian, and American ancestries.
Variants meeting these criteria were further annotated based on their presence and pathogenicity information in Human Gene Mutation Database (HGMD; Available online: http://www.hgmd.cf.ac.uk (accessed on 23 November 2020)), the Deafness Variation Database (DVD) (Available online: deafnessvariationdatabase.org (accessed on 23 November 2020)), and ClinVar (Available online: http://www.ncbi.nlm.nih.gov/clinvar/ (accessed on 23 November 2020)). In the final step, all variants were re-classified based on the American College of Medical Genetics and Genomics (ACMG) and Association for Molecular Pathology (AMP) guidelines [66], together with the variant interpretation guidelines for genetic hearing loss as published by Oza et al., 2018 [67]. These guidelines recommend the use of specific standard terminology for DNA variants in five categories to include pathogenic, likely pathogenic, uncertain significance, likely benign, and benign. They describe criteria using evidence from population data, computational data, functional data, and segregation data for variant interpretation. Copy number variation (CNV) calling was performed using an R-based tool [68]. This method normalizes read-depth data by sample batch and compares median read-depth ratios using a sliding-window approach.
Sanger sequencing was used for the confirmation of variant calls and PCR for the CNVs. Family members, when available, were used for segregation, de novo status, and trans configuration of biallelic variants. During the interpretation, we also considered phenotypic correlations between the gene variants and their reported phenotypes.
In silico, 3D modelling was performed to investigate and predict the possible effects of the mutations on the protein product.
3. Results
3.1. Family Pedigrees
From a previous study involving 184 congenitally deaf indigenous African individuals from the Limpopo province of South Africa, all negative for GJB2 mutations, the GJB6-D13S1830 deletion and four mitochondrial mutations, A1555G, A3243G, A7511C and A7445G [69], a cohort of 94 individuals representing 92 families was assessed for mutations in 180 deafness genes by capture targeted massively parallel sequencing (CTMPS) using the MiamiOtogene panel [70].
Eight MYO7A variants were detected in ten individuals (including two sibling pairs) from this cohort. All were adults between 26 and 37 years of age.
Two new families with deaf family members identified in the course of family tracing agreed to be included in the study and were recruited. These families are branches of Family TS074/TS093, and are designated as Branch B and Branch C, respectively, on the pedigree charts.
All families demonstrated co-segregation of the MYO7A mutations with hearing loss. (Figure 2, Table 1 and Table 2).
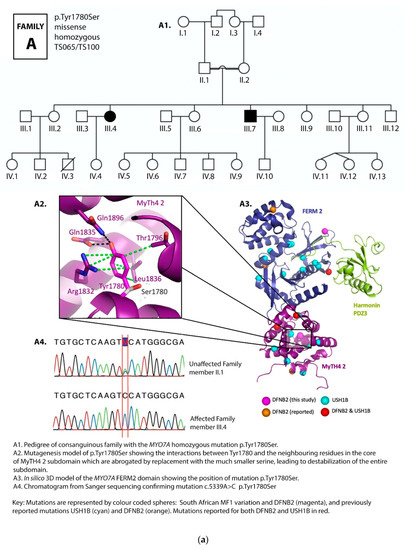
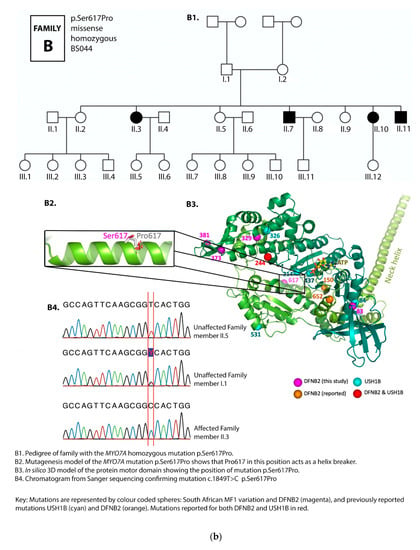
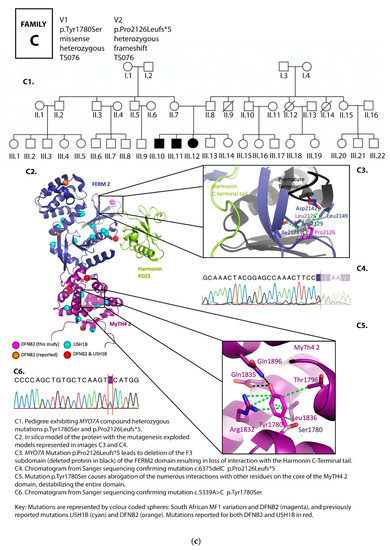
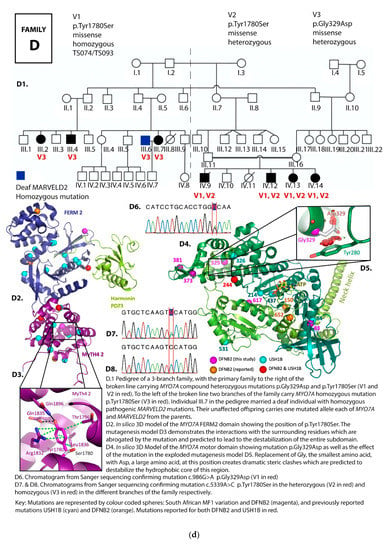
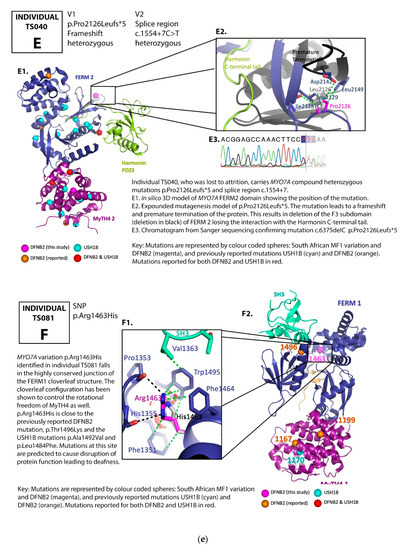
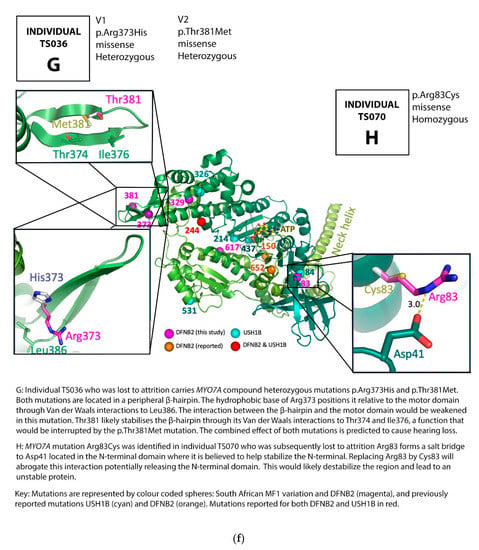
Figure 2.
(a) Pedigree and mutation profile of DFNB2 in Family A; (b) pedigree and mutation profile of DFNB2 in Family B; (c) pedigree and mutation profile of DFNB2 in Family C; (d) pedigree and mutation profile of DFNB2 in Family D; (e) mutation profile and variant of DFNB2 in individuals TS040 and TS081; (f) mutation profiles of DFNB2 in individuals TS036 and TS070.
