Differential Expression of a Panel of Ten CNTN1-Associated Genes during Prostate Cancer Progression and the Predictive Properties of the Panel towards Prostate Cancer Relapse
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Western Blot Analysis
2.3. RNA Sequencing Analysis
2.4. Analysis of CNTN1-Associated Genes
2.5. Generation of CRPC in Animal Models
2.6. Patient Populations
2.7. Assignment of Signature Scores to Patients/Tumors and Cutoff Point Estimation
2.8. Statistical Analysis
3. Results
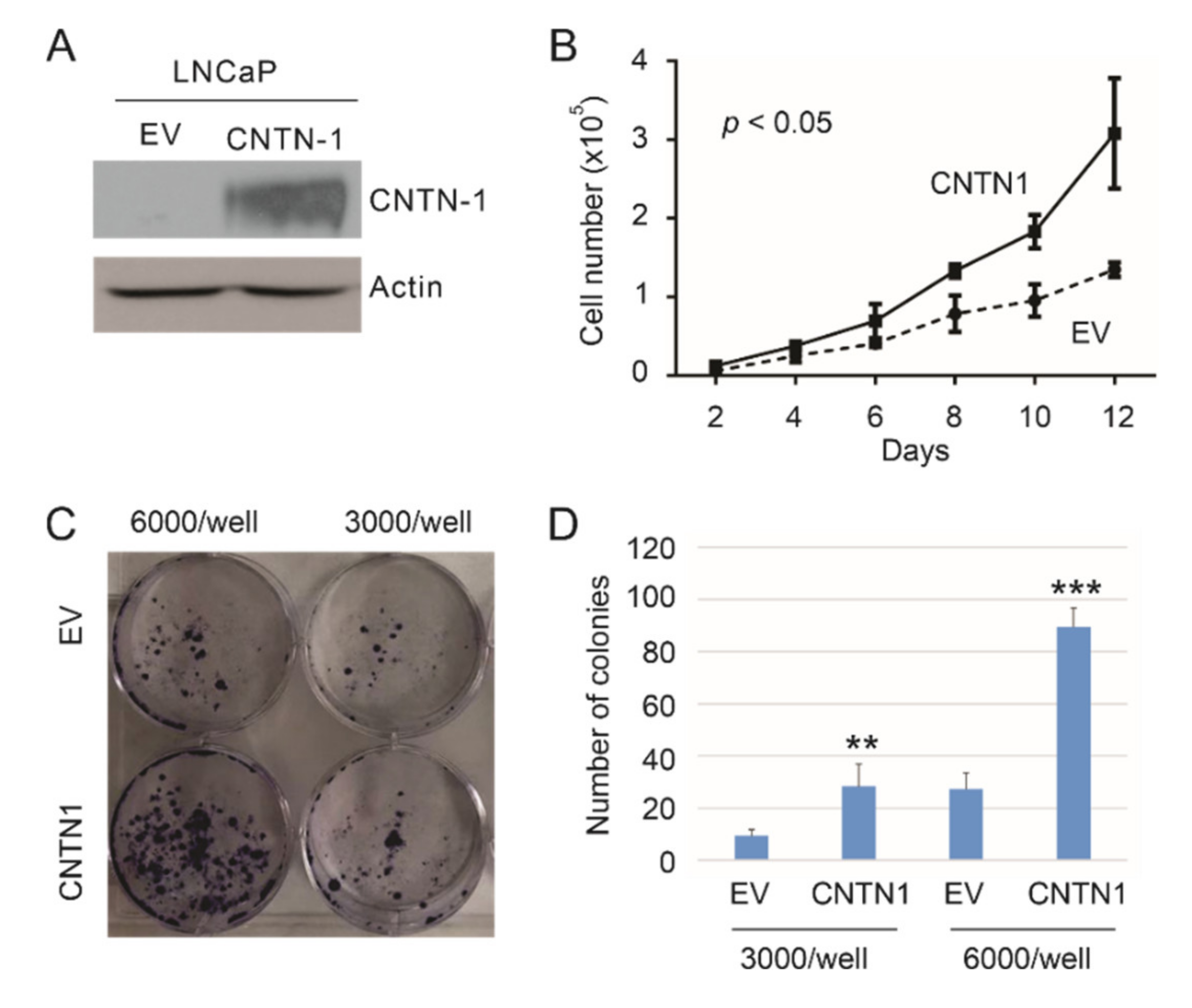
3.1. CNTN1-Mediated Promotion of LNCaP Cell Proliferation and Effects on Gene Expression
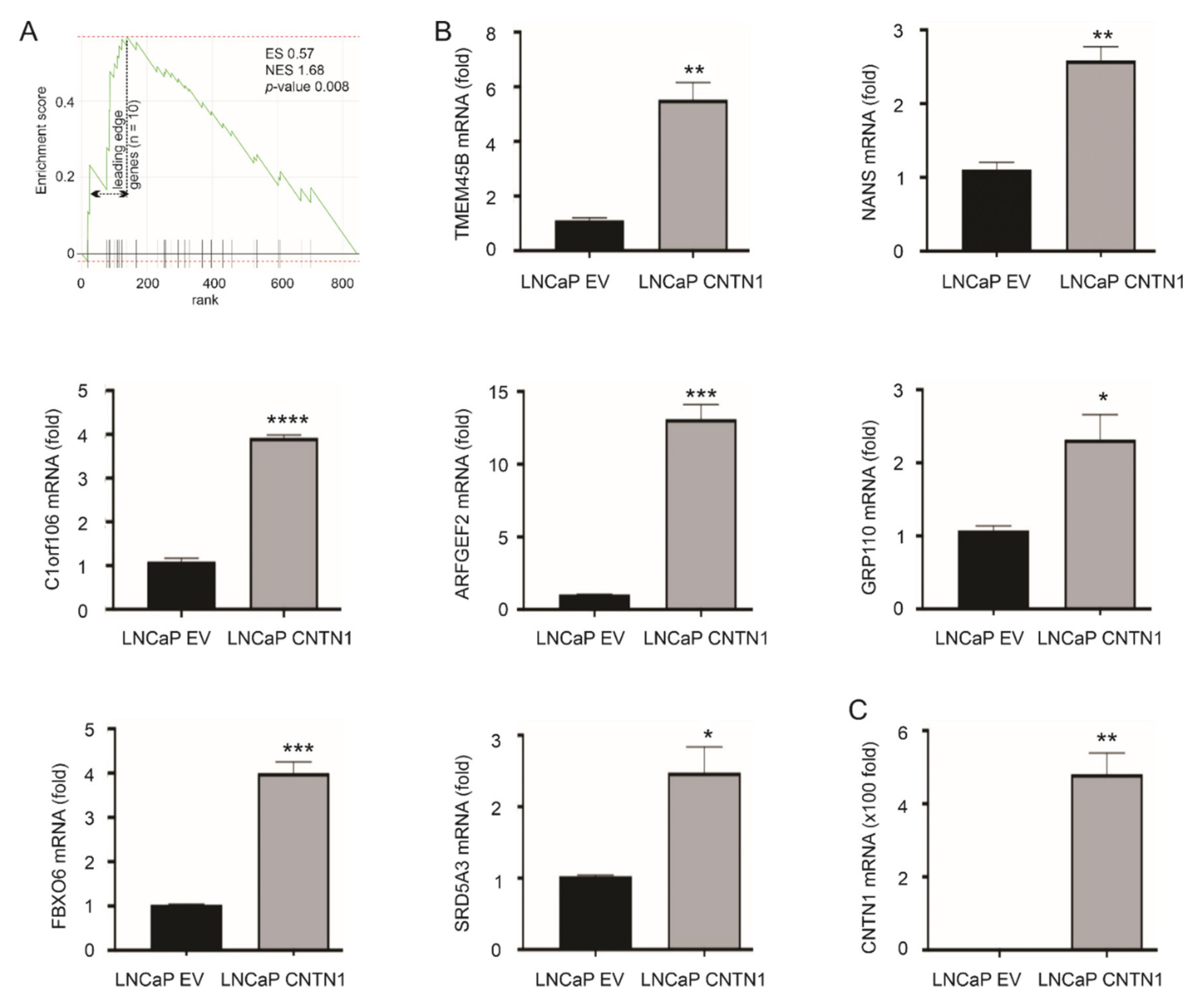
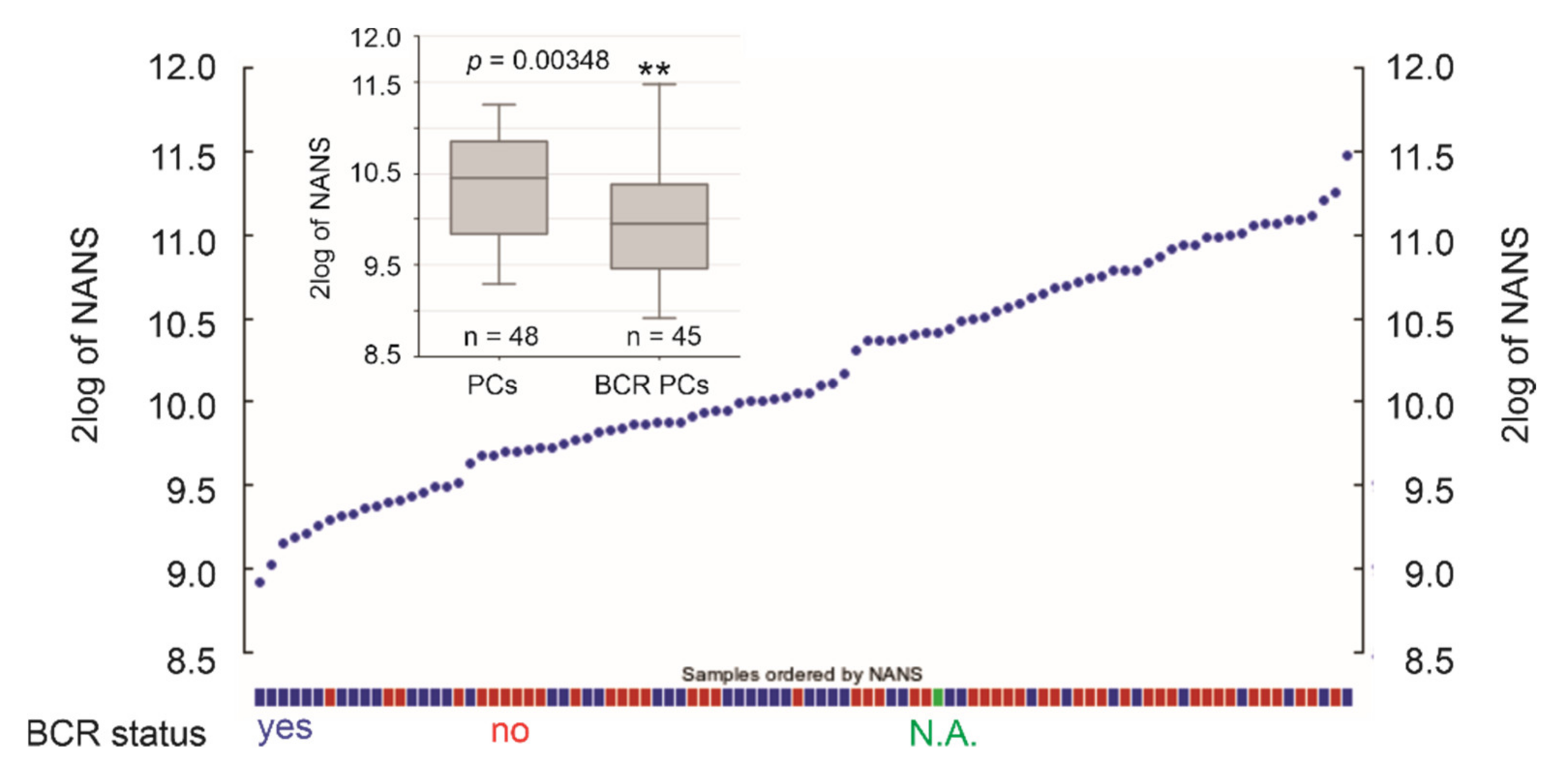
3.2. Differentional Expression of the 10 Leading-Edge Genes Following PC Progression
| Gene | Gene Details | Log2 1 | p-Value | Function in PC | Refs |
|---|---|---|---|---|---|
| TMEM45B | transmembrane protein 45B | 12.2 | 0.0015 ** | a biomarker of PC progression and metastasis | [47] |
| NANS | N-acetylneuraminic acid synthase | 11.9 | 0.0428 * | an androgen-responsive gene | [48] |
| C1orf106 | chromosome 1 open reading frame 106 | 10.2 | 0.0428 * | unknown | NA |
| LY75 | lymphocyte antigen 75 | 9.8 | 0.0164 * | not clear | NA |
| ARFGEF2 | ADP-ribosylation factor guanine nucleotide-exchange factor 2 | 9.5 | 0.0101 * | protein detected in PC bone metastasis | [52] |
| RNF144A | ring finger protein 144A | 3.2 | 0.0028 ** | not clear | NA |
| GPR110 | G protein-coupled receptor 110 | 3.0 | 0.0152 * | an oncogene in PC | [49] |
| FBXO6 | F-box protein 6 | 2.9 | 1.06 × 10−5 *** | unknown | NA |
| PTPN3 | protein tyrosine phosphatase, non-receptor type 3 | 2.6 | 0.0065 ** | unknown | NA |
| SRD5A3 | steroid 5 α-reductase 3 | 2.3 | 0.0027 ** | sustaining androgen biosynthesis in CRPC | [50,51] |
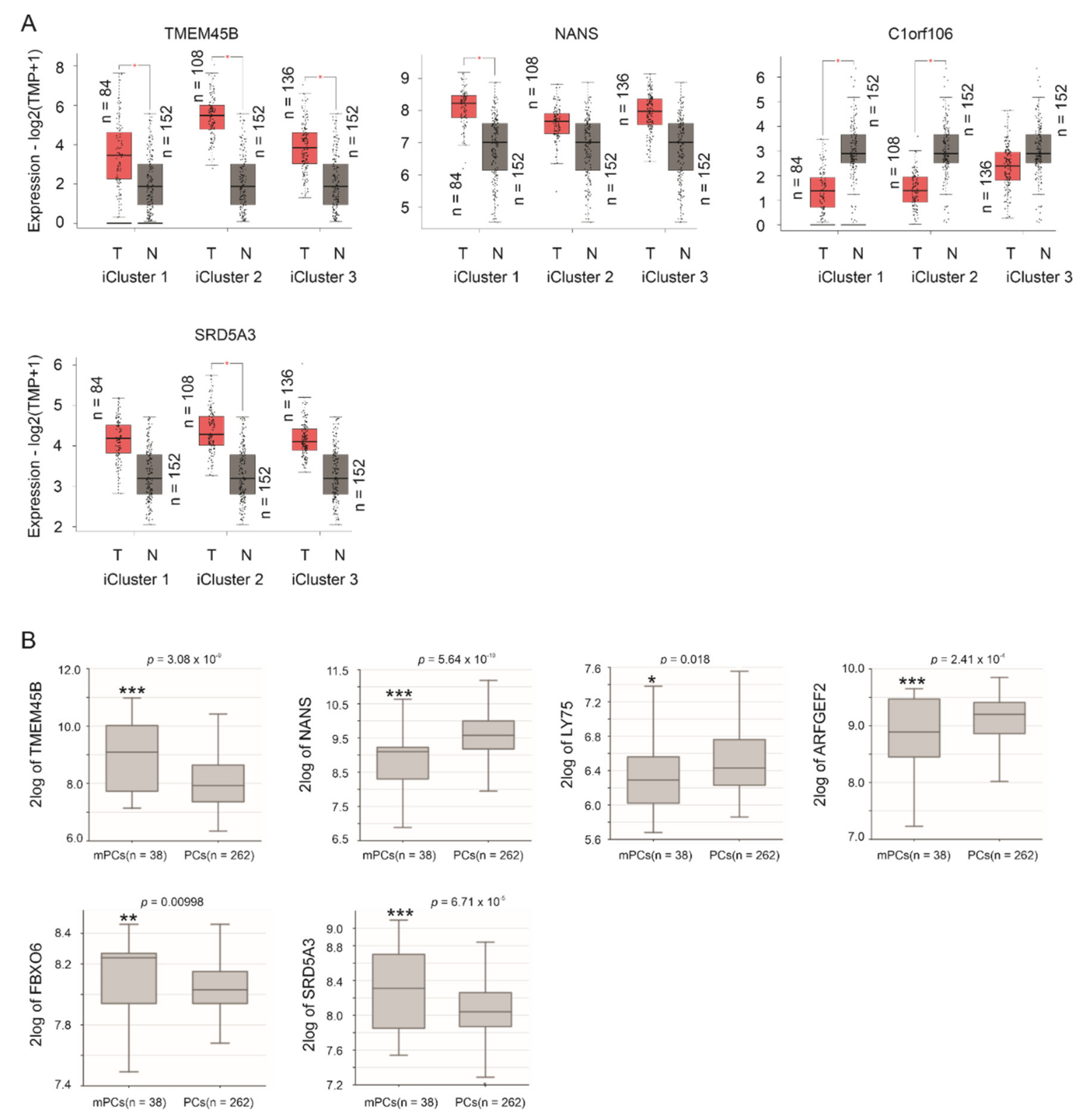
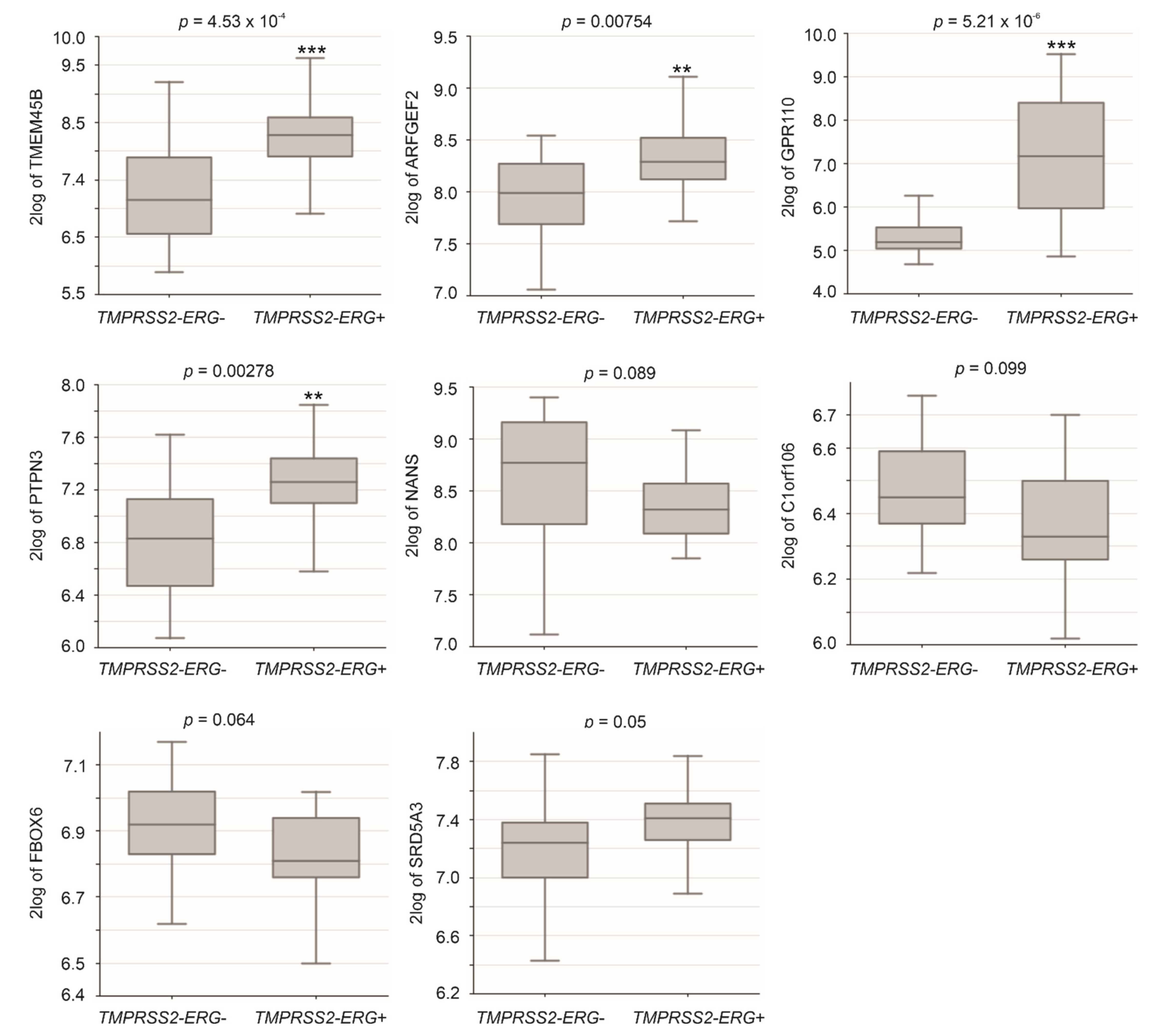
3.3. Differentional Expression of CNTN1 and LE Genes in CRPC
3.4. The LE Genes Predicting PC Relapse
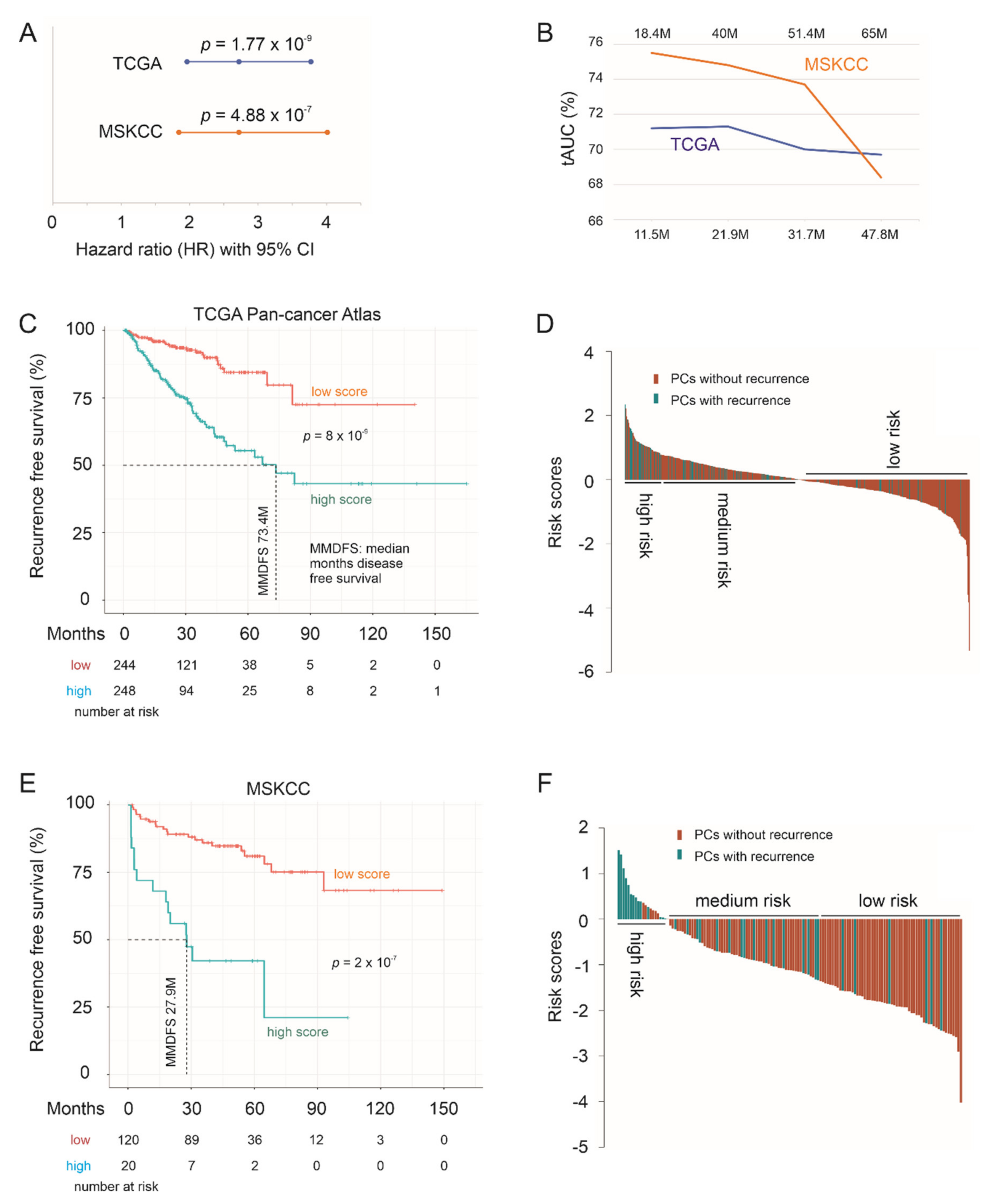
3.5. Effective Prediction of PC Recurrence Using LE Genes as a Multigene Panel
3.6. The LE Panel as an Independent Risk Factor of PC Recurrence
3.7. LE Gene Panel Predicts Poor Overall Survival (OS) in Renal Cell Carcinoma (RCC) and Bladder Carcinoma
3.8. LE Gene Panel Predicts Poor OS in CNTN1-Associated Cancer Types
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ferlay, J.; Soerjomataram, I.; Dikshit, R.; Eser, S.; Mathers, C.; Rebelo, M.; Parkin, D.M.; Forman, D.; Bray, F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in globocan 2012. Int. J. Cancer 2015, 136, E359–E386. [Google Scholar] [CrossRef]