3.2. MYO7A Mutations
The eight MYO7A variants, including a splice site variant, identified in the South African cohort were ascertained among the respective family members. Out of thirty-two family members assessed, variants presenting both in the homozygous and in the compound heterozygous states were identified among seventeen adult individuals. All seventeen affected individuals demonstrated bilateral profound sensorineural hearing loss but no visual defects. One deaf individual, a 58-year old female, exhibited a subjective mild vestibular defect. Although she was not available for vestibular assessment, we were able to interview her via telephone. She, however, had normal vision and visual fields according to her medical records. All five available families demonstrated perfect co-segregation between the variants and the phenotype, with only the individuals carrying homozygous or compound heterozygous states exhibiting nonsyndromic hearing loss.
Four novel mutations, p.Gly329Asp, p.Arg373His, p.Tyr1780Ser, and p.Pro2126Leufs*5, as well as reported mutations p.Ser617Pro and p.Thr381Met, were identified (Table 1 and Table 2; Figure 2a–f, Figure 3).
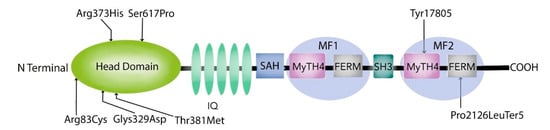
Figure 3.
Diagrammatic representation of Myosin 7A indicating the mutations in DFNB2 South African families.
Mutation p.Tyr1780Ser was the most common variant, found in both homozygous and compound heterozygous states in four families. Three families carried the p.Tyr1780Ser mutation in the homozygous state, while it was presented in the compound heterozygous state in one family. In families D1–D3, it was found in both the homozygous and compound heterozygous genotypes among different branches of the family (Table 1, Figure 2d). The spectrum of mutations identified in the current study were compared to known DFNB2 and USH1B mutations (Table 3). These results are further referred to in Section 4.

Table 3.
Spectrum of MYO7A mutations reported on ClinVar among DFNB2 and USH1B families.
4. Discussion
Current understanding, based on years of the cumulative research on myosin VIIA, reveals that, in the inner ear, defective myosin VIIA will affect not only the morphology and function of hair cell stereocilia, but the whole MET process of the hair cells, leading to hearing loss. Studies have demonstrated (as confirmed by electron microscopy) that the wild type, full-length human myosin VIIA is a monomer that demonstrates tail-dependent inhibition, and that, in its compact structure, the protein tail domain is bent back towards the head-neck domain [28,39]. Furthermore, at high ionic strength and in the presence of Ca2+, the molecule is extended. The researchers also concluded that the release of this tail-dependent inhibition enables the wild type full-length human myosin VIIA to transport its cargo molecules, such as USH1 proteins. Current understanding also states that, though DNFB2-associated alleles retain some residual function in localizing accurately to the hair cell stereocilia, the mutant unconventional myosin VIIA proteins are unable to support normal hearing function [19].
In silico 3D-modelling in the current study demonstrated the interruption or abrogation of multiple interactions between the mutant and neighbouring residues.
The mutations in the myosin VIIA motor domain identified in our study, p.Arg83Cys, p.Gly329Asp, p.Arg373His, p.Thr381Met, p.Ser617Pro, are predicted to interfere with the conformational properties of the protein through the disruption of the numerous ionic interactions between the mutated and the neighbouring residues (Figure 2b,d,f and Figure 3). These effects could cause hearing loss through any one of the following mechanisms: functional disruption of the movement of the myosin VIIA motor head, the protein’s cargo-carrying function, as well as the formation and function of the protein complexes at the MET apparatus.
Though the function of the N-terminal domain is currently unknown, arginine 83, a positively charged amino acid, forms a salt bridge to aspartate 41 (Asp41) located in the N-terminal domain, and probably stabilizes the N-terminal. Replacing Arginine 83 by a cysteine (a neutral amino acid) will abrogate this interaction potentially releasing the N-terminal domain (Figure 2f) and, in so doing, destabilizing the region and leading to an unstable protein, and, we predict, through this, causing hearing loss.
Glycine is the smallest amino acid with a side chain consisting of hydrogen while aspartate is a large amino acid. Glycine 329 closely packs on to tyrosine 280. Replacing Glycine 329 by aspartate (Asp329) will create dramatic steric clashes (Figure 2d) which will destabilize the whole hydrophobic core of the region and affect the folding of the protein. The function of the entire protein will likely be abolished.
Arginine 373 is located in a peripheral β-hairpin (Figure 2f). Its hydrophobic base positions the β-hairpin relative to the motor domain through van der Waals interactions to leucine 386. Replacing arginine 373 by histidine (Arg373His) will weaken the interaction between the β-hairpin and the motor domain. Although the function of the β-hairpin is unknown, threonine 381 is located in the same peripheral β-hairpin as arginine 373, where it stabilizes the β-hairpin by van der Waals interactions to threonine 374 and isoleucine 376. Replacing threonine 381 by methionine may affect the positioning and function of the β-hairpin. In the current study, one of the families carries both β-hairpin mutations Arg373His and Thr381Met in the compound heterozygous state, which, from our 3D model results, we predict to be pathological when combined together at these sites (Figure 2f, Table 1 and Supplementary Table S1).
Serine 617 is located in the middle of an α-helix. Replacing serine 617 by proline—a known “helix breaker”—will cause a kink or bend in the α-helix, interfering with its folding and the conformation of the protein, which will result in defective protein function (Figure 2b). This, in turn, would lead to hearing loss. The family with this mutation exhibited segregation of hearing loss with the homozygous genotype (Figure 2b), demonstrating the severe effect of the mutation as a helix breaker.
The two mutations in the C-terminal MyTH4-FERM domain, p.Tyr1780Ser and p.Pro2126Leufs*5, both show interesting characteristics on 3D modelling.
Tyrosin 1780 extensively interacts with numerous other side chains through van der Waals interactions, hydrogen bonds, as well as π π-stacking interactions. Replacing it with serine (Ser1780) leaves the pocket in this subdomain empty (Figure 2a,c,d). The effect is a transformation of the entire domain leading to severe disruption of the conformational properties of the protein, and we predict, through this, to hearing loss. In the large pedigree with three affected branches of the family, this mutation occurred both in the homozygous and compound heterozygous states, segregating clearly with deafness in all cases.
Proline 2126 (Pro2126) is located in the second MyTH7 [89], the last subdomain of the FERM 2 domain (Figure 2c,e). The mutation p.Pro2126Leufs*5 leads to destabilization of this domain in two ways. First, it leads to a frameshift and premature termination of the myosin VIIA protein five amino acids into the new reading frame (Figure 2c,e). This abolishes the critical site for the interaction between the tail and the motor domain, a site essential for the autoregulation of the protein and critical for protein function [39]. Secondly, the mutation is close to the harmonin PDZ3 recognition and binding site. Pro2126Leufs*5 deletes the F3 lobe of FERM 2 (deletion in black, Figure 2c,e), losing the interaction with the Harmonin C-terminal tail.
Mutations in the second MyTH4-FERM (MF2) domain, p.Tyr1780Ser and p.Pro2126Leufs*5, occurred in more than one family in this South African cohort (Table 1, Supplementary Table S1).
Following the original alignments by Chen et al., 1996 [3], subsequent studies demonstrated that the crystal structures of FERM domains consist of three lobes (subdomains) organized in a cloverleaf pattern, each containing 100 amino acids [27,90,91,92]. The sequences immediately following the first two subdomains are highly conserved among myosin VIIAs, myosin VII, and myosin VIIBs, and are as conserved as or more highly conserved than the two lobes of the FERM domain that directly precede them (Supplementary Table S2). Due to this, the 100-amino-acid stretch following FERM 1 and the 75-amino-acid stretch following FERM 2 domains have been referred to as the first and second MyTH7 domains, respectively.
Characterization studies (Supplementary Table S2) on the second MyTH7 subdomain of myosin 7a confirmed important mechanisms of action in its role [39,89]. These studies demonstrated that it is the second FERM domain in the myosin 7a tail that binds with actin filaments, and, through cosedimentation experiments, the researchers further revealed that the second FERM domain bound to actin in a saturable manner. They also demonstrated that truncated and unregulated myosin 7a-TD3, in which the C-terminal FERM/MyTH7 domain is missing, was extended with a single clearly discernible motor domain strongly angled to the rest of the molecule, confirming that the MyTH7 subdomain is required to effect the folded conformation [39,89]. Moreover, through MgATPase assays, the conserved SH3-binding motif in the MyTH7 subdomain was shown not to be involved in the regulation of myosin 7a. Finally, when the highly conserved residues Arg2140 and Lys2143, located in the second MyTH7 subdomain, were mutated through point mutations to alanines (Ala2140 and Ala2143 respectively), the mutant remained extended and resembled the myosin 7a-TD3 construct, even in the presence of ATP at low ionic strength, demonstrating a loss of autoregulation. This structure was confirmed by electron microscopy [39,89]. Together, these findings confirmed that it is the second MyTH7 subdomain that is essential for the conformation and the regulation of the protein.
The MYO7A C-terminal MyTH4-FERM domain (MF2) recognizes harmonin through the extended PDZ3 domain (Figure 1) of USH1C (harmornin). Several reported Myo7a deafness mutants that mapped to the surface of MF2 were shown to disrupt harmonin binding, revealing the molecular basis for the disruption of the tripartite complex assembly and of mechanotransduction [27,29,93].
Proline 2126 (Pro2126) is located in the second MyTH7 [89], the last subdomain of the FERM 2 domain (Figure 2c,e). As mentioned above, this subdomain has been shown to be critical for the interaction with the Harmonin PDZ3 domain [29,93]. The C-terminus of Harmonin extends into a deep pocket created by the myosin VIIA FERM 2 domain. Yang et al., 2009 [39] found that point mutations in the MyTH7 subdomain carried a similar enzymatic signature as larger truncations, including ones that even deleted all of the tail. Furthermore, evidence from the reported human truncating mutation in myosin VIIA linked to USH1B [94] that results in a myosin molecule similar in length to the TD3 construct reported by Yang et al., 2009 [39] leads us to predict that our mutant too is unregulated (Supplementary Table S2). The combined effect of the p.Pro2126Leufs*5 mutation identified in South African DFNB2 families on the myosin molecule would likely lead to a non-functional, unregulated protein that fails to form functional tripartite complexes in the hair cell stereocilia and the tip-links, leading to the failure of the MET process, and, we predict, through this, result in hearing loss.
DFNB2 versus Usher 1B Syndrome
Wu et al., 2011 [27] mapped and analyzed 17 missense mutations (non-truncating or deletion mutations were excluded) reported in Usher syndrome onto a crystal structure of myosin VIIA MyTH4-FERM domains. They noted that 1 was in the SH3, 9 were located in FERM, while 7 were located in MyTH4 domains. They further subdivided the mutations into classes depending on location. They predicted that the mutations in the folding core of MyTH4 and FERM domains (Class 1) negatively affected the folding of the MyTH4-FERM supramodule, while mutations in the second class would likely affect interactions between domains or interactions among the tripartite protein complexes, such as between myosin VIIA and Sans. They demonstrated that Glu1349, which is closely involved in myosin VIIA binding to Sans CEN1, when mutated to Lys1349, weakened the binding by 20-fold [27]. The third mutation class involved the conserved solvent exposed residues in F1 and F3. These were predicted to interfere with interactions of the protein with other molecules. The fourth mutation class consisted of residues outside the prediction of the MFS-CEN structure. These were analyzed through sequence analysis. They found a conserved R-X-X-X-P-X-(X)-X-E motif in all MyTH4 domains in loops a9/a10 and a10. Furthermore, they noted that the orientation of a9 and a10 was determined by the salt bridges between the Arg and Glu residues, while the turn structure specifically of the a9/a10 loop was facilitated by the Pro residue (Pro1244). They therefore predicted that, with regard to myosin VIIA MyTH4-FERM tandems in humans, mutations in Arg or Pro in this particular motif would cause deaf-blindness.
Looking at the reported myosin VIIA mutations in published literature and the affected families, and comparing them to the results of the current study, there does not seem to be much difference in the type, site, or spatial distribution pattern of the mutations causing DFNB2 in comparison to USH1B (Table 3, Figure 4 and Figure 5). The DFNB2 mutations identified in the current study in the MyTH4-FERM domains fall into class 1 or class 2 according to the above classification. These mutations are either in the core or partial solvent exposed (Figure 4 and Figure 5). We further mapped the reported USH1B and DFNB2 mutations from published literature to investigate whether there was any significant difference in spatial distribution between the phenotypes and found overlapping results (Figure 4 and Figure 5). Both characteristics were demonstrated for mutations causing DFNB2 and USH1B. Of note, four identical mutations are reported for both USH1B and DFNB2 in published literature. These are p.Arg244Pro, p.Arg1873Trp, p.Pro1887Leu and p.Gly2163Ser (Table 3). Although we did not specifically search for it in the current study, we are yet to identify any possible vision-sparing factor in the South African DFNB2 cohort. We can only conclude that there may well be a modifying factor or gene that influences the phenotypic outcome.
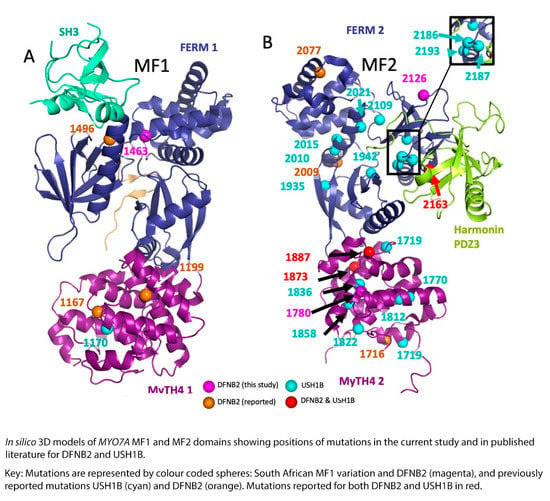
Figure 4.
Mapping of DFNB2 and USH1B mutations from the current study and published literature located in the MYO7A gene MF domains. Subfigure A represents MyTH4-FERM 1 domain (MF1) and subfigure B represents MyTH4-FERM 2 domain (MF2).