- Egevad, L.; Delahunt, B.; Srigley, J.R.; Samaratunga, H. International society of urological pathology (isup) grading of prostate cancer-an isup consensus on contemporary grading. APMIS Acta Pathol. Microbiol. Immunol. Scand. 2016, 124, 433–435. [Google Scholar] [CrossRef] [PubMed]
- Gordetsky, J.; Epstein, J. Grading of prostatic adenocarcinoma: Current state and prognostic implications. Diagn. Pathol. 2016, 11, 25. [Google Scholar] [CrossRef]
- Epstein, J.I.; Zelefsky, M.J.; Sjoberg, D.D.; Nelson, J.B.; Egevad, L.; Magi-Galluzzi, C.; Vickers, A.J.; Parwani, A.V.; Reuter, V.E.; Fine, S.W.; et al. A contemporary prostate cancer grading system: A validated alternative to the gleason score. Eur. Urol. 2016, 69, 428–435. [Google Scholar] [CrossRef] [PubMed]
- The Cancer Genome Atlas Research Network. The molecular taxonomy of primary prostate cancer. Cell 2015, 163, 1011–1025. [Google Scholar] [CrossRef] [PubMed]
- Shen, R.; Olshen, A.B.; Ladanyi, M. Integrative clustering of multiple genomic data types using a joint latent variable model with application to breast and lung cancer subtype analysis. Bioinformatics 2009, 25, 2906–2912. [Google Scholar] [CrossRef]
- Kumar-Sinha, C.; Tomlins, S.A.; Chinnaiyan, A.M. Recurrent gene fusions in prostate cancer. Nat. Rev. Cancer 2008, 8, 497–511. [Google Scholar] [CrossRef] [PubMed]
- Zoni, E.; Karkampouna, S.; Thalmann, G.N.; Kruithof-de Julio, M.; Spahn, M. Emerging aspects of microrna interaction with tmprss2-erg and endocrine therapy. Mol. Cell. Endocrinol. 2018, 462, 9–16. [Google Scholar] [CrossRef]
- Zaorsky, N.G.; Raj, G.V.; Trabulsi, E.J.; Lin, J.; Den, R.B. The dilemma of a rising prostate-specific antigen level after local therapy: What are our options? Semin. Oncol. 2013, 40, 322–336. [Google Scholar] [CrossRef] [PubMed]
- Shipley, W.U.; Seiferheld, W.; Lukka, H.R.; Major, P.P.; Heney, N.M.; Grignon, D.J.; Sartor, O.; Patel, M.P.; Bahary, J.P.; Zietman, A.L.; et al. Radiation with or without antiandrogen therapy in recurrent prostate cancer. N. Engl. J. Med. 2017, 376, 417–428. [Google Scholar] [CrossRef]
- Semenas, J.; Allegrucci, C.; Boorjian, S.A.; Mongan, N.P.; Persson, J.L. Overcoming drug resistance and treating advanced prostate cancer. Curr. Drug Targets 2012, 13, 1308–1323. [Google Scholar] [CrossRef]
- Ojo, D.; Lin, X.; Wong, N.; Gu, Y.; Tang, D. Prostate cancer stem-like cells contribute to the development of castration-resistant prostate cancer. Cancers 2015, 7, 2290–2308. [Google Scholar] [CrossRef]
- Tanaudommongkon, I.; Tanaudommongkon, A.; Prathipati, P.; Nguyen, J.T.; Keller, E.T.; Dong, X. Curcumin nanoparticles and their cytotoxicity in docetaxel-resistant castration-resistant prostate cancer cells. Biomedicines 2020, 8, 253. [Google Scholar] [CrossRef] [PubMed]