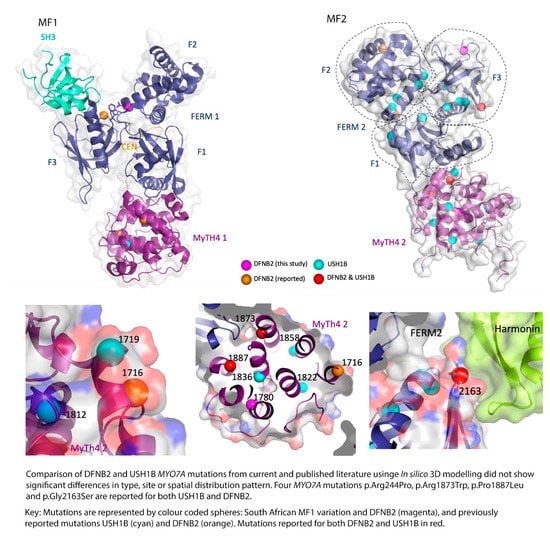
Figure 5.
In silico models comparing location of DFNB2 and USH1 mutations.
We also identified an interesting heterozygous variation, Arg1463His, which falls in the highly conserved junction of the FERM1 clover leaf structure, in a South African deaf proband (Table 1, Figure 2e). It is close to the previously reported DFNB2 mutation, p.Thr1496Lys, and the USH1B mutations p.Ala1492Val and p.Leu1484Phe. The cloverleaf configuration is known to control not only the three lobes of the FERM domain but the rotational freedom of the MyTH4 as well. Mutations at this site would be predicted to cause disruption of the protein function and lead to deafness if identified in the homozygous or compound heterozygous states. This particular South African proband, however, did not demonstrate a second variation to warrant classification as DFNB2. We will watch out for this variation in future studies in the population.
5. Conclusions
The search for prevalent deafness genes among sub-Saharan African populations has been long and fraught with challenges due to the extreme heterogeneity of hearing loss and the tendency of population-based clustering of deafness genes [95]. We have now identified MYO7A as a key deafness gene among the indigenous sub-Saharan African populations from the Limpopo Province of South Africa. This was made possible by the use of Next Generation Sequencing technologies. Using a 180 gene panel, capture targeted massively parallel sequencing yielded MYO7A variations in 10 out of 94 probands, resulting in the identification of nine DFNB2 families in the study population, a relatively high yield that underlines the importance of MYO7A in this African population.
When DFNB2 was proposed as a distinct form of NSHL in 1997, families were sought to confirm the phenotype. Most reported families have come from populations of Middle Eastern, Chinese and North African descent. We have added nine families to the published 14 DFNB2 families worldwide. This is the first report of DNFB2 in sub-Saharan African families. With perfect cosegregation, the spectrum of MYO7A mutations in this South African population confirms DFNB2 as a specific entity that may occur in a homozygous or in a compound heterozygous state.
Although some of the families had reported consanguinity, this is not a widespread cultural practice. In fact, the practice is widely discouraged among the sub-Saharan African populations. The cause of the recurring mutations, especially Pro1780Ser, in this cohort could be due to a founder effect, but allele estimates would first have to be made to confirm this.
Our study did not identify a factor, be it a type of mutation or domain or subdomain location of mutation, responsible for the preservation of vision among DFNB2 individuals when compared to USH1B individuals. We believe a modifying gene is the likely reason. Further studies are scheduled to look into this.
Supplementary Materials
The following are available online at https://www.mdpi.com/2073-4425/12/2/274/s1, Table S1: Summary of MYO7A allele and genotype distribution in the sub-Saharan South African DFNB2 families, Table S2: Alignment of the conserved second MyTH7 subdomain in different species and against p.Pro2126Leufs*5, Figure S1: Sanger sequencing electropherograms of MYO7A mutations among South African DFNB2 families.
Author Contributions
Conceptualization of research, R.I.K.; Recruitment and clinical assessment of probands and families, R.I.K.; DNA extraction and mutation analysis, R.I.K.; Data analysis, R.I.K., W.-D.S., C.L., D.Y. and S.H.B.; Writing up of the manuscript, R.I.K.; 3D modelling, W.-D.S.; Editing of manuscript, W.-D.S., C.L., D.Y., S.H.B., M.S.P. and X.Z.L.; Sequencing, C.L.; D.Y.; Funding, C.L.; M.S.P. and X.Z.L.; Supervisor, M.S.P.; Supervision of research, X.Z.L. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported through the Fulbright Senior Research Scholar Award and the University of Pretoria RDP funding to RI Kabahuma; R01 DC05575 and R01 DC012115 from the National Institutes of Health/National Institute on Deafness and Other Communication Disorders to X.Z.L.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the Ethics Committee of the University of Pretoria, South Africa, Approval number 395/2014, February 2015. It was also approved by the University of Miami Institutional Review Board.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
Data is contained within the article and supplementary material, as well as in our previously published papers [69,70,95].
Acknowledgments
We appreciate the involvement of the participants and their families.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Weil, D.; Blanchard, S.; Kaplan, J.; Guilford, P.; Gibson, F.; Walsh, J.; Mburu, P.; Varela, A.; Levilliers, J.; Weston, M.D.; et al. Defective myosin VIIA gene responsible for Usher syndrome type 1B. Nature 1995, 374, 60–61. [Google Scholar] [CrossRef] [PubMed]
- Lévy, G.; Levi-Acobas, F.; Blanchard, S.; Gerber, S.; Larget-Piet, D.; Chenal, V.; Liu, X.Z.; Newton, V.; Steel, K.P.; Brown, S.D.; et al. Myosin VIIA gene: Heterogeneity of the mutations responsible for Usher syndrome type IB. Hum. Mol. Genet. 1997, 6, 111–116. [Google Scholar] [CrossRef]
- Chen, Z.Y.; Hasson, T.; Kelley, P.M.; Schwender, B.J.; Schwartz, M.F.; Ramakrishnan, M.; Kimberling, W.J.; Mooseker, M.S.; Corey, D.P. Molecular cloning and domain structure of human myosin-VIIa, the gene product defective in Usher syndrome 1B. Genomics 1999, 36, 440–448. [Google Scholar] [CrossRef]
- Adato, A.; Vreugde, S.; Joensuu, T.; Avidan, N.; Hamalainen, R.; Belenkiy, O.; Olender, T.; Bonne-Tamir, B.; Ben-Asher, E.; Espinos, C.; et al. USH3A transcripts encode clarin-1, a four-transmembrane-domain protein with a possible role in sensory synapses. Eur. J. Hum. Genet. 2002, 10, 339–350. [Google Scholar] [CrossRef] [PubMed]
- Karolyi, I.J.; Probst, F.J.; Beyer, L.; Odeh, H.; Dootz, G.; Cha, K.B.; Martin, D.M.; Avraham, K.B.; Kohrman, D.; Dolan, D.F.; et al. Myo15 function is distinct from Myo6, Myo7a and pirouette genes in development of cochlear stereocilia. Hum Mol Genet. 2003, 12, 2797–2805. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Corey, D.P.; Hudspeth, A.J. Kinetics of the receptor current in bullfrog saccular hair cells. J. Neurosci. 1983, 3, 962–976. [Google Scholar] [CrossRef] [PubMed]
- Pickles, J.O.; Comis, S.D.; Osborne, M.P. Cross-links between stereocilia in the guinea pig organ of Corti, and their possible relation to sensory transduction. Hear. Res. 1984, 15, 103–112. [Google Scholar] [CrossRef]
- Pickles, J.O.; Brix, J.; Comis, S.D.; Gleich, O.; Köppl, C.; Manley, G.A.; Osborne, M.P. The organization of tip links and stereocilia on hair cells of bird and lizard basilar papillae. Hear. Res. 1989, 41, 31–41. [Google Scholar] [CrossRef]
- Sakaguchi, H.; Tokita, J.; Müller, U.; Kachar, B. Tip links in hair cells: Molecular composition and role in hearing loss. Curr. Opin. Otolaryngol. Head Neck Surg. 2009, 17, 388–393. [Google Scholar] [CrossRef]
- Hudspeth, A.J. Integrating the active process of hair cells with cochlear function. Nat. Rev. Neurosci. 2014, 15, 600–614. [Google Scholar] [CrossRef] [PubMed]
- Pepermans, E.; Petit, C. The tip-link molecular complex of the auditory mechano-electrical transduction machinery. Hear. Res. 2015, 330, 10–17. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Pan, L.; Zhang, C.; Zhang, M. Large protein assemblies formed by multivalent interactions between cadherin23 and harmonin suggest a stable anchorage structure at the tip link of stereocilia. J. Biol. Chem. 2012, 287, 33460–33471. [Google Scholar] [CrossRef] [PubMed]
- Krendel, M.; Mooseker, M.S. Myosins: Tails (and Heads) of Functional Diversity. Physiology 2005, 20, 239–251. [Google Scholar] [CrossRef] [PubMed]
- Woolner, S.; Bement, W.M. Unconventional myosins acting unconventionally. Trends Cell Biol. 2009, 19, 245–252. [Google Scholar] [CrossRef]
- Kros, C.J.; Marcotti, W.; van Netten, S.M.; Self, T.J.; Libby, R.T.; Brown, S.D.; Richardson, G.P.; Steel, K.P. Reduced climbing and increased slipping adaptation in cochlear hair cells of mice with Myo7a mutations. Nat. Neurosci. 2002, 5, 41–47. [Google Scholar] [CrossRef]
- Self, T.; Mahony, M.; Fleming, J.; Walsh, J.; Brown, S.D.; Steel, K.P. Shaker-1 mutations reveal roles for myosin VIIA in both development and function of cochlear hair cells. Development 1998, 125, 557–566. [Google Scholar] [PubMed]
- El-Amraoui, A.; Petit, C. Usher I syndrome: Unravelling the mechanisms that underlie the cohesion of the growing hair bundle in inner ear sensory cells. J. Cell Sci. 2005, 118, 4593–4603. [Google Scholar] [CrossRef] [PubMed]
- Blanco-Sánchez, B.; Clément, A.; Fierro, J., Jr.; Washbourne, P.; Westerfield, M. Complexes of Usher proteins preassemble at the endoplasmic reticulum and are required for trafficking and ER homeostasis. Dis. Models Mech. 2014, 7, 547–559. [Google Scholar] [CrossRef]
- Riazuddin, S.; Nazli, S.; Ahmed, Z.M.; Yang, Y.; Zulfiqar, F.; Shaikh, R.S.; Zafar, A.U.; Khan, S.N.; Sabar, F.; Javid, F.T.; et al. Mutation spectrum of MYO7A and evaluation of a novel nonsyndromic deafness DFNB2 allele with residual function. Hum. Mutat. 2008, 29, 502–511. [Google Scholar] [CrossRef] [PubMed]
- Crawley, S.W.; Shifrin, D.A., Jr.; Grega-Larson, N.E.; McConnell, R.E.; Benesh, A.E.; Mao, S.; Zheng, Y.; Zheng, Q.Y.; Nam, K.T.; Millis, B.A.; et al. Intestinal Brush Border Assembly Driven by Protocadherin-Based Intermicrovillar Adhesion. Cell 2014, 157, 433–446. [Google Scholar] [CrossRef]
- Crawley, S.W.; Weck, M.L.; Grega-Larson, N.E.; Shifrin, D.A., Jr.; Tyska, M.J. ANKS4B Is Essential for Intermicrovillar Adhesion Complex Formation. Dev. Cell 2016, 36, 190–200. [Google Scholar] [CrossRef]
- Senften, M.; Schwander, M.; Kazmierczak, P.; Lillo, C.; Shin, J.B.; Hasson, T.; Géléoc, G.S.; Gillespie, P.G.; Williams, D.; Holt, J.R.; et al. Physical and functional interaction between protocadherin 15 and myosin VIIa in mechanosensory hair cells. J. Neurosci. 2006, 26, 2060–2071. [Google Scholar] [CrossRef]
- Grati, M.; Kachar, B. Myosin VIIa and sans localization at stereocilia upper tip-link density implicates these Usher syndrome proteins in mechanotransduction. Proc. Natl. Acad. Sci. USA 2011, 108, 11476–11481. [Google Scholar] [CrossRef] [PubMed]
- Belyantseva, I.A.; Boger, E.T.; Friedman, T.B. Myosin XVa localizes to the tips of inner ear sensory cell stereocilia and is essential for staircase formation of the hair bundle. Proc. Natl. Acad. Sci. USA 2003, 100, 13958–13963. [Google Scholar] [CrossRef]
- Indzhykulian, A.A.; Stepanyan, R.; Nelina, A.; Spinelli, K.J.; Ahmed, Z.M.; Belyantseva, I.A.; Friedman, T.B.; Barr-Gillespie, P.G.; Frolenkov, G.I. Molecular Remodeling of Tip Links Underlies Mechanosensory Regeneration in Auditory Hair Cells. PLoS Biol. 2013, 11, 1001583. [Google Scholar] [CrossRef]
- Bahloul, A.; Michel, V.; Hardelin, J.P.; Nouaille, S.; Hoos, S.; Houdusse, A.; England, P.; Petit, C. Cadherin-23, myosin VIIa and harmonin, encoded by Usher syndrome type I genes, form a ternary complex and interact with membrane phospholipids. Hum. Mol. Genet. 2010, 19, 3557–3565. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Pan, L.; Wei, Z.; Zhang, M. Structure of MyTH4-FERM domains in myosin VIIa tail bound to cargo. Science 2011, 331, 757–760. [Google Scholar] [CrossRef] [PubMed]
- Sakai, T.; Jung, H.S.; Sato, O.; Yamada, M.D.; You, D.J.; Ikebe, R.; Ikebe, M. Structure and Regulation of the Movement of Human Myosin VIIA. J. Biol. Chem. 2015, 290, 17587–17598. [Google Scholar] [CrossRef]