- De Bono, J.S.; Logothetis, C.J.; Molina, A.; Fizazi, K.; North, S.; Chu, L.; Chi, K.N.; Jones, R.J.; Goodman, O.B., Jr.; Saad, F.; et al. Abiraterone and increased survival in metastatic prostate cancer. N. Engl. J. Med. 2011, 364, 1995–2005. [Google Scholar] [CrossRef]
- Scher, H.I.; Fizazi, K.; Saad, F.; Taplin, M.E.; Sternberg, C.N.; Miller, K.; de Wit, R.; Mulders, P.; Chi, K.N.; Shore, N.D.; et al. Increased survival with enzalutamide in prostate cancer after chemotherapy. N. Engl. J. Med. 2012, 367, 1187–1197. [Google Scholar] [CrossRef] [PubMed]
- Messner, E.A.; Steele, T.M.; Tsamouri, M.M.; Hejazi, N.; Gao, A.C.; Mudryj, M.; Ghosh, P.M. The androgen receptor in prostate cancer: Effect of structure, ligands and spliced variants on therapy. Biomedicines 2020, 8, 422. [Google Scholar] [CrossRef] [PubMed]
- Chaturvedi, S.; Garcia, J.A. Novel agents in the management of castration resistant prostate cancer. J. Carcinogenesis 2014, 13, 5. [Google Scholar]
- Drake, C.G. Prostate cancer as a model for tumour immunotherapy. Nat. Rev. Immunol. 2010, 10, 580–593. [Google Scholar] [CrossRef]
- Mei, W.; Gu, Y.; Jiang, Y.; Major, P.; Tang, D. Circulating cell-free DNA is a potential prognostic biomarker of metastatic castration-resistant prostate cancer for taxane therapy. AME Med. J. 2018, 3, 1–5. [Google Scholar] [CrossRef]
- Rana, Z.; Diermeier, S.; Hanif, M.; Rosengren, R.J. Understanding failure and improving treatment using hdac inhibitors for prostate cancer. Biomedicines 2020, 8, 22. [Google Scholar] [CrossRef]
- Bizzoca, A.; Corsi, P.; Gennarini, G. The mouse f3/contactin glycoprotein: Structural features, functional properties and developmental significance of its regulated expression. Cell Adhes. Migr. 2009, 3, 53–63. [Google Scholar] [CrossRef] [PubMed]
- Falk, J.; Bonnon, C.; Girault, J.A.; Faivre-Sarrailh, C. F3/contactin, a neuronal cell adhesion molecule implicated in axogenesis and myelination. Biol. Cell Under Auspices Eur. Cell Biol. Organ. 2002, 94, 327–334. [Google Scholar] [CrossRef]
- Boyle, M.E.; Berglund, E.O.; Murai, K.K.; Weber, L.; Peles, E.; Ranscht, B. Contactin orchestrates assembly of the septate-like junctions at the paranode in myelinated peripheral nerve. Neuron 2001, 30, 385–397. [Google Scholar] [CrossRef]
- Compton, A.G.; Albrecht, D.E.; Seto, J.T.; Cooper, S.T.; Ilkovski, B.; Jones, K.J.; Challis, D.; Mowat, D.; Ranscht, B.; Bahlo, M.; et al. Mutations in contactin-1, a neural adhesion and neuromuscular junction protein, cause a familial form of lethal congenital myopathy. Am. J. Hum. Genet. 2008, 83, 714–724. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.; Li, T.; Kapoor, A.; Major, P.; Tang, D. Contactin 1: An important and emerging oncogenic protein promoting cancer progression and metastasis. Genes 2020, 11, 874. [Google Scholar] [CrossRef]
- Su, J.L.; Yang, C.Y.; Shih, J.Y.; Wei, L.H.; Hsieh, C.Y.; Jeng, Y.M.; Wang, M.Y.; Yang, P.C.; Kuo, M.L. Knockdown of contactin-1 expression suppresses invasion and metastasis of lung adenocarcinoma. Cancer Res. 2006, 66, 2553–2561. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Chen, S.; Wu, W.; Liu, B.; Shen, W.; Wang, F.; He, X.; Zhang, S. Contactin-1 (cntn-1) overexpression is correlated with advanced clinical stage and lymph node metastasis in oesophageal squamous cell carcinomas. Jpn. J. Clin. Oncol. 2012, 42, 612–618. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.M.; Cao, W.; Ye, D.; Ren, G.X.; Wu, Y.N.; Guo, W. Contactin 1 (cntn1) expression associates with regional lymph node metastasis and is a novel predictor of prognosis in patients with oral squamous cell carcinoma. Mol. Med. Rep. 2012, 6, 265–270. [Google Scholar] [CrossRef]
- Li, G.Y.; Huang, M.; Pan, T.T.; Jia, W.D. Expression and prognostic significance of contactin 1 in human hepatocellular carcinoma. OncoTargets Ther. 2016, 9, 387–394. [Google Scholar] [CrossRef]
- Chen, N.; He, S.; Geng, J.; Song, Z.J.; Han, P.H.; Qin, J.; Zhao, Z.; Song, Y.C.; Wang, H.X.; Dang, C.X. Overexpression of contactin 1 promotes growth, migration and invasion in hs578t breast cancer cells. BMC Cell Biol. 2018, 19, 5. [Google Scholar] [CrossRef] [PubMed]
- Eckerich, C.; Zapf, S.; Ulbricht, U.; Muller, S.; Fillbrandt, R.; Westphal, M.; Lamszus, K. Contactin is expressed in human astrocytic gliomas and mediates repulsive effects. Glia 2006, 53, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Shi, K.; Xu, D.; Yang, C.; Wang, L.; Pan, W.; Zheng, C.; Fan, L. Contactin 1 as a potential biomarker promotes cell proliferation and invasion in thyroid cancer. Int. J. Clin. Exp. Pathol. 2015, 8, 12473–12481. [Google Scholar]
- Liu, Y.C.; Zhao, J.; Hu, C.E.; Gan, J.; Zhang, W.H.; Huang, G.J. Comprehensive analysis of vascular endothelial growth factor-c related factors in stomach cancer. Asian Pac. J. Cancer Prev. APJCP 2014, 15, 1925–1929. [Google Scholar] [CrossRef][Green Version]
- Yan, J.; Ojo, D.; Kapoor, A.; Lin, X.; Pinthus, J.H.; Aziz, T.; Bismar, T.A.; Wei, F.; Wong, N.; de Melo, J.; et al. Neural cell adhesion protein cntn1 promotes the metastatic progression of prostate cancer. Cancer Res. 2016, 76, 1603–1614. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Yang, X.; Zhao, T.; Du, H.; Wang, T.; Zhong, S.; Yang, B.; Li, H. Upregulation of contactin-1 expression promotes prostate cancer progression. Oncol. Lett. 2020, 19, 1611–1618. [Google Scholar] [CrossRef]