- Li, J.; He, Y.; Weck, M.L.; Lu, Q.; Tyska, M.J.; Zhang, M. Structure of Myo7b/USH1C complex suggests a general PDZ domain binding mode by MyTH4-FERM myosins. Proc. Natl. Acad. Sci. USA 2017, 114, 3776–3785. [Google Scholar] [CrossRef] [PubMed]
- Cheney, R.E.; Mooseker, M.S. Unconventional myosins. Curr. Opin. Cell Biol. 1992, 4, 27–35. [Google Scholar] [CrossRef]
- Todorov, P.T.; Hardisty, R.E.; Brown, S.D. Myosin VIIA is specifically associated with calmodulin and microtubule-associated protein-2B (MAP-2B). Biochem. J. 2001, 354, 267–274. [Google Scholar] [CrossRef] [PubMed]
- Udovichenko, I.P.; Gibbs, D.; Williams, D.S. Actin-based motor properties of native myosin VIIa. J. Cell Sci. 2002, 115, 445–450. [Google Scholar]
- Inoue, A.; Ikebe, M. Characterization of the Motor Activity of Mammalian Myosin VIIA. J. Biol. Chem. 2003, 278, 5478–5487. [Google Scholar] [CrossRef]
- Adato, A.; Michel, V.; Kikkawa, Y.; Reiners, J.; Alagramam, K.N.; Weil, D.; Yonekawa, H.; Wolfrum, U.; El-Amraoui, A.; Petit, C. Interactions in the network of Usher syndrome type 1 proteins. Hum. Mol. Genet. 2005, 14, 347–356. [Google Scholar] [CrossRef]
- Zhang, H.; Berg, J.S.; Li, Z.; Wang, Y.; Lang, P.; Sousa, A.D.; Bhaskar, A.; Cheney, R.E.; Stromblad, S. MyosinX provides a motor-based link between integrins and the cytoskeleton. Nat. Cell Biol. 2004, 6, 523–531. [Google Scholar] [CrossRef]
- Liu, L.; Srikakulam, R.; Winkelmann, J. Unc45 activates Hsp90-dependent folding of the myosin motor domain. J. Biol. Chem. 2008, 283, 13185–13193. [Google Scholar] [CrossRef] [PubMed]
- Boëda, B.; El-Amraoui, A.; Bahloul, A.; Goodyear, R.; Daviet, L.; Blanchard, S.; Perfettini, I.; Fath, K.R.; Shorte, S.; Reiners, J.; et al. Myosin VIIa, harmonin and cadherin 23, three Usher I gene products that cooperate to shape the sensory hair cell bundle. EMBO J. 2002, 21, 6689–6699. [Google Scholar] [CrossRef]
- Zhu, X.J.; Wang, C.Z.; Dai, P.G.; Xie, Y.; Song, N.N.; Liu, Y.; Du, Q.S.; Mei, L.; Ding, Y.Q.; Xiong, W.C. Myosin X regulates netrin receptors and functions in axonal path-finding. Nat. Cell Biol. 2007, 9, 184–192. [Google Scholar] [CrossRef]
- Yang, Y.; Baboolal, T.G.; Siththanandan, V.; Chen, M.; Walker, M.L.; Knight, P.J.; Sellers, J.R. A FERM domain autoregulates Drosophila myosin 7a activity. Proc. Natl. Acad. Sci. USA 2009, 106, 4189–4194. [Google Scholar] [CrossRef] [PubMed]
- Pi, X.; Ren, R.; Kelley, R.; Zhang, C.; Moser, M.; Bohil, A.B.; Divito, M.; Cheney, R.E.; Patterson, C. Sequential roles for myosin-X in BMP6-dependent filopodial extension, migration, and activation of BMP receptors. J. Cell Biol. 2007, 179, 1569–1582. [Google Scholar] [CrossRef] [PubMed]
- Umeki, N.; Jung, H.S.; Sakai, T.; Sato, O.; Ikebe, R.; Ikebe, M. Phospholipid-dependent regulation of the motor activity of myosin X. Nat. Struct. Mol. Biol. 2011, 18, 783–788. [Google Scholar] [CrossRef]
- Küssel-Andermann, P.; El-Amraoui, A.; Safieddine, S.; Hardelin, J.P.; Nouaille, S.; Camonis, J.; Petit, C. Unconventional myosin VIIA is a novel A-kinase-anchoring protein. J. Biol. Chem. 2000, 275, 29654–29659. [Google Scholar] [CrossRef]
- Liu, X.Z.; Hope, C.; Walsh, J.; Newton, V.; Ke, X.M.; Liang, C.Y.; Xu, L.R.; Zhou, J.M.; Trump, D.; Steel, K.P.; et al. Mutations in the myosin VIIA gene cause a wide phenotypic spectrum, including atypical Usher syndrome. Am. J. Hum. Genet. 1998, 63, 909–912. [Google Scholar] [CrossRef]
- Gibbs, D.; Kitamoto, J.; Williams, D.S. Abnormal phagocytosis by retinal pigmented epithelium that lacks myosin VIIa, the Usher syndrome 1B protein. Proc. Natl. Acad. Sci. USA 2003, 100, 6481–6486. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Udovichenko, I.P.; Brown, S.D.; Steel, K.P.; Williams, D.S. Myosin VIIa participates in opsin transport through the photoreceptor cilium. J. Neurosci. 1999, 19, 6267–6274. [Google Scholar] [CrossRef]
- Wolfrum, U.; Schmitt, A. Rhodopsin transport in the membrane of the connecting cilium of mammalian photoreceptor cells. Cell Motil. Cytoskelet. 2000, 46, 95–107. [Google Scholar] [CrossRef]
- El-Amraoui, A.; Sahly, I.; Picaud, S.; Sahel, J.; Abitbol, M.; Petit, C. Human Usher 1B/mouse shaker-1: The retinal phenotype discrepancy explained by the presence/absence of myosin VIIA in the photoreceptor cells. Hum. Mol. Genet. 1996, 5, 1171–1178. [Google Scholar] [CrossRef] [PubMed]
- Guilford, P.; Ben Arab, S.; Blanchard, S.; Levilliers, J.; Weissenbach, J.; Belkahia, A.; Petit, C. A non-syndrome form of neurosensory, recessive deafness maps to the pericentromeric region of chromosome 13q. Nat. Genet. 1994, 6, 24–28. [Google Scholar] [CrossRef] [PubMed]
- Weil, D.; Küssel, P.; Blanchard, S.; Lévy, G.; Levi-Acobas, F.; Drira, M.; Ayadi, H.; Petit, C. The autosomal recessive isolated deafness, DFNB2, and the Usher 1B syndrome are allelic defects of the myosin-VIIA gene. Nat. Genet. 1997, 16, 191–193. [Google Scholar] [CrossRef]
- Liu, X.Z.; Walsh, J.; Mburu, P.; Kendrick-Jones, J.; Cope, M.J.; Steel, K.P.; Brown, S.D. Mutations in the myosin VIIA gene cause non-syndromic recessive deafness. Nat. Genet. 1997, 16, 188–190. [Google Scholar] [CrossRef]
- Zina, Z.B.; Masmoudi, S.; Ayadi, H.; Chaker, F.; Ghorbel, A.M.; Drira, M.; Petit, C. From DFNB2 to Usher syndrome: Variable expressivity of the same disease. Am. J. Med Genet. 2001, 101, 181–183. [Google Scholar] [CrossRef] [PubMed]
- Astuto, L.M.; Bork, J.M.; Weston, M.D.; Askew, J.W.; Fields, R.R.; Orten, D.J.; Kimberling, W.J. CDH23 mutation and phenotype heterogeneity: A profile of 107 diverse families with Usher syndrome and nonsyndromic deafness. Am. J. Hum. Genet. 2002, 71, 262–275. [Google Scholar] [CrossRef]