- Jiang, Y.; Lin, X.; Kapoor, A.; He, L.; Wei, F.; Gu, Y.; Mei, W.; Zhao, K.; Yang, H.; Tang, D. Fam84b promotes prostate tumorigenesis through a network alteration. Ther. Adv. Med. Oncol. 2019, 11, 1758835919846372. [Google Scholar] [CrossRef]
- He, L.; Fan, C.; Kapoor, A.; Ingram, A.J.; Rybak, A.P.; Austin, R.C.; Dickhout, J.; Cutz, J.C.; Scholey, J.; Tang, D. Alpha-mannosidase 2c1 attenuates pten function in prostate cancer cells. Nat. Commun. 2011, 2, 307. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Ingram, A.; Rybak, A.P.; Tang, D. Shank-interacting protein-like 1 promotes tumorigenesis via pten inhibition in human tumor cells. J. Clin. Investig. 2010, 120, 2094–2108. [Google Scholar] [CrossRef]
- Wei, F.; Hao, P.; Zhang, X.; Hu, H.; Jiang, D.; Yin, A.; Wen, L.; Zheng, L.; He, J.Z.; Mei, W.; et al. Etoposide-induced DNA damage affects multiple cellular pathways in addition to DNA damage response. Oncotarget 2018, 9, 24122. (in press). [CrossRef] [PubMed]
- Tang, Z.; Kang, B.; Li, C.; Chen, T.; Zhang, Z. Gepia2: An enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019, 47, W556–W560. [Google Scholar] [CrossRef]
- Wong, N.; Major, P.; Kapoor, A.; Wei, F.; Yan, J.; Aziz, T.; Zheng, M.; Jayasekera, D.; Cutz, J.C.; Chow, M.J.; et al. Amplification of muc1 in prostate cancer metastasis and crpc development. Oncotarget 2016, 7, 83115–83133. [Google Scholar] [CrossRef]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cbio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cbioportal. Sci. Signal 2013, 6, pl1. [Google Scholar] [CrossRef]
- Liu, J.; Lichtenberg, T.; Hoadley, K.A.; Poisson, L.M.; Lazar, A.J.; Cherniack, A.D.; Kovatich, A.J.; Benz, C.C.; Levine, D.A.; Lee, A.V.; et al. An integrated tcga pan-cancer clinical data resource to drive high-quality survival outcome analytics. Cell 2018, 173, 400–416. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Creighton, C.J.; Massarweh, S.; Huang, S.; Tsimelzon, A.; Hilsenbeck, S.G.; Osborne, C.K.; Shou, J.; Malorni, L.; Schiff, R. Development of resistance to targeted therapies transforms the clinically associated molecular profile subtype of breast tumor xenografts. Cancer Res. 2008, 68, 7493–7501. [Google Scholar] [CrossRef]
- Luo, F.; Yang, K.; Wang, Y.Z.; Lin, D. Tmem45b is a novel predictive biomarker for prostate cancer progression and metastasis. Neoplasma 2018, 65, 815–821. [Google Scholar] [CrossRef] [PubMed]
- Romanuik, T.L.; Wang, G.; Holt, R.A.; Jones, S.J.; Marra, M.A.; Sadar, M.D. Identification of novel androgen-responsive genes by sequencing of longsage libraries. BMC Genomics 2009, 10, 476. [Google Scholar] [CrossRef] [PubMed]
- Lum, A.M.; Wang, B.B.; Beck-Engeser, G.B.; Li, L.; Channa, N.; Wabl, M. Orphan receptor gpr110, an oncogene overexpressed in lung and prostate cancer. BMC Cancer 2010, 10, 40. [Google Scholar] [CrossRef]
- Uemura, M.; Tamura, K.; Chung, S.; Honma, S.; Okuyama, A.; Nakamura, Y.; Nakagawa, H. Novel 5 alpha-steroid reductase (srd5a3, type-3) is overexpressed in hormone-refractory prostate cancer. Cancer Sci. 2008, 99, 81–86. [Google Scholar] [PubMed]
- Mitsiades, N.; Sung, C.C.; Schultz, N.; Danila, D.C.; He, B.; Eedunuri, V.K.; Fleisher, M.; Sander, C.; Sawyers, C.L.; Scher, H.I. Distinct patterns of dysregulated expression of enzymes involved in androgen synthesis and metabolism in metastatic prostate cancer tumors. Cancer Res. 2012, 72, 6142–6152. [Google Scholar] [CrossRef]
- Iglesias-Gato, D.; Thysell, E.; Tyanova, S.; Crnalic, S.; Santos, A.; Lima, T.S.; Geiger, T.; Cox, J.; Widmark, A.; Bergh, A.; et al. The proteome of prostate cancer bone metastasis reveals heterogeneity with prognostic implications. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2018, 24, 5433–5444. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.; Yang, L.; Tanasa, B.; Hutt, K.; Ju, B.G.; Ohgi, K.; Zhang, J.; Rose, D.W.; Fu, X.D.; Glass, C.K.; et al. Nuclear receptor-induced chromosomal proximity and DNA breaks underlie specific translocations in cancer. Cell 2009, 139, 1069–1083. [Google Scholar] [CrossRef] [PubMed]
- Borno, S.T.; Fischer, A.; Kerick, M.; Falth, M.; Laible, M.; Brase, J.C.; Kuner, R.; Dahl, A.; Grimm, C.; Sayanjali, B.; et al. Genome-wide DNA methylation events in tmprss2-erg fusion-negative prostate cancers implicate an ezh2-dependent mechanism with mir-26a hypermethylation. Cancer Discov. 2012, 2, 1024–1035. [Google Scholar] [CrossRef] [PubMed]
- Taylor, B.S.; Schultz, N.; Hieronymus, H.; Gopalan, A.; Xiao, Y.; Carver, B.S.; Arora, V.K.; Kaushik, P.; Cerami, E.; Reva, B.; et al. Integrative genomic profiling of human prostate cancer. Cancer Cell 2010, 18, 11–22. [Google Scholar] [CrossRef] [PubMed]
- Chandran, U.R.; Ma, C.; Dhir, R.; Bisceglia, M.; Lyons-Weiler, M.; Liang, W.; Michalopoulos, G.; Becich, M.; Monzon, F.A. Gene expression profiles of prostate cancer reveal involvement of multiple molecular pathways in the metastatic process. BMC Cancer 2007, 7, 64. [Google Scholar] [CrossRef]
- Yu, Y.P.; Landsittel, D.; Jing, L.; Nelson, J.; Ren, B.; Liu, L.; McDonald, C.; Thomas, R.; Dhir, R.; Finkelstein, S.; et al. Gene expression alterations in prostate cancer predicting tumor aggression and preceding development of malignancy. J. Clin. Oncol. 2004, 22, 2790–2799. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Mei, W.; Gu, Y.; Lin, X.; He, L.; Zeng, H.; Wei, F.; Wan, X.; Yang, H.; Major, P.; et al. Construction of a set of novel and robust gene expression signatures predicting prostate cancer recurrence. Mol. Oncol. 2018, 12, 1559–1578. [Google Scholar] [CrossRef]