- Hildebrand, M.S.; Thorne, N.P.; Bromhead, C.J.; Kahrizi, K.; Webster, J.A.; Fattahi, Z.; Bataejad, M.; Kimberling, W.J.; Stephan, D.; Najmabadi, H.; et al. Specific isoforms of drosophila shroom define spatial requirements for the induction of apical constriction. Clin. Genet. 2010, 77, 563–571. [Google Scholar] [CrossRef]
- Brownstein, Z.; Abu-Rayyan, A.; Karfunkel-Doron, D.; Sirigu, S.; Davidov, B.; Shohat, M.; Frydman, M.; Houdusse, A.; Kanaan, M.; Avraham, K.B. Novel myosin mutations for hereditary hearing loss revealed by targeted genomic capture and massively parallel sequencing. Eur. J. Hum. Genet. 2014, 22, 768–775. [Google Scholar] [CrossRef]
- Chen, A.; Wayne, S.; Bell, A.; Ramesh, A.; Srisailapathy, C.R.; Scott, D.A.; Sheffield, V.C.; Van Hauwe, P.; Zbar, R.I.; Ashley, J.; et al. New gene for autosomal recessive non-syndromic hearing loss maps to either chromosome 3q or 19p. Am. J. Med Genet. 1997, 71, 467–471. [Google Scholar] [CrossRef]
- Sun, Y.; Chen, J.; Sun, H.; Cheng, J.; Li, J.; Lu, Y.; Lu, Y.; Jin, Z.; Zhu, Y.; Ouyang, X.; et al. Novel missense mutations in MYO7A underlying postlingual high- or low-frequency non-syndromic hearing impairment in two large families from China. J. Hum. Genet. 2011, 56, 64–70. [Google Scholar] [CrossRef]
- Ben-Salem, S.; Rehm, H.L.; Willems, P.J.; Tamimi, Z.A.; Ayadi, H.; Ali, B.R.; Al-Gazali, L. Analysis of two Arab families reveals additional support for a DFNB2 nonsyndromic phenotype of MYO7A. Mol. Biol. Rep. 2014, 41, 193–200. [Google Scholar] [CrossRef] [PubMed]
- Asgharzade, S.; Reiisi, S.; Tabatabaiefar, M.A.; Chaleshtori, M.H. Screening of Myo7A Mutations in Iranian Patients with Autosomal Recessive Hearing Loss from West of Iran. Iran. J. Public Health 2017, 46, 76–82. [Google Scholar]
- Sadeghi, A.; Sanati, M.; Alasti, F.; Hashemzadeh Chaleshtori, M.; Mahmoudian, S.; Ataei, M. Contribution of GJB2 Mutations and Four Common DFNB Loci in Autosomal Recessive Non-Syndromic Hearing Impairment in Markazi and Qom Provinces of Iran. Iran. J. Biotechnol. 2009, 7, 108–111. [Google Scholar]
- Talbi, S.; Bonnet, C.; Riahi, Z.; Boudjenah, F.; Dahmani, M.; Hardelin, J.P.; Wong Jun Tai, F.; Louha, M.; Ammar-Khodja, F.; Petit, C. Genetic heterogeneity of congenital hearing impairment in Algerians from the Ghardaïa province. Int. J. Pediatr. Otorhinolaryngol. 2018, 112, 1–5. [Google Scholar] [CrossRef]
- Bakhchane, A.; Charif, M.; Bousfiha, A.; Boulouiz, R.; Nahili, H.; Rouba, H.; Charoute, H.; Lenaers, G.; Barakat, A. Novel compound heterozygous MYO7A mutations in Moroccan families with autosomal recessive non-syndromic hearing loss. PLoS ONE 2017, 12, e0176516. [Google Scholar] [CrossRef]
- Goebel, J. The Ten-Minute Examination of the Dizzy Patient. Semin. Neurol. 2002, 21, 391–398. [Google Scholar] [CrossRef] [PubMed]
- Tekin, D.; Yan, D.; Bademci, G.; Feng, Y.; Guo, S.; Foster, J., 2nd; Blanton, S.; Tekin, M.; Liu, X. A next-generation sequencing gene panel (MiamiOtoGenes) for comprehensive analysis of deafness genes. Hear Res. 2016, 333, 179–184. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate long-read alignment with burrows-wheeler transform. Bioinformatics 2010, 26, 589–595. [Google Scholar] [CrossRef]
- Garrison, E.; Marth, G. Haplotype-based variant detection from short-read sequencing. arXiv 2012, arXiv:1207.3907. [Google Scholar]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. ACMG Laboratory Quality Assurance Committee (2015). Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. Off. J. Am. Coll. Med Genet. 2015, 17, 405–424. [Google Scholar] [CrossRef]
- Oza, A.M.; DiStefano, M.T.; Hemphill, S.E.; Cushman, B.J.; Grant, A.R.; Siegert, R.K.; Shen, J.; Chapin, A.; Boczek, N.J.; Schimmenti, L.A.; et al. ClinGen Hearing Loss Clinical Domain Working Group Expert specification of the ACMG/AMP variant interpretation guidelines for genetic hearing loss. Hum. Mutat. 2018, 39, 1593–1613. [Google Scholar] [CrossRef]
- Nord, A.S.; Lee, M.; King, M.; Walsh, T. Accurate and exact CNV identification from targeted high-throughput sequence data. BMC Genom. 2011, 12, 184. [Google Scholar] [CrossRef] [PubMed]
- Kabahuma, R.I.; Ouyang, X.; Du, L.L.; Yan, D.; Hutchin, T.; Ramsay, M.; Penn, C.; Liu, X.Z. Absence of GJB2 gene mutations, the GJB6 deletion (GJB6-D13S1830) and four common mitochondrial mutations in nonsyndromic genetic hearing loss in a South African population. Int. J. Pediatr. Otorhinolaryngol. 2011, 75, 611–617. [Google Scholar] [CrossRef][Green Version]
- Yan, D.; Tekin, D.; Bademci, G.; Foster, J., 2nd; Cengiz, F.B.; Kannan-Sundhari, A.; Guo, S.; Mittal, R.; Zou, B.; Grati, M.; et al. Spectrum of DNA variants for non-syndromic deafness in a large cohort from multiple continents. Hum. Genet. 2017, 135, 953–961. [Google Scholar] [CrossRef] [PubMed]
- Ammar-Khodja, F.; Faugère, V.; Baux, D.; Giannesini, C.; Léonard, S.; Makrelouf, M.; Malek, R.; Djennaoui, D.; Zenati, A.; Claustres, M.; et al. Molecular screening of deafness in Algeria: High genetic heterogeneity involving DFNB1 and the Usher loci, DFNB2/USH1B, DFNB12/USH1D and DFNB23/USH1F. Eur. J. Med Genet. 2009, 52, 174–179. [Google Scholar] [CrossRef] [PubMed]
- Adato, A.; Weil, D.; Kalinski, H.; Pel-Or, Y.; Ayadi, H.; Petit, C.; Korostishevsky, M.; Bonne-Tamir, B. Mutation profile of all 49 exons of the human myosin VIIA gene, and haplotype analysis, in Usher 1B families from diverse origins. Am. J. Hum. Genet. 1997, 61, 813–821. [Google Scholar] [CrossRef] [PubMed]
- Jaijo, T.; Aller, E.; Beneyto, M.; Najera, C.; Graziano, C.; Turchetti, D.; Seri, M.; Ayuso, C.; Baiget, M.; Moreno, F.; et al. MYO7A mutation screening in Usher syndrome type I patients from diverse origins. J. Med. Genet. 2007, 44, e71. [Google Scholar] [CrossRef]
- Le Guédard-Méreuze, S.; Vaché, C.; Baux, D.; Faugère, V.; Larrieu, L.; Abadie, C.; Janecke, A.; Claustres, M.; Roux, A.F. Tuffery-Giraud, S. Ex vivo splicing assays of mutations at noncanonical positions of splice sites in USHER genes. Hum. Mutat. 2010, 31, 347–355. [Google Scholar] [CrossRef] [PubMed]