- Cohen, H.T.; McGovern, F.J. Renal-cell carcinoma. N. Engl. J. Med. 2005, 353, 2477–2490. [Google Scholar] [CrossRef]
- Aguirre-Gamboa, R.; Gomez-Rueda, H.; Martinez-Ledesma, E.; Martinez-Torteya, A.; Chacolla-Huaringa, R.; Rodriguez-Barrientos, A.; Tamez-Pena, J.G.; Trevino, V. Survexpress: An online biomarker validation tool and database for cancer gene expression data using survival analysis. PLoS ONE 2013, 8, e74250. [Google Scholar] [CrossRef]
- Lin, X.; Kapoor, A.; Gu, Y.; Chow, M.J.; Xu, H.; Major, P.; Tang, D. Assessment of biochemical recurrence of prostate cancer (review). Int. J. Oncol. 2019, 55, 1194–1212. [Google Scholar] [CrossRef]
- Maleki, F.; Ovens, K.; Hogan, D.J.; Kusalik, A.J. Gene set analysis: Challenges, opportunities, and future research. Front. Genet. 2020, 11, 654. [Google Scholar] [CrossRef]
- Das, S.; McClain, C.J.; Rai, S.N. Fifteen years of gene set analysis for high-throughput genomic data: A review of statistical approaches and future challenges. Entropy 2020, 22, 427. [Google Scholar] [CrossRef]
- Kohler, C.U.; Walter, M.; Lang, K.; Plottner, S.; Roghmann, F.; Noldus, J.; Tannapfel, A.; Tam, Y.C.; Kafferlein, H.U.; Bruning, T. In-vitro identification and in-vivo confirmation of DNA methylation biomarkers for urothelial cancer. Biomedicines 2020, 8, 233. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Liu, C.; Yang, D.; Song, J.; Zhang, J.; Wang, T.; Wang, M.; Xu, W.; Li, X.; Ding, S.; et al. C1orf106, an innate immunity activator, is amplified in breast cancer and is required for basal-like/luminal progenitor fate decision. Sci. China Life Sci. 2019, 62, 1229–1242. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.W.; Brognard, J.; Coughlin, C.; You, Z.; Dolled-Filhart, M.; Aslanian, A.; Manning, G.; Abraham, R.T.; Hunter, T. The f box protein fbx6 regulates chk1 stability and cellular sensitivity to replication stress. Mol. Cell 2009, 35, 442–453. [Google Scholar] [CrossRef]
- Flynn, R.L.; Zou, L. Atr: A master conductor of cellular responses to DNA replication stress. Trends Biochem. Sci. 2011, 36, 133–140. [Google Scholar] [CrossRef] [PubMed]
- Wengner, A.M.; Scholz, A.; Haendler, B. Targeting DNA damage response in prostate and breast cancer. Int. J. Mol. Sci. 2020, 21, 8273. [Google Scholar] [CrossRef] [PubMed]
- Sherr, C.J. Tumor surveillance via the arf-p53 pathway. Genes & Dev. 1998, 12, 2984–2991. [Google Scholar]
- Bartek, J.; Bartkova, J.; Lukas, J. DNA damage signalling guards against activated oncogenes and tumour progression. Oncogene 2007, 26, 7773–7779. [Google Scholar] [CrossRef]


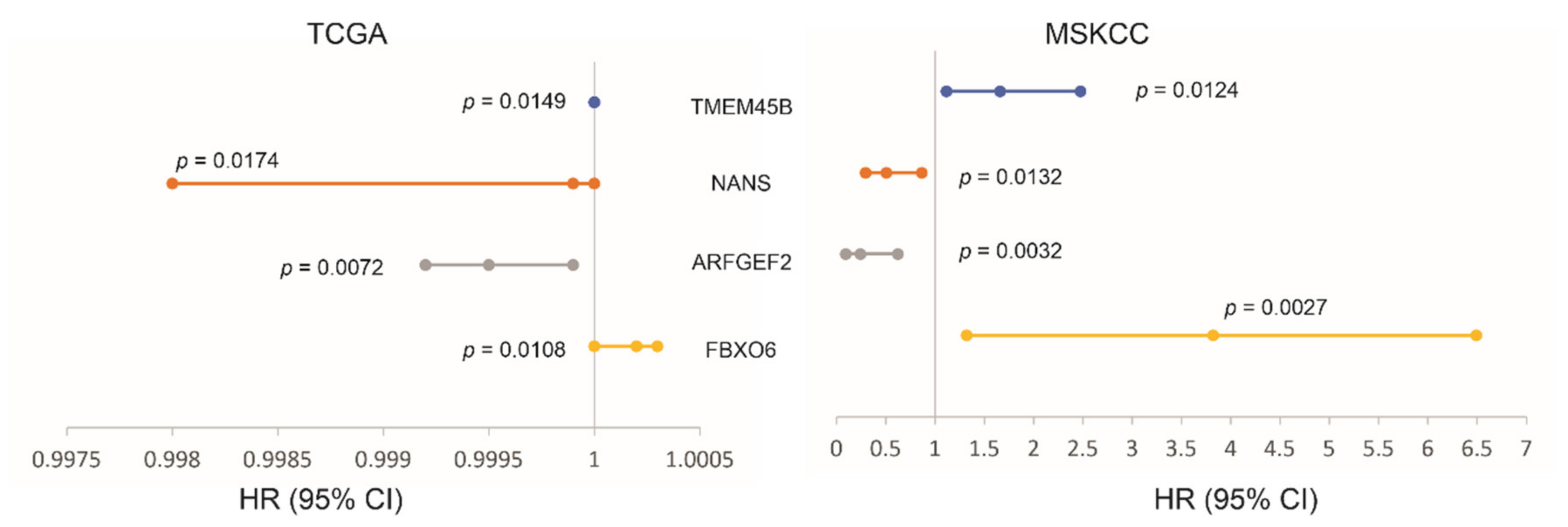
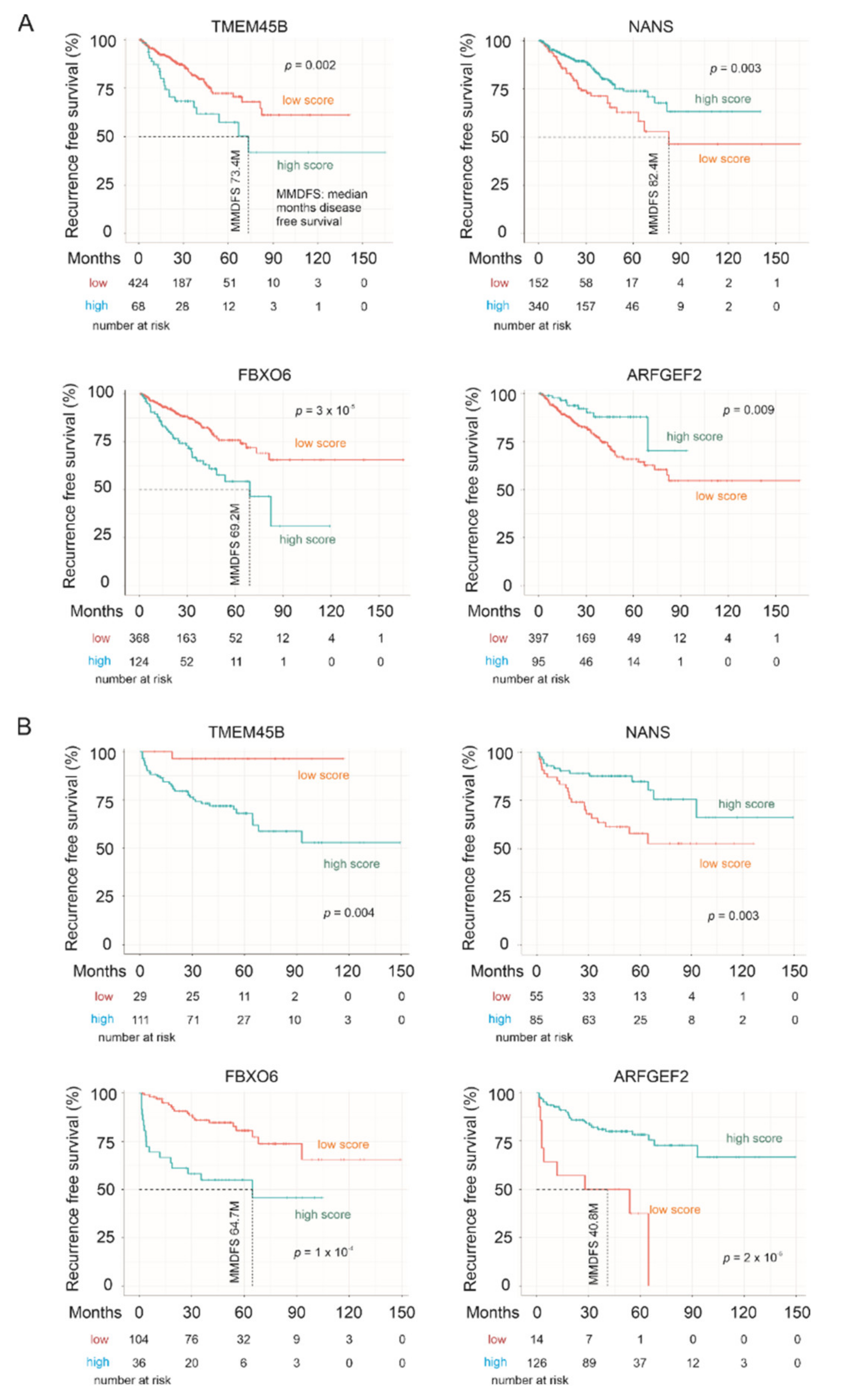
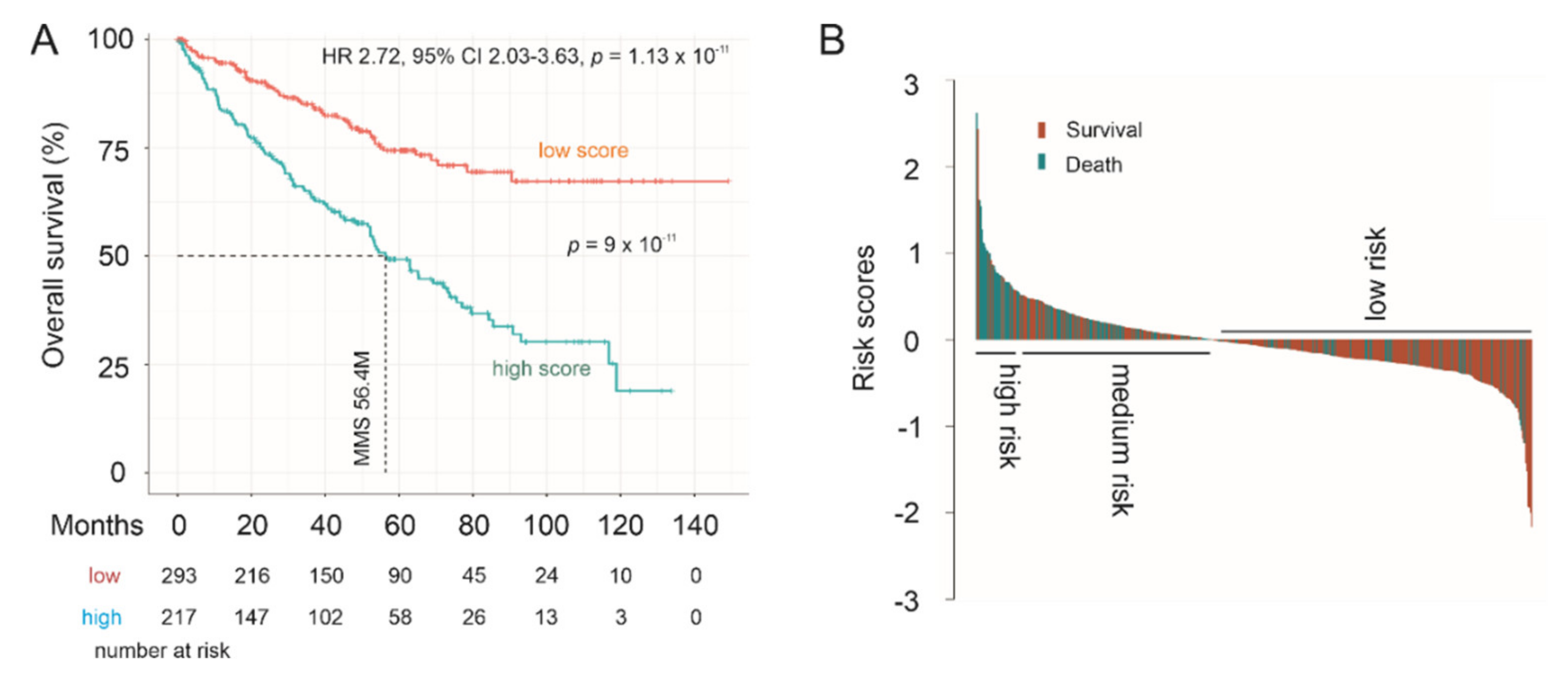