- Cuevas, J.M.; Espinós, C.; Millan, J.M.; Sanchez, F.; Trujillo, M.J.; Ayuso, C.; Beneyto, M.; Najera, C. Identification of three novel mutations in the MYO7A gene. Hum. Mutat. 1999, 14, 181. [Google Scholar] [CrossRef]
- Duzkale, H.; Shen, J.; McLaughlin, H.; Alfares, A.; Kelly, M.A.; Pugh, T.J.; Funke, B.H.; Rehm, H.L.; Lebo, M.S. A systematic approach to assessing the clinical significance of genetic variants. Clin. Genet. 2013, 84, 453–463. [Google Scholar] [CrossRef]
- Janecke, A.R.; Meins, M.; Sadeghi, M.; Grundmann, K.; Apfelstedt-Sylla, E.; Zrenner, E.; Rosenberg, T.; Gal, A. Twelve novel myosin VIIA mutations in 34 patients with Usher syndrome type I: Confirmation of genetic heterogeneity. Hum. Mutat. 1999, 13, 133–140. [Google Scholar] [CrossRef]
- Vaché, C.; Besnard, T.; Blanchet, C.; Baux, D.; Larrieu, L.; Faugère, V.; Mondain, M.; Hamel, C.; Malcolm, S.; Claustres, M.; et al. Nasal epithelial cells are a reliable source to study splicing variants in Usher syndrome. Hum. Mutat. 2010, 31, 734–741. [Google Scholar] [CrossRef]
- Nájera, C.; Beneyto, M.; Blanca, J.; Aller, E.; Fontcuberta, A.; Millán, J.M.; Ayuso, C. Mutations in myosin VIIA (MYO7A) and usherin (USH2A) in Spanish patients with Usher syndrome types I and II, respectively. Hum. Mutat. 2002, 20, 76–77. [Google Scholar] [CrossRef]
- Jaijo, T.; Aller, E.; Oltra, S.; Beneyto, M.; Nájera, C.; Ayuso, C.; Baiget, M.; Carballo, M.; Antiñolo, G.; Valverde, D.; et al. Mutation profile of the MYO7A gene in Spanish patients with Usher syndrome type I. Hum. Mutat. 2006, 27, 290–301. [Google Scholar] [CrossRef] [PubMed]
- Roux, A.F.; Faugère, V.; Vaché, C.; Baux, D.; Besnard, T.; Léonard, S.; Blanchet, C.; Hamel, C.; Mondain, M.; Gilbert-Dussardier, B.; et al. Four-year follow-up of diagnostic service in USH1 patients. Investig. Ophthalmol. Vis. Sci. 2011, 52, 4063–4071. [Google Scholar] [CrossRef]
- Bharadwaj, A.K.; Kasztejna, J.P.; Huq, S.; Berson, E.L.; Dryja, T.P. Evaluation of the myosin VIIA gene and visual function in patients with Usher syndrome type I. Exp. Eye Res. 2000, 71, 173–181. [Google Scholar] [CrossRef]
- Cremers, F.P.; Kimberling, W.J.; Külm, M.; de Brouwer, A.P.; van Wijk, E.; te Brinke, H.; Cremers, C.W.; Hoefsloot, L.H.; Banfi, S.; Simonelli, F.; et al. Development of a genotyping microarray for Usher syndrome. J. Med. Genet. 2007, 44, 153–160. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Roux, A.F.; Faugère, V.; Le Guédard, S.; Pallares-Ruiz, N.; Vielle, A.; Chambert, S.; Marlin, S.; Hamel, C.; Gilbert, B.; Malcolm, S.; et al. French Usher Syndrome Collaboration. Survey of the frequency of USH1 gene mutations in a cohort of Usher patients shows the importance of cadherin 23 and protocadherin 15 genes and establishes a detection rate of above 90%. J. Med. Genet. 2006, 43, 763–768. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ouyang, X.M.; Yan, D.; Du, L.L.; Hejtmancik, J.F.; Jacobson, S.G.; Nance, W.E.; Li, A.R.; Angeli, S.; Kaiser, M.; Newton, V.; et al. Characterization of Usher syndrome type I gene mutations in an Usher syndrome patient population. Hum. Genet. 2005, 116, 292–299. [Google Scholar] [CrossRef]
- Duman, D.; Sirmaci, A.; Cengiz, F.B.; Ozdag, H.; Tekin, M. Screening of 38 genes identifies mutations in 62% of families with nonsyndromic deafness in Turkey. Genet. Test. Mol. Biomarks 2011, 15, 29–33. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Dong, C.; Wang, Q.; Zhong, Z.; Qi, Y.; Ke, X.; Liu, Y. Targeted Next-Generation Sequencing Successfully Detects Causative Genes in Chinese Patients with Hereditary Hearing Loss. Genet. Test. Mol. Biomarkers 2016, 20, 660–665. [Google Scholar] [CrossRef]
- Bonnet, C.; Grati, M.; Marlin, S.; Levilliers, J.; Hardelin, J.P.; Parodi, M.; Niasme-Grare, M.; Zelenika, D.; Délépine, M.; Feldmann, D.; et al. Complete exon sequencing of all known Usher syndrome genes greatly improves molecular diagnosis. Orphanet J. Rare Dis. 2011, 6, 21. [Google Scholar] [CrossRef]
- Kiehart, D.P.; Franke, J.D.; Chee, M.K.; Montague, R.A.; Chen, T.L.; Roote, J.; Ashburner, M. Drosophila crinkled, mutations of which disrupt morphogenesis and cause lethality, encodes fly myosin VIIA. Genetics 2004, 168, 1337–1352. [Google Scholar] [CrossRef] [PubMed]
- Pearson, M.A.; Reczek, D.; Bretscher, A.; Karplus, P.A. Structure of the ERM protein moesin reveals the FERM domain fold masked by an extended actin binding tail domain. Cell 2000, 101, 259–270. [Google Scholar] [CrossRef]
- Smith, D.A.; Geeves, M.A. Cooperative regulation of myosin-actin interactions by a continuous flexible chain II: Actin-tropomyosin-troponin and regulation by calcium. Biophys. J. 2003, 84, 3168–3180. [Google Scholar] [CrossRef]
- Hamada, K.; Shimizu, T.; Yonemura, S.; Tsukita, S.; Tsukita, S.; Hakoshima, T. Structural basis of adhesion-molecule recognition by ERM proteins revealed by the crystal structure of the radixin-ICAM-2 complex. EMBO J. 2003, 22, 502–514. [Google Scholar] [CrossRef] [PubMed]
- Yu, I.M.; Planelles-Herrero, V.J.; Sourigues, Y.; Moussaoui, D.; Sirkia, H.; Kikuti, C.; Stroebel, D.; Titus, M.A.; Houdusse, A. Myosin 7 and its adaptors link cadherins to actin. Nat. Commun. 2017, 8, 15864. [Google Scholar] [CrossRef] [PubMed]
- Vastinsalo, H.; Isosomppi, J.; Aittakorpi, A.; Sankila, E.M. Two Finnish USH1B patients with three novel mutations in myosin VIIA. Mol. Vis. 2006, 12, 1093–1097. [Google Scholar]
- Rudman, J.R.; Kabahuma, R.I.; Bressler, S.E.; Feng, Y.; Blanton, S.H.; Yan, D.; Liu, X.Z. The genetic basis of deafness in populations of African descent. J. Genet. Genom. 2017, 44, 285–294. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).