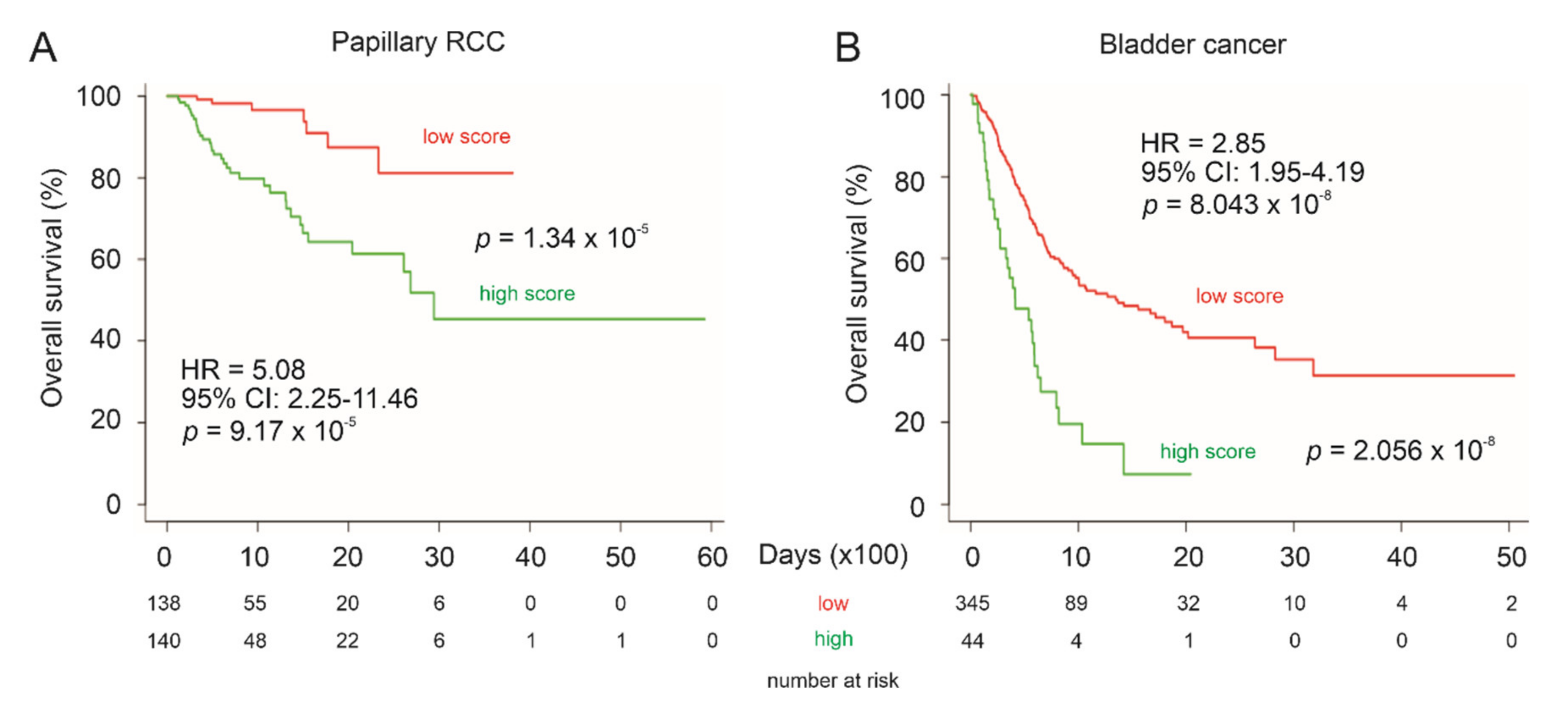
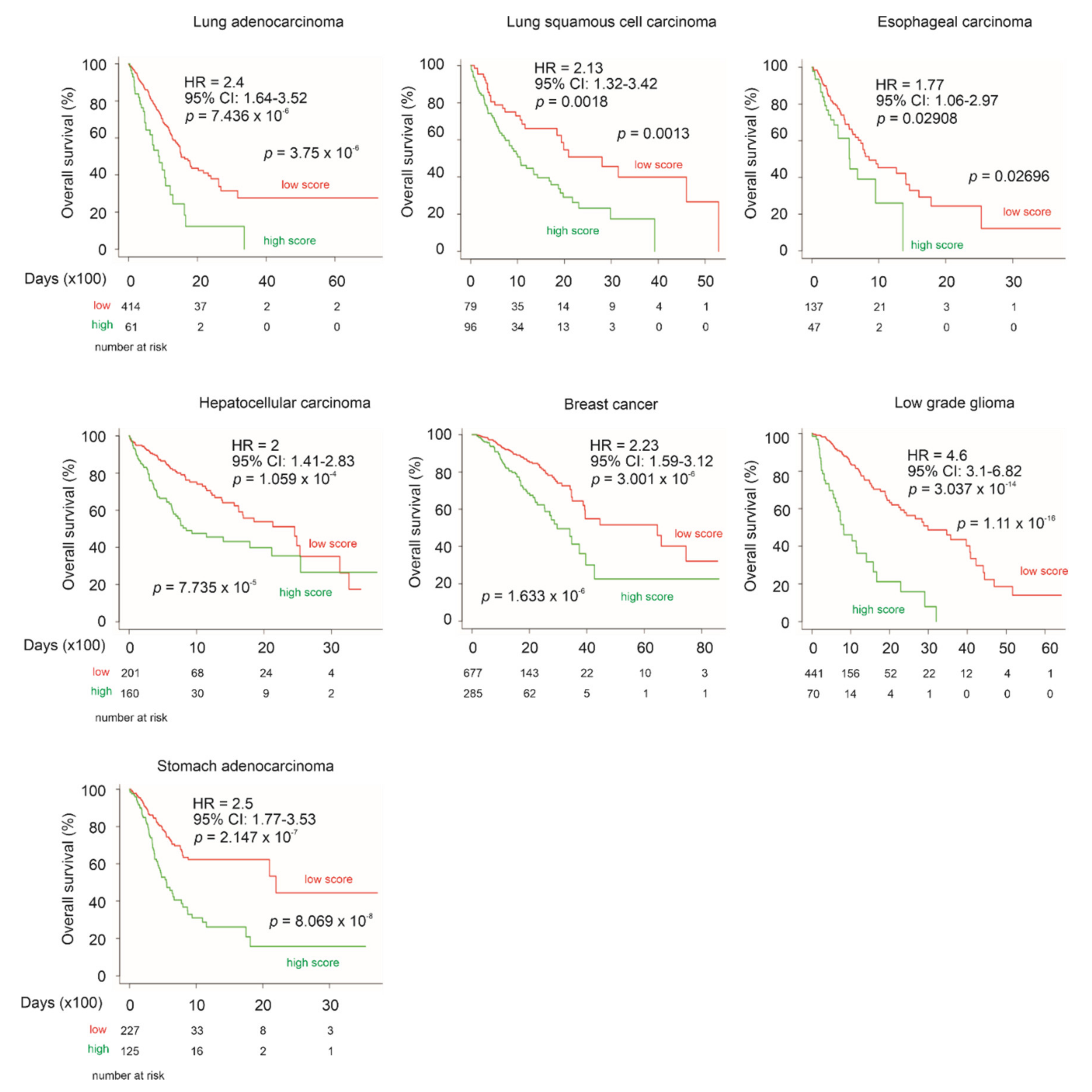
| Factors | Univariate Cox Analysis | Multivariate Cox Analysis | ||||
|---|---|---|---|---|---|---|
| HR | 95% CI | p-Value | HR | 95% CI | p-Value | |
| LE Panel 1 | 2.72 | 1.96–3.37 | 1.77 × 10−9 *** | 1.91 | 1.37–2.671 | 0.000132 *** |
| Age 2 | 1.02 | 0.99–1.05 | 0.189 | 0.99 | 0.96–1.02 | 0.6201 |
| WHO IV 3 | 9.76 | 1.28–74.6 | 0.0282 * | 6.32 | 0.81–49.55 | 0.079158 |
| WHO V 3 | 21.38 | 2.96–154.5 | 0.00241 ** | 10.28 | 1.36–77.77 | 0.024047 * |
| Tstage 1 4 | 3.69 | 2.08–6.52 | 7.45 × 10−6 *** | 1.61 | 0.84–3.09 | 0.151878 |
| Margin 1 5 | 2.30 | 1.52–3.48 | 8.1 × 10−5 *** | 1.32 | 0.84–2.08 | 0.228092 |
| Factors | Univariate Cox Analysis | Multivariate Cox Analysis | ||||
|---|---|---|---|---|---|---|
| HR | 95% CI | p-Value | HR | 95% CI | p-Value | |
| LE Panel 1 | 2.72 | 2.03–3.63 | 1.13 × 10−11 *** | 1.67 | 1.18–2.35 | 0.00341 ** |
| Age 2 | 1.03 | 1.02–1.04 | 2.78 × 10−6 *** | 1.03 | 1.02–1.05 | 3.68 × 10−5 *** |
| Sex 3 | 0.96 | 0.70–1.31 | 0.793 | 0.96 | 0.696–1.329 | 0.81224 |
| Stage III 4 | 2.80 | 1.84–4.23 | 1.28 × 10−6 *** | 2.05 | 1.32–3.16 | 0.00127 ** |
| Stage IV 4 | 6.83 | 4.60–10.12 | <2 × 10−16 *** | 4.53 | 2.87–7.13 | 7.13 × 10−11 *** |
| Grade 3 5 | 1.94 | 1.32–2.86 | 0.000753 *** | 1.43 | 0.96–2.13 | 0.08017 |
| Grade 4 5 | 5.74 | 3.59–8.05 | 3.06 × 10−16 *** | 2.01 | 1.25–3.23 | 0.00383 ** |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gu, Y.; Chow, M.J.; Kapoor, A.; Lin, X.; Mei, W.; Tang, D. Differential Expression of a Panel of Ten CNTN1-Associated Genes during Prostate Cancer Progression and the Predictive Properties of the Panel towards Prostate Cancer Relapse. Genes 2021, 12, 257. https://doi.org/10.3390/genes12020257
Gu Y, Chow MJ, Kapoor A, Lin X, Mei W, Tang D. Differential Expression of a Panel of Ten CNTN1-Associated Genes during Prostate Cancer Progression and the Predictive Properties of the Panel towards Prostate Cancer Relapse. Genes. 2021; 12(2):257. https://doi.org/10.3390/genes12020257
Chicago/Turabian StyleGu, Yan, Mathilda Jing Chow, Anil Kapoor, Xiaozeng Lin, Wenjuan Mei, and Damu Tang. 2021. "Differential Expression of a Panel of Ten CNTN1-Associated Genes during Prostate Cancer Progression and the Predictive Properties of the Panel towards Prostate Cancer Relapse" Genes 12, no. 2: 257. https://doi.org/10.3390/genes12020257
APA StyleGu, Y., Chow, M. J., Kapoor, A., Lin, X., Mei, W., & Tang, D. (2021). Differential Expression of a Panel of Ten CNTN1-Associated Genes during Prostate Cancer Progression and the Predictive Properties of the Panel towards Prostate Cancer Relapse. Genes, 12(2), 257. https://doi.org/10.3390/genes12020257







